Abstract
Virus-induced gene silencing identified the Avr9/Cf-9 RAPIDLY ELICITED gene ACRE189 as essential for the Cf-9– and Cf-4–mediated hypersensitive response (HR) in Nicotiana benthamiana. We report a role for ACRE189 in disease resistance in tomato (Solanum lycopersicum) and tobacco (Nicotiana tabacum). ACRE189 (herein renamed Avr9/Cf-9–INDUCED F-BOX1 [ACIF1]) encodes an F-box protein with a Leu-rich-repeat domain. ACIF1 is widely conserved and is closely related to F-box proteins regulating plant hormone signaling. Silencing of tobacco ACIF1 suppressed the HR triggered by various elicitors (Avr9, Avr4, AvrPto, Inf1, and the P50 helicase of Tobacco mosaic virus [TMV]). ACIF1 is recruited to SCF complexes (a class of ubiquitin E3 ligases), and the expression of ACIF1 F-box mutants in tobacco compromises the HR similarly to ACIF1 silencing. ACIF1 affects N gene–mediated responses to TMV infection, including lesion formation and salicylic acid accumulation. Loss of ACIF1 function also reduced confluent cell death induced by Pseudomonas syringae pv tabaci. ACIF1 silencing in Cf9 tomato attenuated the Cf-9–dependent HR but not Cf-9 resistance to Cladosporium fulvum. Resistance conferred by the Cf-9 homolog Cf-9B, however, was compromised in ACIF1-silenced tomato. Analysis of public expression profiling data suggests that Arabidopsis thaliana homologs of ACIF1 (VFBs) regulate defense responses via methyl jasmonate– and abscisic acid–responsive genes. Together, these findings support a role of ACIF1/VFBs in plant defense responses.
INTRODUCTION
Plants are frequently attacked by pathogens and have evolved a multilayer defense system. An important layer of defense involves resistance (R) genes encoding receptors that allow recognition of pathogens carrying corresponding avirulence (Avr) genes (Jones and Dangl, 2006). R protein–mediated pathogen recognition can be direct or indirect via the action of effectors (Avr proteins) on host targets (Jones and Dangl, 2006). R gene–based disease resistance is generally referred to as gene-for-gene resistance. A hallmark of this type of resistance is the hypersensitive response (HR), comprising localized cell death around the infection site.
The fungus Cladosporium fulvum (syn. Passalora fulva) is a biotrophic plant pathogen causing leaf mold in tomato (Solanum lycopersicum) (Rivas and Thomas, 2005; Thomma et al., 2005). Several tomato R genes against C. fulvum have been cloned, including Cf-2, Cf-4, and Cf-9 (Rivas and Thomas, 2005). Cf genes form a distinct class of R genes encoding receptor-like membrane-anchored glycoproteins with extracellular leucine-rich repeats (LRRs) and a short cytoplasmic region (Fritz-Laylin et al., 2005; van der Hoorn et al., 2005). Cf-9 is the best characterized of the Cf genes, and it confers strong resistance to C. fulvum strains expressing Avr9. Cf-9B (Hcr9-9B) is a paralog of Cf-9 present at the same locus, but Cf-9B confers only partial resistance to C. fulvum in mature plants by recognition of an unknown elicitor (Panter et al., 2002).
To study Cf-9 function, a heterologous system was developed in tobacco (Nicotiana tabacum) (Hammond-Kosack et al., 1998; Piedras et al., 1998). Cf-9–containing tobacco plants and derived cell cultures respond to Avr9 infiltration in the extracellular space with a rapid HR in a strict gene-for-gene–dependent manner, indicating that all components required for Cf-9 function are conserved in tobacco. This heterologous system was used to identify signaling components and mechanisms downstream of Cf-9, revealing a role for ion fluxes (Blatt et al., 1999), reactive oxygen species (Piedras et al., 1998), and mitogen-activated and calcium-dependent protein kinases (Romeis et al., 1999, 2000, 2001; Ludwig et al., 2005).
Tobacco Cf-9 cell cultures were used to identify Avr9/Cf-9 RAPIDLY ELICITED (ACRE) genes (15 to 30 min) using cDNA/amplified fragment length polymorphism (AFLP) (Durrant et al., 2000). Many ACRE genes encode putative signaling components and thus may play pivotal roles in the initial development of defense responses. To identify ACRE genes that are essential for Cf-9/Cf-4–mediated HR, a virus-induced gene silencing (VIGS) screen was performed using 43 cloned ACRE fragments (Rowland et al., 2005; Gonzalez-Lamothe et al., 2006; Yang et al., 2006). Four genes (ACRE74, ACRE189, ACRE264, and ACRE276) were found to be essential for both Cf-9– and Cf-4–mediated HR. Three of these four ACRE genes (ACRE74, ACRE264, and ACRE276) were also essential for Cf-9–mediated resistance to C. fulvum. ACRE264 encodes a protein kinase renamed ACIK1 (for Avr9/Cf-9–INDUCED KINASE1) that can phosphorylate Cf-9–INTERACTING THIOREDOXIN1 in planta (Rowland et al., 2005; Nekrasov et al., 2006). This thioredoxin interacts with the cytoplasmic tail of Cf-9 and negatively regulates Cf-9 function (Rivas et al., 2004). ACRE74/Nt CMPG1 and ACRE276 both encode U-box proteins that function as ubiquitin E3 ligases targeting proteins for degradation by the 26S proteasome (Gonzalez-Lamothe et al., 2006; Yang et al., 2006). These two U-box proteins were required for multiple resistance responses in different plant species.
The fourth gene (ACRE189) is the subject of this study. ACRE189 encodes an F-box protein (renamed Avr9/Cf-9–INDUCED F-BOX1 [ACIF1]). F-box proteins constitute one of the largest superfamilies (>600 members) in plants (Gagne et al., 2002; Kuroda et al., 2002; Jain et al., 2007). F-box proteins are part of SCF (for SKP1/CUL1/F-box) complexes that function as ubiquitin E3 ligases (Willems et al., 2004; Petroski and Deshaies, 2005; Lechner et al., 2006). The core of the SCF complex is formed by Rbx1 (RING-box 1), CUL1 (Cullin1), and SKP1 (S-phase kinase–related protein1). The Arabidopsis thaliana genome contains 21 SKP1 homologs (called ASKs). SKP1/ASKs recruit F-box proteins to SCF complexes via a direct interaction with the F-box domain. F-box proteins are responsible for substrate recognition and substrate recruitment to SCF complexes. Arabidopsis F-box proteins have been shown to be essential for auxin (Ruegger et al., 1998; Dharmasiri et al., 2005a, 2005b; Tan et al., 2007), methyl jasmonate (Xie et al., 1998; Xu et al., 2002; Chini et al., 2007; Thines et al., 2007), gibberellin (Strader et al., 2004), and ethylene (Guo and Ecker, 2003; Potuschak et al., 2003) signaling. In addition, F-box proteins regulate the circadian clock, photomorphogenesis (Büche et al., 2000; Marrocco et al., 2006), senescence (Stirnberg et al., 2007), floral development, self-incompatibility, and responses to (a)biotic stresses (Lechner et al., 2006). In Arabidopsis, four ACIF1 homologs were identified, which were recently named VIER F-BOX PROTEINE1 (VFB1) to VFB4 (Navarro et al., 2004; Schwager et al., 2007). Knockdown of the entire VFB gene family resulted in the repression of auxin-responsive genes and cell wall metabolic genes, stunted growth, and reduced lateral root formation.
Several lines of evidence suggest that SCF complexes play a role in disease resistance. For instance, VIGS of SKP1, SGT1 (for SUPPRESOR OF G-2 ALLELE OF SKP1), and COI1 (for CORONATINE-INSENSITIVE1) compromised N gene–mediated resistance to Tobacco mosaic virus (TMV) in Nicotiana benthamiana (Liu et al., 2002a, 2004). VIGS of SGT1 also impaired the HR mediated by R genes, including Cf-9 and Cf-4 (Peart et al., 2002). Finally, the Arabidopsis F-box protein SUPPRESOR OF NIM1-1 negatively regulates plant defense responses to virulent Hyaloperonospora parasitica and Pseudomonas syringae pv tomato DC3000 independently of systemic acquired resistance and salicylic acid (SA) accumulation (Kim and Delaney, 2002).
Here, we show that the F-box domain of ACIF1 interacts with Arabidopsis SCF subunits and that mutations in the F-box domain abolish these interactions. Silencing of ACIF1 not only compromises the HR mediated by Cf genes but also the HR mediated by R genes belonging to the NB-LRR class (van Ooijen et al., 2007). Tobacco ACIF1 plays a role in N-mediated TMV lesion formation and in cell death triggered by Pseudomonas syringae pv tabaci (Pst). Expression of ACIF1 F-box mutant protein in tobacco results in a dominant-negative phenotype similar to ACIF1 silencing. In tomato, ACIF1 silencing compromises Cf-9B–mediated resistance to C. fulvum. Gene expression data obtained in an Arabidopsis knockdown mutant of the ACIF1 homologs (VFBs) (Schwager et al., 2007) suggest a novel role for this gene family in regulating methyl jasmonate (MeJA)– and abscisic acid (ABA)–dependent responses. The G-box DNA binding site and its derivatives are overrepresented in the promoter of these MeJA- and ABA-responsive genes. Together, these data support a role for ACIF1/VFBs in the regulation of plant defense responses.
RESULTS
ACRE189 Encodes a Widely Conserved F-Box/LRR Protein
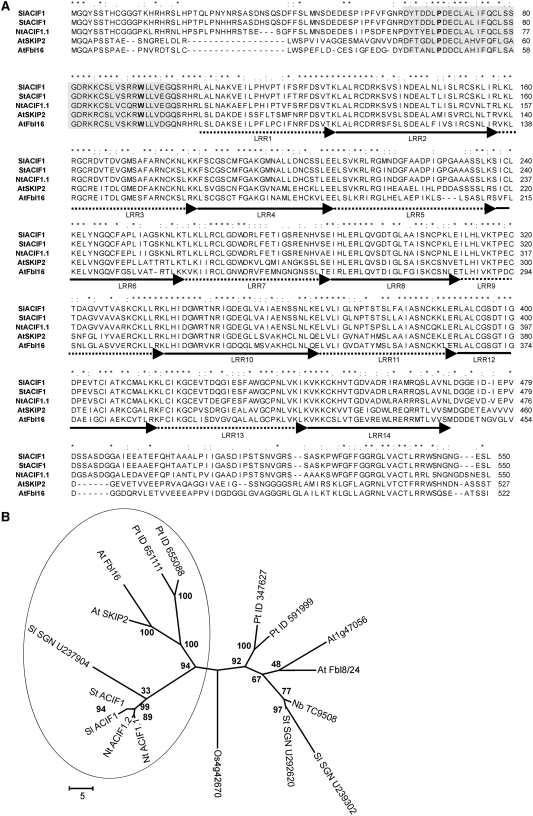
VIGS using the ACRE189 cDNA fragment compromised Cf-9– and Cf-4–mediated HR in N. benthamiana (Rowland et al., 2005). This ACRE189 fragment was used to screen a cDNA library derived from Avr9-elicited tobacco cells to obtain several independent full-length cDNA clones. Two highly similar full-length cDNA clones (96% identical at the protein and DNA levels) were identified. Each clone encodes a 550-residue protein consisting of a 39-residue F-box motif (residues 58 to 96) followed by 14 putative LRRs (residues 99 to 469) of variable length (∼24 to 29 residues) (Figure 1A; see Supplemental Figure 1 online). Based on the F-box motif, ACRE189 was renamed ACIF1.
Figure 1.
Sequence Analysis of ACIF1 Homologs in Different Plant Species.
(A) Protein sequence alignment of ACIF1 homologs from tobacco (Nt), tomato (Sl), potato (St), and Arabidopsis (SKIP2 and Fbl16). The gray box denotes the F-box motif, while the putative LRRs are delineated by arrows. Identical amino acids are indicated by asterisks, and similar residues are marked by colons or dots. The conserved residues P64 and W90 of tobacco ACIF1 are highlighted in boldface.
(B) Phylogenetic tree of ACIF1 homologs identified in tobacco, tomato, potato, N. benthamiana, Arabidopsis, rice (Os), and poplar (Pt). The circle denotes putative ACIF1 orthologs, while the others likely represent ACIF1 paralogs. Bootstrap values are shown at the nodes.
A BLAST search in the tomato (Solanum lycopersicum) EST databases SOL Genomics Network (SGN; www.sgn.cornell.edu/) and The Institute for Genomic Research (TIGR; www.tigr.org/) revealed two tomato ACIF1 homologs. Alignment of these tomato EST contigs with ACIF1 showed that contig SGN-U218766/TIGR-TC164112 was the putative tomato ortholog of ACIF1, sharing 88% identity at the protein level. The other contig (SGN-U237904) covered only 40% of the tobacco ACIF1 sequence (mostly the 3′ part), with 73% identity to tobacco ACIF1 at the protein level. The Sl ACIF1/SGN-U218766 cDNA encodes a predicted protein of 550 residues with a molecular mass of 60 kD (Figure 1A). We also identified a putative potato (Solanum tuberosum) ortholog of ACIF1 by combining three overlapping EST contigs (SGN-U273676, SGN-U273677, and SGN-U282566), resulting in a contig that covers the full length of ACIF1 with 99/87% identity to the Sl/Nt ACIF1 protein, respectively. In addition, other ACIF1 homologs in tomato, potato and N. benthamiana (Sl SGN-U239302, St SGN-U292620, and Nb TC9508) shared ∼61% identity with tobacco ACIF1 (at the protein level), but none of these were represented by EST contigs that covered a complete cDNA; therefore, they were excluded from further studies.
Analysis of the Arabidopsis genome revealed a small gene family of four ACIF1 homologs (SKIP2/VFB4 [for ASK1-INTERACTING PROTEIN2/VFB4], Fbl16/VFB2, Fbl8/Fbl24/VFB3, and VFB1) (Navarro et al., 2004). Phylogenetically, these homologs belong to the C3/C4 clades of the F-box protein superfamily (Gagne et al., 2002). The phylogenetic tree of the Arabidopsis F-box family (Gagne et al., 2002) was based on the F-box protein motif of these F-box proteins, but intron–exon structure and other protein domains encoded by these genes formed additional support for the resulting tree (e.g., genes belonging to the combined C3/C4 clade also often encode a LRR region in addition to the F-box domain). Most gene members of this C3/C4 clade also are conserved between monocots and dicots, and their gene functions appear to be conserved (e.g., COI1 and GID2/SLEEPY1). This combined clade includes the well-characterized Arabidopsis F-box proteins TIR1/AFB1-3 (for TRANSPORT INHIBITOR RESPONSE1/TIR1 and its homologs, designated AUXIN SIGNALING F-BOX1 to -3], EFB1/EFB2 [EIN3 BINDING F-BOX1 and -2], EID1 [EMPFINDLICHER IM DUNKELROTEN LICHT1], ORE9/MAX2, and SKP2;1/SKP2;2. Moreover, this clade is not under positive selection pressure and rapid birth and death of genes, as was reported for large numbers of Arabidopsis F-box genes (Thomson, 2006). This would indicate that the function of ACIF1 is evolutionarily conserved. Indeed, we found in the draft genome of poplar (Populus trichocarpa) (version 1.0; www.jgi.doe.gov/poplar) four homologous sequences of ACIF1 (i.e., protein identifiers 347627, 591999, 655088, and 651111, with BLAST E-values = −90). Finally, we and Jain et al. (2007) identified one rice (Oryza sativa) homolog of ACIF1 (Os4g42670) by searching the TIGR rice EST and genome databases. All of the retrieved protein sequences were aligned (see Supplemental Table 1 online), and this alignment was used to build a phylogenetic tree. The phylogenetic tree suggests that the ACIF1 preancestor gene has undergone several independent gene duplication events in the different branches (i.e., we found distinct branches for poplar, Arabidopsis, and Solanaceae sequences) (Figure 1B). Therefore, it is difficult to annotate ACIF1 orthologs outside the Solanaceae family based solely on protein sequence similarity.
Tobacco and Tomato ACIF1 Are Upregulated upon Avr9 Elicitation and Wounding
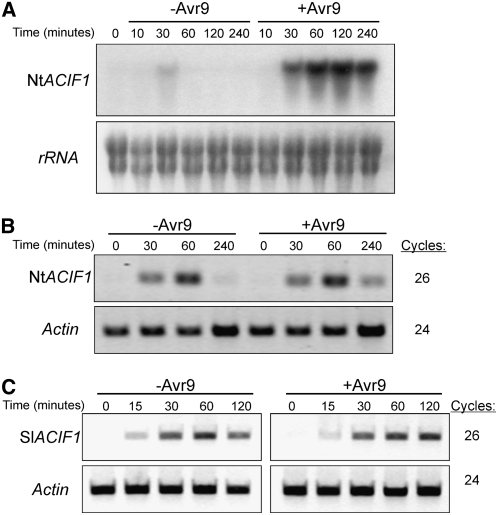
To confirm that the cloned genes corresponded to the initial AFLP fragment, we examined ACIF1 gene expression following Avr9 elicitation. Intracellular fluid (IF) containing Avr9 or IF without Avr9 was added to Cf-9 tobacco cell cultures, and ACIF1 transcript levels were analyzed over 4 h using RNA gel blot analysis (Figure 2A). ACIF1 was first induced at 30 min after adding IF+Avr9, expression reached maximum levels after 2 h, and it remained elevated until 4 h. Mock-treated cell cultures (IF−Avr9) showed only transient ACIF1 expression at 30 min, and expression levels were significantly reduced compared with IF+Avr9–treated samples (Figure 2A). Using RT-PCR, we analyzed ACIF1 expression levels in mature tobacco and tomato leaves after Avr9 elicitation. When mature tobacco leaves were infiltrated with IF+Avr9, ACIF1 was induced in a manner similar to that in tobacco cell suspensions. However, ACIF1 transcript levels also reached similar levels in tobacco leaves after infiltrating IF without Avr9 (Figure 2B). This most likely reflects wound- or stress-specific gene induction, which is triggered by leaf flooding. Similar observations were reported previously for all other tested ACRE genes (Durrant et al., 2000; Rowland et al., 2005; Gonzalez-Lamothe et al., 2006; Yang et al., 2006). The expression pattern of the tomato ACIF1 ortholgue did not differ from that of tobacco ACIF1 after IF infiltration (+Avr9 and −Avr9) in leaves (Figure 2C). Based on this overlap in gene expression and the high sequence homology, we propose that the cloned tomato ACIF1 gene is the tomato ortholog of the tobacco ACIF1 gene.
Figure 2.
Expression Pattern of ACIF1 after Avr9 Elicitation and Wounding.
(A) ACIF1 expression in Cf-9 tobacco cell cultures after adding IF with or without Avr9. Following treatment, cultured suspension cells were harvested at the indicated times and total RNA was isolated for RNA gel blot analysis. The blot was hybridized with a probe specific for the 3′ part of ACIF1 (top panel).
(B) ACIF1 expression in Cf-9 tobacco leaves after infiltration with IF+Avr9 or IF−Avr9. Leaf tissue samples were collected at the indicated times, and total RNA was extracted for RT-PCR analysis. Actin transcript levels are shown as a control for cDNA loading. The number of cycles used during PCR amplification is shown.
(C) ACIF1 expression in tomato (cv Moneymaker Cf9) leaves after infiltration with IF+Avr9 or IF−Avr9. Leaf tissue samples were collected at the indicated times, and total RNA was extracted for RT-PCR analysis. Actin transcript levels are shown as a control for cDNA loading. The number of cycles used during PCR amplification is shown.
Tobacco ACIF1 Forms an SCF Complex with ASK1/2 and CUL1
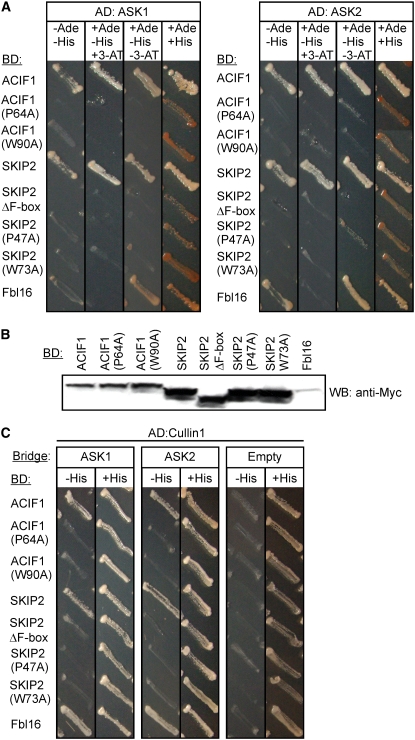
Using yeast two-hybrid interactions (Y2H), we tested whether tobacco ACIF1 interacts with subunits of the SCF complex. ACIF1 and two Arabidopsis homologs of ACIF1 (SKIP2 and Fbl16) were fused to the GAL4 binding domain (BD). These BD-F-box protein fusions were coexpressed in yeast with Arabidopsis ASK1 or ASK2 (both fused to the GAL4 activation domain [AD]). The full-length BD-F-box fusion proteins interacted with AD-ASK1 and AD-ASK2 fusion proteins, as indicated by selective growth on medium lacking Ade or His (3-amino-1′,2′,4′-triazole was added to suppress leaky HIS3 expression) (Figure 3A). These interactions between ACIF1/SKIP2 and the ASK proteins were also noted when yeast was grown on limiting amounts of Ade (+Ade/+His) (i.e., transformants expressing full-length ACIF1 or SKIP2 did not accumulate red pigment, which is indicative of impaired adenine biosynthesis in the yeast strain used). The Ade levels, however, were sufficient to allow yeast growth for several days. The interaction between Fbl16 and ASK1 is sensitive to 3-amino-1′,2′,4′-triazole, suggesting leaky HIS3 expression, but specific growth was detected on the Ade selection marker for the interaction with ASK2. Moreover, Fbl16 accumulated to lower levels than ACIF1 or SKIP2 in yeast (Figure 3B). Y2H had previously demonstrated that SKIP2 and Fbl16 both interact with different ASKs (Farras et al., 2001; Gagne et al., 2002; Schwager et al., 2007). The Y2H with ASK1 and ASK2 proved to be F-box–dependent, as deletion of the F-box domain resulted in the loss of interaction with ASK1 and ASK2 in Y2H (ΔF-box) (Figure 3A; see Supplemental Figure 2 online). We also separately mutated two conserved residues in ACIF1 (and SKIP2) that are presumed to be located at the interaction surface between the F-box domain and the ASK proteins based on the crystal structure of the yeast SCF complex, where these two residues were shown to be essential for this interaction (Schulman et al., 2000; Zheng et al., 2002). As expected, the mutant ACIF1 (P64A and W90A) and mutant SKIP2 (P47A and W73A) proteins did not interact with ASK1 or ASK2 when applying stringent (growth on medium –Ade/–His) or moderate (growth on medium –His alone) selection. The loss of interaction was not the result of altered expression levels, as mutant ACIF1 or SKIP2 was expressed at similar levels as wild-type ACIF1 or SKIP2 in yeast (Figure 3B).
Figure 3.
ACIF1, SKIP2, and Fbl16 Interact with Subunits of the SCF Complex in the Y2H Assay.
(A) Y2H assay of interactions between ASK1 or ASK2 and the F-box proteins ACIF1, SKIP2, and Fbl16. Proteins with a deletion of (ΔF-box) or mutations in [ACIF1(P64A)/(W90A) and SKIP2(P47A)/(W73A)] the F-box motif were also tested. Yeast are shown after growth for 5 d on moderate (−His medium) or stringent (−Ade/−His medium) selection for protein–protein interactions.
(B) Protein gel blot analysis demonstrating expression of the Myc-tagged BD-F-box fusion proteins in the yeast strains used in (A).
(C) Y3H assay of ACIF1 interactions with CUL1 via ASK1 or ASK2. One yeast mating type was cotransformed with an AD-CUL1 construct and the different F-box-BD fusion constructs. These transformants were mated with yeast carrying either the empty bridge vector or the bridge vector harboring ASK1 or ASK2. Yeast was grown for 5 d on moderate (−His medium) selection for protein–protein interactions.
We also examined whether ACIF1 can interact indirectly with Arabidopsis CUL1 via ASK1 or ASK2 as a bridge in a yeast three-hybrid (Y3H) interaction (Figure 3C). The BD-F-box fusions and the AD-CUL1 construct were cotransformed into the yeast mating type MATa, while the bridge vector (pVT102-U) carrying ASK1 or ASK2 was transformed in the yeast mating type MATα. After mating, interactions were tested by growing yeast for 5 d on moderate (−His) or stringent (−Ade/−His) selection for protein–protein interactions. Growth was noted only on moderate selection (−His) for yeast expressing AD-CUL1, any of the three wild-type BD-F-box protein fusions (ACIF1, SKIP2, or Fbl16), and ASK1 or ASK2. All other prey–bait combinations (including empty vector controls) tested negative for protein–protein interactions. We conclude from this analysis that ACIF1 can form a trimeric SCFACIF1 complex in yeast consisting of ASK1/2, CUL1, and ACIF1.
Knockdown of Tobacco ACIF1 Compromises the HR Triggered by Different Elicitors
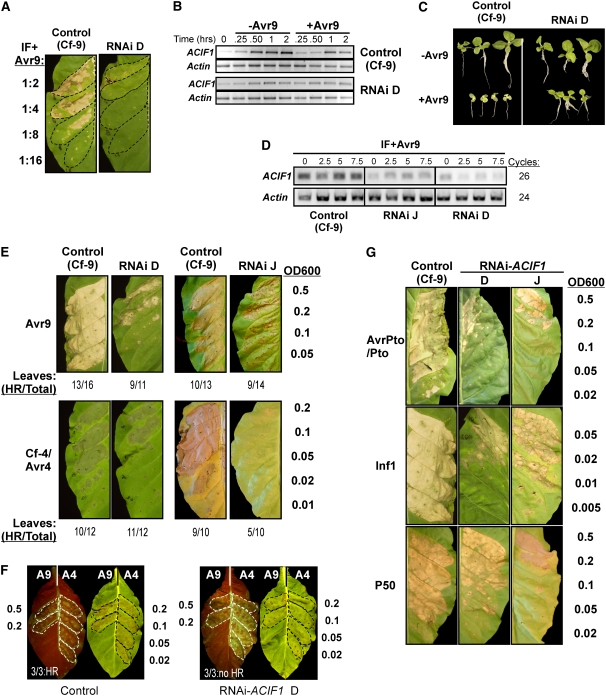
To study ACIF1 function, we engineered an RNA interference (RNAi) construct to silence ACIF1 in Cf-9 tobacco (using an intron-containing self-complementary hairpin construct that triggers double-stranded RNA–mediated degradation of the ACIF1 mRNA). After plant transformation, 14 independent lines were obtained. Two lines (D and J) showed a reduced HR after infiltrating different dilutions of IF+Avr9 between the lateral veins of mature leaves (Figure 4A). These lines also showed reduced ACIF1 expression after IF infiltration compared with the control (Cf-9 tobacco) (Figure 4B). This indicates that RNAi-mediated silencing of ACIF1 compromises Avr9-triggered HR, as seen previously with VIGS of ACIF1 in N. benthamiana (Rowland et al., 2005). In addition, seedling growth inhibition assays indicated that these two lines were less responsive to Avr9 (i.e., RNAi-ACIF1 seedlings [lines D and J] developed lateral roots, showed continued root elongation, and did not have chlorotic primary leaves when these seedlings were grown on medium containing IF+Avr9) (Figure 4C; see Supplemental Figure 3 online). By contrast, control seedlings (Cf-9 tobacco) showed stunted growth, poor lateral root development, and chlorotic leaves in the presence of IF+Avr9. In the absence of Avr9, both the RNAi-ACIF1 and the parental Cf-9 seedlings had similar growth, thus indicating that the observed growth deficiencies are strictly caused by Avr9 recognition. RNA was extracted from these seedlings at 21 d after transfer to medium containing IF, and ACIF1 mRNA levels were determined using RT-PCR. ACIF1 transcript levels were reduced in both the RNAi-ACIF1 lines compared with control seedlings (Cf-9 tobacco) (Figure 4D). The other 12 T1 lines did not show reduced Avr9 sensitivity in this seedling assay and were not analyzed further. The two lines that showed ACIF1 silencing cosegregated 1:3 (T1) for BASTA resistance and the growth inhibition phenotype, which indicates a single insertion event. Homozygous lines were selected for further phenotypic analysis.
Figure 4.
RNAi-Mediated Silencing of ACIF1 in Tobacco Compromises the HR Triggered by Different Elicitor/R Protein Combinations.
(A) Avr9-dependent HR is compromised in RNAi-ACIF1 (line D) and not in the control after infiltrating different dilutions of IF+Avr9. The photograph was taken after 4 d, when the HR had completely developed.
(B) ACIF1 expression levels in leaves of RNAi-ACIF1 (line D) and the control after infiltrating IF with or without Avr9. Leaf samples were taken at the indicated time points, and RT-PCR was used to determine ACIF1 and actin (as a control) mRNA levels.
(C) Cf-9 seedlings show poor root development, stunted growth, and leaf chlorosis in the presence of 5 μL IF+Avr9/mL medium. RNAi-ACIF1 seedlings (line D) show less growth inhibition in response to similar Avr9 concentrations, as indicated by intermediate growth, no leaf chlorosis, and sustained root development. Photographs were taken at 21 d after transfer to medium containing IF.
(D) ACIF1 expression levels are reduced in RNAi-ACIF1 seedlings (lines D and J) compared with the control after 21 d of growth on medium containing Avr9 (0, 2.5, 5, and 7.5 μL IF+Avr9/mL medium). RT-PCR was used to determine ACIF1 and actin (as a control) mRNA levels.
(E) Assay of the HR to transient expression of Avr9 or Cf-4/Avr4 in RNAi-ACIF1 tobacco leaves. The leaf areas between the lateral veins received the indicated Agrobacterium strain at the labeled OD600. The experiment was performed four times with three to four plants per line per experiment. Photographs were taken at 4 d after agroinfiltration (HR, leaves with similar HR levels as depicted; Total, total number of leaves tested).
(F) RNAi-ACIF1 and Cf-9 (control) tobacco plants were crossed with Cf-4 tobacco. Leaves of the resulting F1 plants were infiltrated with Agrobacterium carrying Avr9 (left leaf half/A9) or Avr4 (right leaf half/A4). Leaves are shown at 4 d after agroinfiltration (autofluorescence of the same leaf is shown at left in each panel). Highlighted areas of each leaf were infiltrated with the indicated OD600. Three plants were tested for each cross with similar results, as indicated at bottom in each panel.
(G) As in (E), except that Agrobacterium was used to transiently deliver Pto/AvrPto, Inf1, or the P50 helicase of TMV.
To examine whether RNAi-mediated silencing of ACIF1 also compromises the Cf-4–mediated HR, we transiently coexpressed Avr4 and Cf-4 in ACIF1-silenced tobacco leaves by infiltrating different dilutions of an Agrobacterium tumefaciens suspension between the lateral veins (Avr9 was transiently expressed in the other leaf half using agroinfiltration as well). This dose-dependent elicitor expression assay revealed again that RNAi-ACIF1 lines are compromised in Avr4- and Avr9-triggered HR. Symptoms varied from chlorosis to patchy necrosis instead of the confluent necrosis that was seen with the nonsilenced control at similar dilutions (Figure 4E). A complete collapse of the infiltrated area was obtained consistently only when the OD600 of the infiltrated Agrobacterium suspensions was increased by 5 to 10 times for the RNAi-ACIF1 lines. We also crossed RNAi-ACIF1 line D with Cf-4 tobacco and again performed a dose-dependent elicitor expression experiment in the F1 crosses. The RNAi-ACIF1 × Cf-4 F1 plants also showed compromised HR when transiently expressing Avr4 or Avr9 in planta, confirming the dominant phenotype caused by silencing ACIF1 (Figure 4F). The areas that did not turn necrotic displayed autofluorescence, most likely caused by the accumulation of phenolic compounds (Nicholson and Hammerschmidt, 1992). This is associated with defense responses and indicates that RNAi-ACIF1 tobacco still partially responds to agroinfiltration.
Similarly, we tested whether ACIF1 expression is required for cell death triggered by the following non-Cf-like R/Avr gene pairs: (1) Pto/AvrPto, (2) N gene/the P50 helicase of TMV, and (3) the general elicitor Inf1 from Phytophthora infestans. We noted a dose-dependent compromise of the HR in response to the expression of each of these elicitors in the RNAi-ACIF1 lines (Figure 4G). Depending on the tested elicitor/R protein combination, the OD600 of the infiltrated Agrobacterium suspensions needed to be increased by 5 to 10 times to induce similar levels of HR in the RNAi-ACIF1 lines and in the nonsilenced control. These findings indicate that ACIF1 silencing attenuates the HR triggered by various elicitors.
Wild-Type ACIF1 Protein Is Degraded in an SCF- and 26S Proteasome–Dependent Manner
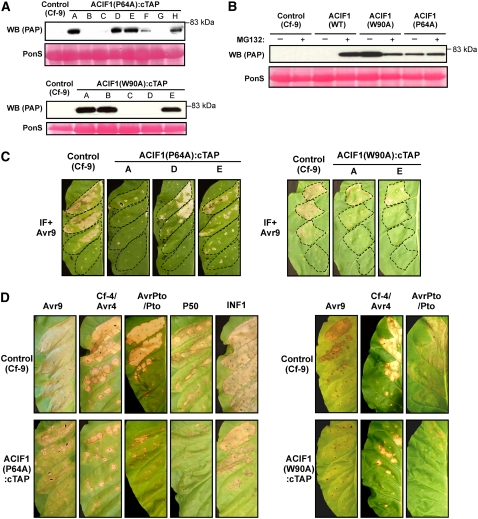
To study protein function, we created transgenic tobacco expressing C-terminal TAP-tagged (for tandem-affinity purification tag) (Rigaut et al., 1999; Rohila et al., 2004) ACIF1. In parallel, we created transgenic tobacco expressing mutant ACIF1:cTAP. Transformation of Cf-9 tobacco resulted in 15, 8, and 5 independent BASTA-resistant T1 lines for wild-type ACIF1:cTAP, ACIF1(P64A):cTAP, and ACIF1(W90A):cTAP, respectively. Surprisingly, we could not detect wild-type ACIF1:cTAP in the soluble protein fraction obtained from mature leaves in any of the transgenic lines. However, both ACIF1(P64A):cTAP and ACIF1(W90A):cTAP (∼80 kD) were detected readily in the soluble protein fraction of several transgenic lines (Figure 5A). The lines that show accumulation of mutant ACIF1 protein (P64A, lines A, D, and E; W90A, lines A and E) all segregated 1:3 for the BASTA resistance, which indicates a single insertion event. In yeast, the recruitment of F-box proteins to SCF complexes can trigger autoubiquitination of the F-box proteins when they do not cointeract with their targets (Cope and Deshaies, 2006). This leads to degradation of the F-box proteins by the 26S proteasome. Accumulation of the mutant F-box ACIF1 proteins in our tobacco lines thus suggests that the mutations each disrupt the interaction with the SCF complex in planta and increase the stability of the protein. This hypothesis predicts that wild-type ACIF1:cTAP is expressed in some lines but that the protein is degraded due to autoubiquitination. To test this, 3-week-old seedlings were incubated for 4 h with the 26S proteasome inhibitor MG132, which effectively blocks 26S proteasome–dependent protein degradation. Treatment with MG132 indeed allowed the accumulation of wild-type ACIF1 (Figure 5B). Together, these findings argue that wild-type ACIF1:cTAP protein is degraded in an SCF complex– and 26S proteasome–dependent manner.
Figure 5.
Expression of ACIF1(P64A) or ACIF1(W90A) Compromises the HR Triggered by Different Elicitor/R Protein Combinations.
(A) Mutant ACIF1 protein accumulates in several independent transgenic tobacco lines expressing ACIF1(P64A):cTAP or ACIF1(W90A):cTAP. Total protein was extracted from mature leaves, and immunoblotting was used to detect TAP-tagged protein. Ponceau S staining confirmed equal protein loading for all samples (bottom panels). The parental line (Cf-9 tobacco) is loaded in the first lane.
(B) Tobacco seedlings expressing wild-type ACIF1 (line M), ACIF1(P64A) (line E), or ACIF1(W90A) (line A) were incubated for 4 h with the 26S proteasome inhibitor MG132 or mock-treated (+DMSO) prior to protein extraction. Immunoblotting was used to detect TAP-tagged protein. Ponceau S staining confirmed equal protein loading for all samples (bottom panel).
(C) Expression of ACIF1 F-box mutants (P64A or W90A) in Cf-9 tobacco compromises the Avr9-triggered HR. Three transgenic lines were tested for tobacco expressing ACIF1(P64A) (lines A, D, and E; left) and two lines for tobacco expressing ACIF1(W90A) (lines A and E; right). Photographs were taken at 4 d after infiltration with IF+Avr9 at the following dilutions (top to bottom): 1:2, 1:4, 1:8, and 1:16 in the highlighted leaf area.
(D) Tobacco expressing ACIF1(P64A) (line A) or ACIF1(W90A) (line A) showed a dose-dependent reduction of the HR after transient expression of different elicitors and their corresponding R proteins (when needed). Inf1 and the P50 helicase of TMV were only tested for tobacco expressing ACIF1(P64A). The infiltrated OD600 dilution series for the different elicitors was the same as that in Figure 4. Photographs were taken at 4 d after infiltration.
Expression of Mutant ACIF1 Confers a Dominant-Negative Phenotype
Accumulation of mutant ACIF1 protein might suppress native ACIF1 function by sequestering target proteins that are normally ubiquitinated by SCF complexes. This would result in a dominant-negative phenotype (less HR) similar to silencing ACIF1. Indeed, we found that infiltration of IF+Avr9 at concentrations that caused full necrosis in control leaves triggered only responses ranging from chlorosis to necrotic patches in leaves of plants accumulating ACIF1(P64A) or ACIF1(W90A) protein (Figure 5C). Also, when we transiently expressed (using agroinfiltration) Avr9 or other elicitor/R gene combinations (Cf-4/Avr4, Pto/AvrPto, N/P50, and Inf1) in tobacco expressing ACIF1(P64A), we noticed that the concentration of the Agrobacterium suspension needed to be increased by approximately fivefold in order to obtain similar levels of HR as in control leaves (Cf-9 tobacco) (Figure 5D). Similarly, for tobacco expressing ACIF1(W90A), we noticed that the infiltrated OD600 could be increased five times for the Cf-9/Avr9, Cf-4/Avr4, and Pto/AvrPto combinations. These experiments were repeated at least four times with similar results. This dominant-negative phenotype (reduced HR) indicates that the accumulation of mutant ACIF1 protein interferes with endogenous ACIF1 function.
ACIF1 Promotes N Gene–Mediated Resistance Responses to TMV Infection
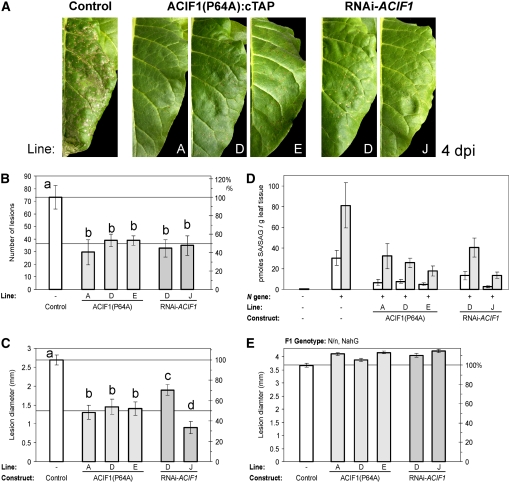
To test for N gene–mediated resistance to TMV, ACIF1-transgenic tobacco plants were rub-inoculated with sap containing infectious TMV. TMV lesions first became visible at 2 d after inoculation (DPI), and the number of lesions formed remained constant from 4 to 7 DPI. Lesion expansion was linear over this period as well (for all plants). The number of lesions formed on the RNAi-ACIF1 plants, however, was decreased by ∼50% compared with the control (Cf-9 tobacco) (Figures 6A and 6B). The average lesion diameter was also reduced on the RNAi-ACIF1 plants (i.e., reductions of ∼30 and 67% in lesion diameter were seen in lines D and J, respectively) (Figure 6C). Also, three different lines of tobacco expressing ACIF1(P64A) developed ∼50% fewer lesions than the control, and these lesions remained ∼50% smaller than in the control. Reduced TMV lesion formation and growth could be a consequence of delayed HR or an increased threshold for P50-triggered HR. Consequently, the delayed lesion formation may lead to viral escape from the initial infection sites, allowing systemic viral spread. However, we never observed mosaic-like symptoms in uninfected leaves or main vein necrosis on TMV-infected ACIF1 transgenic tobacco plants, two classic symptoms observed on TMV-infected (partially) susceptible plants. Systemic spread of TMV was also tested at 21 DPI (in the late stages of infection) by rub-inoculation of resistant tobacco leaves with undiluted sap prepared from the top leaves of infected plants. None of the plants developed TMV lesions, whether they were infected with sap from the resistant control or undiluted sap obtained from the TMV-infected ACIF1 transgenic plants. This indicates that despite reduced lesion growth, reduced ACIF1 function does not lead to systemic viral spread.
Figure 6.
ACIF1 Is Required for N Gene–Mediated Resistance Responses to TMV Infection.
(A) Photographs of tobacco leaves at 4 d after rub-inoculation with TMV. Cf-9 control plants, as well as three lines expressing ACIF1(P64A) and two lines with RNAi silencing of ACIF1, are shown.
(B) Mean (±se) number of TMV lesions formed at 4 DPI on RNAi-ACIF1, ACIF1(P64A), and the parental control (Cf-9) tobacco. At least two independent transgenic lines were examined per construct (eight plants per line), and the experiment was repeated more than four times with similar results. Means were compared by Student's t test. Different letters above the bars indicate significantly different means (P < 0.05).
(C) Mean (±se) TMV lesion diameter at 7 DPI on RNAi-ACIF1, ACIF1(P64A), and the parental control (Cf-9) tobacco. Means were compared by Student's t test. Different letters above the bars indicate significantly different means (P < 0.05). Per line, we quantified at least 80 lesions.
(D) Mean (±se; six biologically independent samples per line) SA (white bars) and SAG (gray bars) levels in the primary infected leaf of the indicated lines at 7 d after TMV infection. Tobacco cv Petite Havana was included as a susceptible control (lacking the N gene) and Cf-9 tobacco as a resistant control (carrying the N gene).
(E) Mean (±se) TMV lesion diameter in F1 crosses between NahG tobacco (nn; lacking the N gene) and RNAi-ACIF1, ACIF1(P64A), and the parental control (Cf-9) tobacco plants (homozyogous for the N gene; NN) at 7 d after TMV inoculation. Comparison of the means with Student's t test indicated that none of the means was significantly different (40 lesions measured per plant line).
Also, VIGS of ACIF1 in N. benthamiana carrying the N gene did not result in TMV:GFP (for green fluorescent protein) accumulation (examined by UV illumination) in leaf areas infiltrated with Agrobacterium carrying TMV:GFP, although the N gene–mediated HR was partially lost due to ACIF1 silencing (see Supplemental Figure 4 online). SA accumulation is important for both lesion formation and the inhibition of viral replication around the initial infection zone to prevent systemic viral spread (Mur et al., 1997; Murphy et al., 1999). We determined SA and conjugated SA (SAG) levels in the primary infected leaves at 6 DPI (Figure 6D; similar data were obtained for 5 DPI). The SA/SAG levels in plants expressing mutant ACIF1(P64A) protein or RNAi-ACIF1 plants were only ∼20 to 35% of the levels in the resistant control (Cf-9 tobacco), but the SA/SAG levels were still increased compared with the susceptible control (cv Petite Havana). Thus, the lesion phenotype is reflected by reduced SA/SAG levels at 5 to 6 DPI. Despite the reduced SA/SAG levels, accumulation of SA/SAG could still prove to be essential for the residual TMV resistance found in the RNAi-ACIF1 and ACIF1(P64A) plants. To test this, we crossed tobacco RNAi-ACIF1 and ACIF1(P64A) with tobacco expressing NahG (Brading et al., 2000). NahG converts SA to catechol, which results in ∼1.5 times increased TMV lesion diameters on primary infected leaves (cf. Figures 6C and 6E) and allows systemic viral spread, as manifested by vein necrosis (Gaffney et al., 1993). The F1 crosses between NahG and the ACIF1 transgenic tobacco developed TMV lesions of similar size as the NahG control, which were ∼1.5 times increased in diameter relative to the resistant non-NahG control (Figures 6C and 6E). Also, the number of lesions that developed did not differ between the NahG F1 crosses and the NahG control. This indicates that the residual SA levels contribute to the RNAi-ACIF1–triggered reduction of TMV lesions and the reduction in lesion size.
ACIF1 Is Required for Necrotic Responses following Infection with Pst
To examine ACIF1 function during bacterial infection, we used Pst as our model system. For an incompatible interaction, we used Pst expressing AvrPto (Ronald et al., 1992; Thilmony et al., 1995). First, we conducted an inoculum titration experiment to determine whether the dose-dependent cell death response was transgenic changed in transgenic tobacco expressing RNAi-ACIF1, ACIF1(P64A), or ACIF1(W90A) (Figure 7A). Symptoms were monitored over time, and the infected leaves were photographed at 6 DPI. To trigger confluent cell death in the infiltrated area as seen at lower doses in the control, we needed to increase the infiltrated dose of Pst by 1 to 2 orders of magnitude in the tobacco expressing RNAi-ACIF1, ACIF1(P64A), or ACIF1(W90A). In the case of Pst+AvrPto (Figure 7B), an increase of 2 to 3 orders of magnitude was required for tobacco expressing RNAi-ACIF1, ACIF1(P64A), or ACIF1(W90A). These results agree with our previous observation when transiently expressing AvrPto/Pto in tobacco. We also monitored bacterial growth of Pst and Pst+AvrPto in mature tobacco plants in time after infiltration of a low bacterial dose (2 × 104 colony-forming units/mL). Pst growth was not significantly different at 1 to 4 DPI between control, RNAi-ACIF1, ACIF1(P64A), or ACIF1(W90A) tobacco (see Supplemental Figure 5 online). However, Pst disease symptoms remained largely absent on RNAi-ACIF1 tobacco and tobacco expressing mutant ACIF1 after infection, while the control developed confluent cell death in the infiltrated area beginning at 2 to 3 DPI. In the case of the incompatible interaction (Pst+AvrPto), bacterial growth also did not differ significantly between any of the tested plants at any stage, but again we noted reduced cell death in the infiltrated leaf areas of tobacco expressing RNAi-ACIF1, ACIF1(P64A), or ACIF1(W90A) compared with the control. Similar results were obtained when tomato was infected with P.s. pv tomato strain DC3000, strain T1, or T1+AvrPto (Ronald et al., 1992): bacterial growth was similar between RNAi-ACIF1 tomato and the control from 0 to 4 DPI (see Supplemental Figure 6 online). Also, when we infected tomato RNAi-ACIF1 with a virulent strain (P.s. pv tomato DC3000), we noted reduced lesion formation compared with control plants (tomato cv Moneymaker Cf9). All of these observations support the conclusion that ACIF1 could negatively regulate a suppressor of Pst-triggered cell death in both tobacco and tomato, but ACIF1 appears not to be required to restrict bacterial growth.
Figure 7.
ACIF1 Function Is Required for AvrPto-Induced HR and Disease-Associated Cell Death Induced by Virulent Pst.
(A) Fully expanded leaves of 6-week-old tobacco [RNAi-ACIF1, ACIF1(P64A), ACIF1(W90A), and Cf-9 control] at 6 d after infiltration of different dilutions of Pst (empty vector) between the lateral veins.
(B) The same as in (A), except that Pst+AvrPto was infiltrated.
RNAi-ACIF1 Leads to Loss of Cf-9B, but Not Cf-9, Resistance to C. fulvum in Tomato
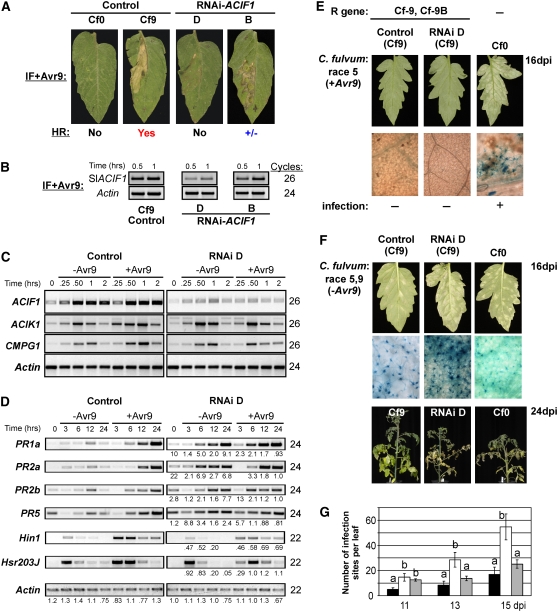
To examine the role of ACIF1 in resistance to C. fulvum, Cf9 tomato was transformed with the Nt ACIF1 hairpin construct to establish RNAi-mediated silencing of Sl ACIF1. The hairpin sequence is 87% identical to the tomato target sequence, sharing three stretches of identity of >21 nucleotides. This means that the tobacco ACIF1 sequence may effectively cross-silence the tomato ACIF1 ortholog. A search for potential tomato off-targets using the small interfering RNA (siRNA) scan tool (blasting the hairpin sequence against the tomato EST database [Xu et al., 2006]) did not reveal any potential off-targets in tomato. However, the program did predict 10 effective siRNAs that all target tomato ACIF1 based on a set of defined rules (Ui-Tei et al., 2004). Five independent primary kanamycin-resistant transformants were obtained. Two of these T1 lines (lines B and D) showed a compromised HR after challenge with different concentrations of IF+Avr9 (Figure 8A). Moreover, silencing of the tomato ACIF1 gene was most effective in line D, as deduced from the low levels of ACIF1 transcript (Figures 8B and 8C). This line also showed the most profound compromise in HR to infiltration of IF+Avr9. In corroboration, VIGS of ACIF1 in tomato also did not compromise Cf-9 resistance to C. fulvum in tomato, while the Avr9-dependent HR was compromised (see Supplemental Figure 7 online).
Figure 8.
RNAi-Mediated Silencing of Tomato ACIF1 Leads to the Loss of Cf-9B, but Not Cf-9, Resistance to C. fulvum.
(A) HR at 2 d after infiltration of IF+Avr9 in leaves of tomato RNAi-ACIF1 (lines B and D) compared with the negative and positive control plants (MM Cf0 and MM Cf9 tomato, respectively).
(B) RT-PCR analysis of ACIF1 transcript levels in tomato RNAi-ACIF1 (lines B and D) and a control after IF+Avr9 infiltration (0.5 and 1 h after infiltration). Tomato actin transcript levels are shown as a control for equal cDNA levels. The number of cycles used during PCR amplification is shown.
(C) RT-PCR analysis of leaf material at the indicated time points after infiltration with IF+Avr9 or IF−Avr9 (mock). Transcript levels were determined for ACIF1, ACIK1, and CMPG1. Actin transcript levels are shown as a control for equal cDNA levels. The number of PCR cycles is indicated at right for each primer combination.
(D) RT-PCR of PR genes (PR1A, PR2A, PR2B, and PR5) and HR marker genes (Hin1 and Hsr203J) in tomato RNAi-ACIF1 (line D) and the control after infiltration with IF−Avr9 or IF+Avr9. The relative difference in expression levels between the control and the RNAi D line are indicated below the panel for the different time points (the number signifies the fold change in signal intensity).
(E) Typical leaves (top panels) of 4-week-old tomato plants (cv MM; susceptible Cf0, resistant Cf9, and Cf9 transgenic for RNAi-ACIF1 D) at 16 DAI with C. fulvum race 5 (+Avr9). Conidiophores (blue threads, bottom panel) emerged through the stomata only in the case of a successful infection (Cf0), as microscopically confirmed after trypan blue staining.
(F) The same as in (E) except that plants were infected with C. fulvum race 5.9 (−Avr9). The plants are also shown at 24 DPI. This experiment was repeated four times with similar results.
(G) Mean (±se) number of infection sites macroscopically observed at 11, 13, and 15 DPI after infection with race 5.9 on Cf0 (gray bars), Cf9 (black bars), or RNAi-ACIF1 (white bars). Six plants were infected per line, and we counted the number of infection patches on two leaves for all plants per time point. Means were compared by Student's t test. Different letters above two columns indicate significantly different means at a single time point (P < 0.05). This experiment was independently repeated four times with similar results.
ACIF1 silencing did not affect the expression pattern of two other tomato ACRE genes (ACIK1 and CMPG1) in response to wounding (caused by infiltration) or Avr9 elicitation (Figure 8C). These two ACRE genes were shown to be essential for Cf-9 resistance (Rowland et al., 2005; Gonzalez-Lamothe et al., 2006). We also analyzed the transcriptional activation of defense-related genes using RT-PCR. Hin1 and Hsr203J were used as gene expression markers for HR execution (Gopalan et al., 1996; Hiraga et al., 2000), whereas acidic PR1 (PR1A), acidic β-1,3-glucanase (PR2A), basic β-1,3-glucanase (PR2B), and PR5 (Niki et al., 1998; Rivas et al., 2004; Gonzalez-Lamothe et al., 2006) were analyzed as markers for PR gene activation (Figure 8D). RNAi-ACIF1 tomato showed less induction of these two HR marker genes after infiltration of IF+Avr9 than did control plants. This supports our observation that the Avr9-triggered HR is compromised as a result of ACIF1 silencing. Strikingly, after infiltration with IF-Avr9, expression of the tested PR genes occurred earlier and was elevated in RNAi-ACIF1 tomato compared with the control plants. In fact, the expression levels of these genes in RNAi-ACIF1 without Avr9 reached the same levels obtained after infiltration of IF+Avr9. This indicates that ACIF1 silencing in tomato derepresses PR gene expression during the wound response.
To test for Cf-9-mediated disease resistance, RNAi-ACIF1 tomato was infected with C. fulvum race 5. At 16 DPI, leaves were microscopically inspected for fungal growth after trypan blue staining. The resistant Cf9 plants showed no clear infection sites after inoculation with C. fulvum race 5 (Figure 8E). By contrast, the susceptible Cf0 plants showed a large number of infection patches on the abaxial leaf surface with conidiophores emerging through the stomata, which confirmed that the infection itself was successful. Similar to the parental Cf9 line, RNAi-ACIF1 (line D) plants inoculated with C. fulvum race 5 showed no fungal infection sites, indicating that Cf-9 resistance is not lost in RNAi-ACIF1 tomato despite a compromised HR. We also tested Cf-9B–mediated resistance triggered by an unknown elicitor of C. fulvum race 5.9 (lacking Avr9). Cf-9B resistance is characterized by extended hyphal growth between mesophyll cells in comparison with Cf-9 resistance, and each inoculated leaf shows a few successful macroscopic fungal infection patches (Hammond-Kosack and Jones, 1994; Panter et al., 2002) (Figure 8F). The number of macroscopic infection patches was increased significantly on RNAi-ACIF1 tomato infected with C. fulvum race 5.9 (Figure 8G). Microscopic examination of these infection patches after trypan blue staining confirmed fungal infection at these sites, as shown by the conidiophores emerging through stomata (Figure 8F). At 24 DPI, the RNAi-ACIF1 plants showed severe wilting, tissue collapse, and enhanced leaf chlorosis, which closely resembled the susceptible Cf0 plants at the same infection stage. On the other hand, Cf9 tomato showed little wilting and chlorosis at this stage. Overall, ACIF1 silencing compromised the weak resistance mediated by the Cf-9B resistance gene, allowing full colonization of the plant by the fungus.
Knockdown of Arabidopsis ACIF1 Homologs (VFBs) Induces a Subset of MeJA- and ABA-Responsive Genes
The Arabidopsis genome contains a small family of four genes named VFBs that are homologs of ACIF1. This family appears to act redundantly (Schwager et al., 2007), as only gene knockdown of the entire gene family (vfb1-1 vfb2-1 vfb3-1 triple knockout/RNAi-VFB4) resulted in stunted plants and reduced lateral root formation. These phenotypes were not observed in single mutants (Schwager et al., 2007). Genome-wide gene expression analysis of the triple knockout/RNAi vfb mutant revealed that subsets of auxin-responsive genes and genes encoding cell wall metabolic enzymes are repressed in this mutant (Schwager et al., 2007).
We reexamined the expression data of the triple knockout/RNAi vfb mutant to evaluate whether this VFB gene family regulates defense-responsive genes and whether this regulation involves particular hormone signaling responses. First, we noted that marker genes for the MeJA responses were upregulated in the mutant (see Supplemental Table 2 online). These marker genes include MeJA biosynthetic genes and wound-responsive genes. Moreover, putative PR genes were either induced or repressed in the triple knockout/RNAi vfb mutant. MeJA is well known to differentially regulate PR gene expression. We examined whether the gene expression profile of the triple knockout/RNAi vfb mutant showed overlap with gene expression patterns obtained after specific hormone treatments (Nemhauser et al., 2006). Venn diagrams representing the overlap in gene expression between a single hormone treatment (at 30, 60, and 180 min) and the triple knockout/RNAi vfb line revealed no clear correlation (see Supplemental Figure 8A online and Supplemental Tables 3 to 5 online). Nevertheless, the highest overlap was found for ABA and MeJA treatments.
We also performed gene clustering to identify genes that are coregulated by VFB function and specific hormone treatments in wild-type Arabidopsis (ecotype Columbia [Col-0]). In total, we obtained 29 gene clusters (see Supplemental Figures 8B and 8C online and Supplemental Table 6 online). Seven gene clusters showed both specific gene expression after ABA treatment (180 min) and gene expression in the vfb knockdown mutant. In addition, we found five gene clusters that mostly represent genes that are progressively induced by MeJA (30, 60, and 180 min) and induced in the vfb knockdown mutant. These data indicate that ACIF1/VFBs repress ABA- and MeJA-responsive genes.
We also analyzed the promoters (1 kb upstream) of the genes that are differentially expressed in the vfb knockdown mutant for overrepresented regulatory cis-elements (Table 1). The DNA binding site of the auxin response factor (ARF) is significantly overrepresented among repressed genes in the vfb mutant, and the target binding site of WRKY transcription factors (W-box) is overrepresented among induced genes. WRKY transcription factors are well-known regulators of defense gene responses (Navarro et al., 2004). This favors a role for VFBs and ACIF1 in plant defense responses. However, the most compelling observation is the overrepresentation of the G-box (CACGTG) and related variants such as the ABRE(-like) binding site (Jakoby et al., 2002) and the MYC binding site (Toledo-Ortiz et al., 2003). The core G-box is overrepresented in the MeJA-specific cluster and is the target binding site of MYC2/JIN1 (for JASMONATE-INSENSITIVE1). The ABRE(-like) derivative is overrepresented in the gene clusters that show specific gene activity at 180 min after ABA treatment. These ABRE(-like) motifs are likely to be binding sites for basic domain/leucine zipper (bZIP) transcription factors (e.g., ABI5 and bZIP10) (Jakoby et al., 2002; Kaminaka et al., 2006). The promoter analysis thus supports the gene clustering analysis. Together, our microarray analysis reveals coregulation of late ABA-responsive genes, genes progressively induced by MeJA, and VFB-regulated genes. Therefore, we propose that the observed phenotypes are most likely established via the direct or indirect regulation of transcription factors that bind to the G-box or variants of this motif. Our data link VFB function to (a)biotic stress responses via the regulation of ABA- and MeJA-responsive genes.
Table 1.
Analysis of Overrepresented DNA cis-Elements in the Promoter Region (Maximum 1 kb Upstream) of Genes Differentially Regulated in the vfb Knockdown Mutant Background
| VFB Induced/Repressed (948 Genes Total)a
|
Induced (477 Genes)b | Repressed (471 Genes)c | ||||
|---|---|---|---|---|---|---|
| cis-Element | Core Sequence | G | N | P < | ||
| MeJA-responsive | ||||||
| G-box | CACGTG | 195 | 504 | 1.00E-10 | + | − |
| MYC AtERD1 | CATGTG | 393 | 564 | 1.00E-10 | − | + |
| At MYC2 RD22 | CACATG | 393 | 564 | 1.00E-10 | − | + |
| ABA-responsive | ||||||
| ABRE-like | (C/G/T)ACGTG(G/T)(A/C) | 254 | 402 | 1.00E-10 | + | − |
| ABRE | (C/T)ACGTGGC | 63 | 67 | 1.00E-03 | + | − |
| GA-DOWN | ACGTGTC | 102 | 126 | 1.00E-03 | + | − |
| ABRE A2OSEM | ACGTG(G/T)C | 181 | 243 | 1.00E-07 | + | − |
| Defense | ||||||
| W-box | TTGAC(C/T) | 615 | 1119 | 1.00E-05 | + | − |
| Auxin/ARF TFs | ||||||
| ARF binding site | TGTCTC | 337 | 428 | 1.00E-03 | − | + |
| Light response | ||||||
| I-box | GATAAG | 399 | 536 | 1.00E-10 | + | + |
All genes induced or repressed constitutively in the triple knockout/RNAi vfb mutant. G, number of promoters with the indicated cis-element among the set of genes constitutively induced/repressed in vfb knockdown plants; N, number of predicted cis-elements found in the promoter region of the set of genes induced/repressed in vfb knockdown plants; P, probability that the cis-element is enriched (P < 1E-3 is considered significantly enriched).
cis-element significantly overrepresented among induced genes (P < 1E-3).
cis-element significantly overrepresented among repressed genes (P < 1E-3).
DISCUSSION
ACIF1 Encodes an F-Box Protein Required for Elicitor-Triggered HR and Is Upregulated after Elicitor Recognition
ACIF1 (ACRE189) gene silencing (via RNAi or VIGS) and the expression of mutant ACIF1(P64A) or ACIF1(W90A) each reduced the HR mediated by several R proteins. The tested R proteins included members of the receptor-like protein class (Cf-4 and Cf-9), R proteins containing a nucleotide binding domain fused to LRRs (NB-LRR class; Prf and N), and an unknown protein that recognizes Inf1. Expression of ACIF1 is induced following the recognition of Avr9 by Cf-9, but ACIF1 expression is also induced by leaf flooding in tomato and tobacco. This leaf flooding/wounding response has been observed for all other ACRE genes characterized to date (Durrant et al., 2000; Rowland et al., 2005; Gonzalez-Lamothe et al., 2006). ACIF1 interacts in an F-box–dependent manner with subunits of the SCF complex (ASK1/ASK2/CUL1). Deletion of the F-box domain or the mutation of either of two conserved residues (P64A or W90A) in the F-box domain disrupted these interactions in Y2H. Y2H has been used widely to demonstrate the interaction between Arabidopsis F-box proteins and SCF complexes, including the Arabidopsis ACIF1 homologs/VFBs (Gray et al., 1999; Farras et al., 2001; del Pozo et al., 2002; Devoto et al., 2002; Gagne et al., 2002; Guo and Ecker, 2003; Potuschak et al., 2003; Marrocco et al., 2006; Schwager et al., 2007). The two mutated residues are part of the interaction surface between F-box domains and the SKP1 partner protein (Schulman et al., 2000; Zheng et al., 2002), and mutation of either of these residues abolishes the interaction. The corresponding residues were also essential for this interaction in the case of the Arabidopsis F-box proteins COI1 and TIR1 (Gray et al., 1999; Devoto et al., 2002).
SCF complexes act as ubiquitin E3 ligases that target specific substrates for degradation by the 26S proteasome (Lechner et al., 2006). We found that ACIF1(P64A) and ACIF1(W90A) accumulated in planta in the absence of the 26S proteasome inhibitor MG132. Wild-type ACIF1:cTAP was detected only after the incubation of seedlings with MG132, meaning that wild-type ACIF1:cTAP is degraded by the 26S proteasome. For yeast F-box proteins, this was caused by SCF-mediated autoubiquitination of F-box proteins followed by degradation by the 26S proteasome (Cope and Deshaies, 2006). Likewise, wild-type ACIF1:cTAP might be autoubiquitinated by the SCF complex and degraded. It is unclear whether endogenous ACIF1 is targeted for degradation or whether only overexpression of TAP-tagged ACIF1 triggers autodegradation. Conceivably, endogenous ACIF1 is targeted for degradation prior to defense activation, but more plausibly, ACIF1 and its bound targets become marked for ubiquitin-mediated degradation. The increased stability of the F-box ACIF1 mutant proteins would confirm that both mutations disrupt the interaction with the SCF complex in planta. The expression of ACIF1(P64A) or ACIF1(W90A) in tobacco leads to a dominant-negative phenotype similar to ACIF1 silencing (e.g., a compromised HR). This indicates that the accumulation of mutant ACIF1 protein suppresses ACIF1 function, most likely by sequestering target proteins, because the intact C-terminal part of ACIF1 is implicated in target binding, as indicated by the crystal structure of the related TIR1 F-box protein and its substrate (Tan et al., 2007). Closely related Arabidopsis F-box proteins (i.e., F-box proteins that also belong to the C3/C4 clade of the F-box superfamily) (Gagne et al., 2002) are predominantly nucleus-localized, and known targets of these plant F-box proteins to date are all transcription regulators (Gray et al., 1999, 2001; del Pozo et al., 2002; Guo and Ecker, 2003; Potuschak et al., 2003; Fu et al., 2004; Dharmasiri et al., 2005a, 2005b; Chini et al., 2007; Stirnberg et al., 2007; Thines et al., 2007). Strikingly, the Arabidopsis ACIF1 homologs (VFBs) appear to be located predominantly in the cytoplasm (Schwager et al., 2007).
ACIF1 Is Also Important for Defense/Wound Responses Other Than the HR
To examine the biological role of ACIF1 further, we examined the role of ACIF1 in N-mediated resistance to TMV, Pto-mediated resistance to P. syringae, and Cf-9– and Cf-9B–mediated resistance to C. fulvum. Knockdown of ACIF1 (via RNAi or the expression of mutant ACIF1 protein) resulted in fewer TMV lesions, and these lesions expanded less than those on resistant plants (N tobacco). Similarly, we found that SA levels were reduced in TMV-infected ACIF1 knockdown plants (6 DPI), although the SA levels were still increased compared with TMV-infected susceptible plants lacking the N gene. Proper SA accumulation is critical for TMV lesion formation and progression in resistant plants (Murphy et al., 1999). Thus, the observed lesion phenotype could be a consequence of reduced SA accumulation shortly after TMV infection. SA accumulation also is essential to inhibit TMV replication and to prevent systemic viral spread (Murphy et al., 1999). We did not observe systemic viral spread in RNAi-ACIF1 tobacco or plants expressing mutant ACIF1. Similarly, VIGS of ACIF1 in N. benthamiana did not compromise N gene resistance to TMV:GFP accumulation, but the N gene–mediated HR was partially lost due to ACIF1 silencing. Thus, the resistance responses downstream of the N gene still accumulate during TMV resistance in ACIF1 knockdown plants, and this could be due to residual SA accumulation. Alternatively, these late defense responses could be activated independently of ACIF1. The phenotypes associated with ACIF1 knockdown were lost in the NahG background (i.e., all plants showed NahG symptoms). NahG converts all SA to catechol, which results in the loss of N resistance and allows systemic viral spread (Gaffney et al., 1993). Therefore, loss of the ACIF1 knockdown phenotypes in the NahG background signifies that the residual SA levels in the ACIF1 knockdown plants prevent viral spread. The Avr9-triggered HR is also SA-dependent, and SA application increases the HR following Avr9 infiltration (Brading et al., 2000). Thus, the suppression of the Avr9-triggered HR due to the loss of ACIF1 function could be a consequence of reduced SA levels in these plants after Avr9 infiltration. SA accumulation, however, is not essential for Cf-9–mediated resistance (Brading et al., 2000).
Either ACIF1 silencing or the expression of mutant ACIF1 in tobacco resulted in reduced and delayed cell death triggered by Pst (+AvrPto or −AvrPto), while bacterial growth was not significantly changed in these plants. Disease symptoms induced by P.s. pv tomato (DC3000) in tomato also were reduced as a result of ACIF1 silencing. Bacterial growth of P.s. pv tomato (strain DC3000 or T1+AvrPto) (Ronald et al., 1992) also was not changed by ACIF1 silencing in tomato. Infiltration of Pst in tobacco leaves generally results in confluent cell death in the infiltrated area, and this cell death is more pronounced with Pst+AvrPto (Thilmony et al., 1995). It is unclear whether this cell death is the result of host manipulation by effectors (such as AvrPto) or of weak recognition of these effectors (Jones and Dangl, 2006). For instance, loss of the P.s. pv tomato effectors HopPtoM and HopPtoN does not lead to reduced bacterial growth in tobacco or tomato, but it does partially suppress cell death (Badel et al., 2003; Lopez-Solanilla et al., 2004). Cell death in these assays can thus be a result of effector recognition or host manipulation. Either way, ACIF1 is required for full cell death induction.
ACIF1 silencing compromises the Cf-4– and Cf-9–mediated HR in tobacco, tomato (both silenced via RNAi; shown in this work), and N. benthamiana (silenced via VIGS [Rowland et al., 2005]). This was also reflected in the reduced expression of two HR-marker genes (Hsr203J and Hin1) following infiltration with IF+Avr9. On the other hand, ACIF1 silencing caused increased PR gene expression after wounding (infiltration with IF−Avr9), reaching levels normally seen after Avr9 elicitation (Rivas et al., 2004). PR gene expression normally is suppressed during the wound response, and MeJA plays a role in this (Niki et al., 1998). Our observations suggest that ACIF1 silencing alters MeJA-dependent responses via an unknown mechanism.
Cf-9–dependent resistance to C. fulvum was not compromised in RNAi-ACIF1 tomato (and not by VIGS of ACIF1 in tomato). In the case of Sl CMPG1/ACRE74, VIGS and RNAi-mediated silencing reduced the Cf-9–mediated HR and allowed similar levels of successful C. fulvum infections (Gonzalez-Lamothe et al., 2006). This indicates that the silencing efficiency is comparable between the two techniques. Transgenic RNAi is preferred, however, because it allows for more homogeneous silencing. ACIF1 silencing compromised the weak resistance mediated by Cf-9B to C. fulvum (race 5.9). Cf-9B–mediated resistance appears to be independent of the HR, as microscopic cell death could not be observed in Cf-9B tomato after infection with race 5.9 (Hammond-Kosack and Jones, 1994; Panter et al., 2002). Possibly, ACIF1 silencing compromises defense responses other than the HR as well. It is not clear whether ACIF1 is involved in resistance mediated by other Cf genes, except that the Avr4/Avr9-triggered HR is reduced. It also is likely that compromised resistance responses would become more apparent when testing weak resistance (Cf-9B) than when testing strong resistance (Cf-9 or Cf-4).
ACIF1 silencing leads to only a partial loss of the HR in tobacco and tomato. This could be explained by incomplete silencing of the target gene, as reported for other genes (Liu et al., 2002a; Brigneti et al., 2004). Alternatively, ACIF1 homologs in these plants could function redundantly, as was shown for the Arabidopsis ACIF1 homologs (VFB gene family) (Schwager et al., 2007) and for other Arabidopsis F-box subfamilies (Guo and Ecker, 2003; Potuschak et al., 2003, Gagne et al., 2004; Strader et al., 2004; Dharmasiri et al., 2005b). In support of this, we identified ACIF1 homologs in tomato, potato, tobacco, and N. benthamiana that appear not to be targeted for silencing by the hairpin or VIGS constructs (i.e., sequences with >22-nucleotide identity to ACIF1). These homologs appear to have evolved independently in poplar, Arabidopsis, and Solanaceae species, but not in rice, based on the different phylogenetic branches (Jain et al., 2007). This could signify unique functions for these homologs in the corresponding dicot families.
We also cannot exclude off-target silencing of unrelated sequences when using RNAi or VIGS (Xu et al., 2006), since full genome sequence information is missing for both tomato and tobacco. Putative off-targets (i.e., sequences with >22-nucleotide identity to ACIF1), however, were not identified using BLAST and the siRNA scan tool with EST databases used here (TIGR and SGN). This means that we largely can exclude off-target silencing, especially for tomato, with the extensive coverage of the its transcriptome in the EST databases.
Knockdown of the Arabidopsis ACIF1 Homologs Induces MeJA- and ABA-Responsive Genes
The Arabidopsis genome contains four ACIF1 homologs named VFBs (Schwager et al., 2007). Previously, it was shown that only SKIP2/VFB4 was induced after elicitation with the general elicitor flg22 (Navarro et al., 2004). SKIP2/VFB4 was also induced (3.2-fold) after ABA treatment (Hoth et al., 2002). Moreover, SKIP2 belongs to a cluster of genes that is universally upregulated during a broad range of stress conditions (cold, osmotic stress, salinity, wounding, and biotic stresses, including treatments with elicitors) (Ma and Bohnert, 2007). We found that ACIF1 is induced during the wound and defense responses as well. Knockdown of the entire VFB F-box subfamily (vfb1-1 vfb2-1 vfb3-1 triple knockout/RNAi-VFB4) resulted in stunted plants with reduced lateral root formation, but these phenotypes were not observed for the single mutants (Schwager et al., 2007). We observed that the tomato RNAi-ACIF1 plants were slightly stunted as well (see Supplemental Figure 9 online). Genome-wide gene expression data obtained for the vfb knockdown plants revealed that a set of auxin-responsive genes and genes encoding cell wall metabolic enzymes are repressed by vfb knockdown (Schwager et al., 2007). We noted that wounding already caused elevated PR gene expression in tomato RNAi-ACIF1. Additional analysis of the microarray expression data from the vfb knockdown revealed that many putative PR genes also are regulated differentially. Moreover, vfb knockdown affects genes that are upregulated late after ABA treatment (180 min) and genes that are induced progressively by MeJA (over 30, 60, and 180 min). The upregulated genes include genes that encode the MeJA biosynthetic pathway, suggesting that MeJA biosynthesis is increased in the vfb knockdown plant.
The G-box and related DNA sequences were significantly overrepresented in the promoter (1 kb upstream) of genes differentially expressed in the triple knockout/RNAi-VFB4 plant. These DNA elements are target binding sites of MYC transcription factors (such as MYC2/JIN1) (Toledo-Ortiz et al., 2003) and bZIP transcription factors (e.g., ABI5 and bZIP10). MYC2 acts as a transcriptional repressor of light signaling (Yadav et al., 2005), an activator of ABA signaling (Abe et al., 2003; Anderson et al., 2004), and a regulator of MeJA responses, promoting wound-responsive genes while repressing certain pathogen-responsive genes (Boter et al., 2004; Lorenzo et al., 2004; Chini et al., 2007). G-box binding bZIP transcription factors are involved in ABA-dependent signaling (Jakoby et al., 2002) but also in basal defense and cell death in Arabidopsis (Kaminaka et al., 2006). SKIP2/VFB4 is also upregulated during the basal defense (after flg22 treatment) (Navarro et al., 2004) and after ABA application. Moreover, upon activation, certain bZIP transcription factors show increased nuclear localization (e.g., bZIP10 and Pc CPRF4a) (Kaminaka et al., 2006). Interestingly, VFBs appear to be localized in the cytoplasm, whereas closely related F-box proteins are localized predominantly in the nucleus. It is tempting, therefore, to speculate that ACIF1/VFBs target a repressor of a bZIP transcription factor, allowing localization to the nucleus and binding to G-box sequences. Together, the full genome transcription data from the vfb knockdown mutant links VFB function not only to auxin-responsive genes but also to ABA and MeJA responses. These effects appear to be regulated via G-box DNA sequences, possibly involving MYC or bZIP transcription factors. These data are consistent with a role for ACIF1 in plant defense in tobacco and tomato.
METHODS
Plant Materials and Growth Conditions
Tobacco (Nicotiana tabacum cv Petite Gerard) and tomato (Solanum lycopersicum cv Moneymaker Cf0 and Cf9) plants were grown in environmentally controlled glasshouses at 22°C with supplementary lighting used to provide a minimum 16-h daylength. Viral inoculations and fungal infections were performed in environmentally controlled growth cabinets at 22°C, with a 16-h-light/8-h-dark cycle and 65% humidity. Transgenic N tobacco carrying either Cf-9 (line 34.1B; Hammond-Kosack et al., 1998) or Cf-4 (line F641; Thomas et al., 2000) was described previously. Tobacco nn (cv Petite Havana, lacking the N gene) carrying NahG was described by Brading et al. (2000). Suspension cultures of Cf-9 tobacco cells derived from line 34.1B were subcultured as described previously (Piedras et al., 1998). Transgenic Nicotiana benthamiana plants expressing the N resistance gene (line 310A) or the Rx resistance gene (line Rx-18) were described previously (Bendahmane et al., 1999; Peart et al., 2002).
Isolation of Full-Length ACIF1 Clones
The 131-bp cDNA-AFLP fragment for ACRE189 was used as probe to screen a cDNA library established from elicited tobacco cells (Durrant et al., 2000). We obtained and sequenced several independent full-length clones, all of which corresponded to two closely related clones (ACIF1.1 and ACIF1.2). ACIF1.1 (pSLJ20243), the clone that matched the exact sequence of the ACRE189 cDNA-AFLP fragment, was used for our further studies in tobacco. The tomato ACIF1 clone was PCR-amplified with the primers F_SlACIF1_EcoRI/R_SlACIF1_BamHI (see Supplemental Table 7 online) using cDNA from Cf9 tomato leaf tissue (cv Moneymaker) after Avr9 elicitation as template DNA.
Sequence Analyses and Phylogenetic Analysis
The Pfam (www.sanger.ac.uk/Software/Pfam/) and SMART (smart.embl-heidelberg.de/) web servers were used for protein motif identification. The F-box domain and the LRRs were aligned using the ClustalX program (version 1.83; Chenna et al., 2003) using default settings and adjusted manually. A phylogenetic tree was constructed by the neighbor-joining method using MEGA software (version 3.1; Kumar et al., 2004). The confidence level of monophyletic groups was estimated by bootstrap analysis of 1000 replicates. The rice (Oryza sativa) homolog was most distant and, therefore, was treated as the phylogenetic outgroup. Trees obtained with maximum-parsimony or minimum-evolution algorithms proved to be similar.
Y2H Experiments
The EcoRI site present in the tobacco ACIF1 cDNA clone was removed using Quikchange site-directed mutagenesis (Stratagene) according to the manufacturer's recommendations (using primers F_QC_NtACIF1_noEcoRI and R_QC_NtACIF1_noEcoRI). The full-length coding sequence of ACIF1 was PCR-amplified using primers F_NtACIF1_EcoRI and R_NtACIF1_BamHI, and this PCR fragment was cloned in the corresponding sites in vector pGBKT7 (Clontech) to produce a protein fusion with the GAL4 DNA BD. The full-length coding sequence of the Arabidopsis thaliana homologs SKIP2 and Fbl16 were PCR-amplified (using primer pairs F_SKIP2_MfeI/R_SKIP2_BamHI and F_Fbl16_EcoRI/R_Fbl16_BamHI, respectively) from Arabidopsis Col-0 cDNA and subcloned in pGemT-easy II (Promega) and sequenced. The template cDNA was obtained by performing a reverse transcriptase reaction (SuperScript II; Invitrogen) using RNA extracted (using Trizol; Sigma-Aldrich) from leaf material that was treated with the flg22 peptide (Navarro et al., 2004). The SKIP2 PCR fragment was subcloned using MfeI and BamHI sites and cloned into EcoRI/BamHI of pGBKT7. The Fbl16 fragment was cloned into pGBKT7 in the same way that ACIF1 was. Point mutations in ACIF1 (P64A and W90A) and SKIP2 (P47A and W73A) cDNA were introduced using Quikchange according to the manufacturer's recommendations using the following primer combinations: F_QC_NtACIF1_P64A/R_QC_NtACIF1_P64A, F_QC_NtACIF1_W90A/R_QC_NtACIF1_W90A, F_QC_SKIP2_P47A/R_QC_SKIP2_P47A, and F_QC_SKIP2_W73A/R_QC_SKIP2_W73A. The EcoRI-BamHI fragment was recloned in pGBKT7. The SKIP2 (ΔF-box) cDNA fragment was obtained by PCR amplification using the primers F_SKIP2_MfeI_deltaFbox and R_SKIP2_BamHI and cloned in BamHI/EcoRI sites of pGBKT7. To produce a protein fusion with the LexA DNA BD in pGilda (Clontech), the full-length ACIF1 and SKIP2 were PCR-amplified using primers F_NtACIF1_NcoI/R_NtACIF1_XhoI and F_SKIP2_NcoI/ R_SKIP2_XhoI, respectively. The resulting PCR fragments were cloned in NcoI-XhoI sites of pGilda. As template, we used the pGBKT7 plasmids. The SKIP2 and ACIF1 (ΔF-box) fragments were obtained by PCR amplification using the primers F_SKIP2_NcoI_deltaFbox/R_SKIP2_Xho and F_ACIF1_NcoI_deltaFbox/R_NtACIF1_XhoI, respectively. The pBD42AD plasmids containing the Arabidopsis ASK1 and ASK2 clones were described previously (Devoto et al., 2002). To produce a protein fusion with the GAL4 AD, both ASK1 and ASK2 cDNA-coding regions were PCR-amplified (using primers F_ASK1_EcoRI/R_ASK1_BamHI and F_ASK2_EcoRI/R_ASK2_BamHI, respectively) and cloned in EcoRI/BamHI of pGADT7. The AD-CUL1 fusion constructs and the yeast expression vector pVT102-U expressing ASK1 and ASK2 proteins are described elsewhere (Stolpe et al., 2005; Marrocco et al., 2006).
The GAL4-based interaction between the BD-F-box proteins and ASK1- and ASK2-AD fusion was tested in the yeast strain AH109 (Clontech). Cotransformed yeast strains were selected on synthetic defined (SD)/−Leu/−Trp medium. Protein–protein interactions were tested using stringent (SD/−Ade/−His) and moderate (SD/−His/+2.5 mM 3-amino-1′,2′,4′-triazole) selection. Y3H assays were performed using the yeast haploid strains PJ69-4A and PJ69-4α (James et al., 1996). The ACIF1, SKIP2, and Fbl16 BD fusion constructs together with the CUL1 AD fusion construct were cointroduced in strain PJ69-4A. The yeast expression vector pVT102-U was transformed in PJ69-4α. Y3H interactions were tested on yeast strains that were selected after mating the two transformed haploid strains on SD/−Ura/−Trp/−Leu medium to select for the introduced plasmids. Yeast transformants carrying both AD-CUL1 and any one of the BD-F-box proteins did not grow when selecting for protein–protein interactions prior to mating. Protein interactions in the Y3H were assayed in the same manner as in the Y2H. The LexA interactions were tested in the yeast strain EGY48(p8op-lacZ) (Clontech). Cotransformed yeast strains carrying pBD42AD/pGilda derivatives were selected on SD/−His/−Trp medium, and protein–protein interactions were subsequently tested after induction of the BD-F-box fusions constructs (+Gal) using two-marker (−Leu and +Leu/+X-Gal) selection. Autoactivation was tested by growing yeast on noninducing medium (+Glu).
Generation of Transgenic Tobacco and Tomato Plants
A 486-bp fragment of the ACRE189 cDNA-AFLP fragment (Rowland et al., 2005) encompassing the LRRs 12 to 14 plus the C terminus was PCR-amplified in the sense and antisense directions (using primers F2_NtACIF_RNAi_XhoI/R2_NtACIF_RNAi_KpnI and F_NtACIF_RNAi_BamHI/R_NtACIF_RNAi_ClaI) and ligated into the pKannibal RNAi vector (Wesley et al., 2001) in the XhoI/KpnI sites and the BamHI/ClaI sites, respectively. The resulting plasmid was digested with NotI to release the Pro35S:ACIF1-hairpin:OCS terminator construct and cloned in the NotI sites of pGreen0029 and pGreen0229 (Hellens et al., 2000), resulting in plasmids pSLJ20786 and pSLJ20239, respectively. Both plasmids were cotransformed with the pSoup plasmid into Agrobacterium tumefaciens strain Agl1 and used for the transformation of tomato cv Moneymaker Cf9 and tobacco Cf-9 plants, respectively (Horsch et al., 1985). Putative transformants were moved onto soil, and the T2 seeds were collected for further characterization.
To generate lines accumulating mutant ACIF1 protein, full-length ACIF1 (lacking the stop codon) was PCR-amplified with primers F_NtACIF1_ClaI and R_NtACIF1_BamHI. The fragment was cloned in the plasmid pGemT-easyII (Promega), and the insert was sequenced. The ClaI-BamHI fragment was finally cloned in the binary vector pEpiGreenB4 (containing a Pro35S:GUS:cTAP cassette) replacing the β-glucuronidase (GUS)-ClaI-BamHI fragment (Gonzalez-Lamothe et al., 2006) and generating a fusion with the TAP tag, driven by Pro35S. Mutations in pGreen:ACIF1 were introduced using Quikchange (the same primer set as above, with conditions according to the manufacturer). Each construct was cotransformed with the pSoup plasmid in Agrobacterium strain Agl1, and stable tobacco transformants were generated (Hammond-Kosack et al., 1998).
Preparation of Protein Extracts, SDS-PAGE, and Immunoblotting
For immunoblot analysis, 1-cm-diameter leaf discs of 6-week-old tobacco plants were homogenized in liquid nitrogen, thawed on ice in 100 μL of a 1:1 mixture of 2× SB (125 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, and 100 mM DTT) and extraction buffer (100 mM Tris-HCl, pH 7.5, 250 mM sucrose, 15 mM EDTA, 5% [w/v] glycerol, 0.5% [w/v] polyvinylpyrrolidone K25, and 1× plant protease inhibitors [Sigma-Aldrich]) and centrifuged for 10 min at 13,000g. SDS-PAGE was performed as described previously (Heese et al., 2005) except that 8% separation gels were used. Immunodetection of the TAP tag was performed as described previously using peroxidase-conjugated anti-peroxidase antibody (Rivas et al., 2002). To extract protein from seedlings, tobacco seeds were germinated on agar Murashige and Skoog (MS) plates. Three-week-old seedlings were transferred to liquid MS medium supplemented with 100 μM MG132 (Sigma-Aldrich) or DMSO (mock). After 4 h, seedlings were homogenized in liquid nitrogen, and total protein was extracted in 2× SB supplemented with 1× plant protease inhibitors. Samples were heated for 10 min at 65°C and centrifuged for 10 min at 13,000g prior to loading.
Protein expression in yeast was established by solubilizing 7.5 OD600 units of yeast cells in 100 μL 8 M urea, 5% SDS, 40 mM Tris-HCl, pH 6.8, 0.1 mM EDTA, 1× protease inhibitors, 5 mM phenylmethylsulfonyl fluoride, and 1% (v/v) β-mercaptoethanol. Samples were heated at 70°C for 10 min followed by vigorous vortexing in the presence of glass beads. The suspensions were spun down at 13,000g for 5 min. The protein samples were size-separated on 8% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes. Immunodetection of the Myc-tagged F-box proteins was performed using a c-Myc 9E11 monoclonal antibody (Santa Cruz), and the LexA domain was recognized by the LexA monoclonal antibody (Clontech).
HR Assay and Agrobacterium-Mediated Transient Expression (Agroinfiltration)
Mature leaves of 6-week-old tobacco plants were assayed for an HR phenotype using intercellular fluid containing Avr9 peptide or by agroinfiltration. IF/+Avr9 was obtained as described previously from 8-week-old wild-type tobacco plants or tobacco expressing the Avr9 peptide (Hammond-Kosack et al., 1998). In the latter case, the Avr9 peptide is secreted in the apoplast of these transgenic tobacco plants. Agrobacterium strains GV3101/C58C1 containing the binary vectors expressing Cf-4/Avr4 (Thomas et al., 2000), Avr9 (Thomas et al., 2000), Pto/AvrPto (Peart et al., 2002), Inf1 (Huitema et al., 2005), and P50 helicase of TMV (Mestre and Baulcombe, 2006) have been described elsewhere. For agroinfiltration, Agrobacterium was grown in 10 mL of L medium at 28°C with antibiotics. Overnight cultures were collected by centrifugation and resuspended in 10 mM MES, pH 5.6, 10 mM MgCl2, and 150 μM acetosyringone at 2 OD600 units. Cultures were left at room temperature for 2 to 3 h prior to infiltration. The cultures were infiltrated between the laterial veins in the following OD600 ranges: Avr9, 0.05 to 0.5; Cf-4/Avr4, 0.01 to 0.2; Inf1, 0.005 to 0.5; P50, 0.02 to 0.5; AvrPto/Pto, 0.02 to 0.5. Growth inhibition assays on 10-d-old tobacco seedlings were performed as described (Gonzalez-Lamothe et al., 2006). Autofluorescence was detected under UV illumination.
Disease Assays
Infections with Cladosporium fulvum race 5, race 4 GUS (Rowland et al., 2005), and race 5.9 (Marmeisse et al., 1993) on 4-week-old tomato plants were performed as described previously (Rowland et al., 2005). Fungal growth was scored at 10 to 24 DPI. Leaves were stained with trypan blue or 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (Rowland et al., 2005). Light microscopy (Axiophot; Zeiss) was used to observe fungal infection structures. For TRV-based VIGS in tomato, a fragment from tomato ACIF1 cDNA was PCR-amplified using the primers F_700_SlACIF1 and R_835_SlACIF1 and cloned in the SmaI site of pTRV-RNA2 vector (Liu et al., 2002b). As a control, the pTRV-RNA2 empty vector and the pTRV-RNA2 vector containing a 150-bp Cf-9 fragment (Rowland et al., 2005) were used. Silencing of tomato plants was performed as before (Rowland et al., 2005). TMV strain U1 was passed through susceptible tobacco plants and stored as sap obtained from infected tissue at −20°C. TMV was mechanically inoculated by rubbing sap dilutions (buffered with 10 mM sodium phosphate buffer, pH 7) on whole leaves of 5-week-old tobacco plants with Carborundum. After inoculation, plants were rinsed with water to remove surface inocula. Mock inoculations were performed with phosphate buffer only. For TRV-based VIGS in N. benthamiana, a fragment of tobacco ACIF1 was PCR-amplified using primers F_167bp_NtACIF1 and R_167bp_NtACIF1 and cloned into pTV00 using SmaI to create TRV:ACIF1. The empty pTV00 vector and pTV00 containing the Nb SGT1.2 fragment (TRV:SGT1) (Peart et al., 2002) were used as controls. TRV-based silencing and infection and detection of TMV:GFP and PVX:GFP were done as described previously (Rowland et al., 2005).
Pseudomonas syringae pv tabaci (Pst) strain 11528 carrying the pPtE6 vector (empty or with AvrPto) (Ronald et al., 1992) was subcultured in King's B medium (King et al., 1954) at 28°C. For cell death induction, bacterial suspensions were infiltrated in 10 mM MgCl2 into the abaxial leaf surfaces. For bacterial growth curves, bacteria were syringe-infiltrated at 2 × 104 colony-forming units/mL in 10 mM MgCl2 and 0.02% Silwet-L77. Bacterial populations were sampled by maceration of leaf discs in 10 mM MgCl2, and serial dilutions were plated on fresh Luria-Bertani agar containing the appropriate antibiotics. At least six samples were taken for each data point. To test for Pto/Prf resistance in tomato, we crossed both RNAi-ACIF1 and Cf9 tomato with the resistant tomato cv RioGrande 76R (carrying Pto and Prf). Tomato plants were vacuum-infiltrated with Pseudomonas syringae pv tomato strains DC3000, T1, and T1+AvrPto (Ronald et al., 1992) at 2 × 104 colony-forming units/mL in 10 mM MgCl and 0.02% Silwet-L77.
RNA Gel Blot Analyses
Total RNA was prepared from ∼100 mg of tissue using Trizol (Sigma-Aldrich). RNA gel blots were prepared according to standard procedures using 10 μg of total RNA and Hybond N+ membranes (GE Healthcare). A tobacco ACRE189 cDNA fragment (amplified with primers F_NtACIF1rnai and R_NtACIF1rnai) was used as a probe after random labeling using the Rediprime labeling system (GE Healthcare) using [γ-32P]dCTP. Prehybridization and hybridization were performed using the ULTRAhyb buffer (Ambion) at 42°C overnight. The membrane was washed twice for 5 min each with 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate)/0.1% SDS and 1× SSC/0.1% SDS and twice for 15 min each with 0.1× SSC/0.1% SDS at 42°C and exposed to x-ray film.
RT-PCR
Total RNA from leaves or from cultured suspension cells was isolated using the Tri Reagent method according to the manufacturer's recommendations (Sigma-Aldrich). First-strand cDNA was synthesized from 2 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen). RT-PCRs were performed as described previously (Romeis et al., 2001) using the following primers for tobacco and tomato: F_NtACIF1_rt/R_NtACIF1_rt and F_SlACIF1_rt/R_SlACIF1_rt. The tobacco and tomato actin transcripts were amplified using primers AC1/AC2. The tomato PR marker gene transcripts were amplified using primer pairs LePR1A, LePR2A LePR2B, and LePR5. The HR marker genes were amplified using the primer pairs LeHin1 and Hsr203J. Relative expression levels were determined using QuantityOne software (Bio-Rad). Expression of tomato ACIK1 and CMPG1 was determined as described previously (González-Lamothe et al., 2006). Gels were stained with ethidium bromide (actin in Figure 8D was stained with SYBR Green I; Invitrogen).
Extraction and Quantification of SA/SAG
SA/SAG levels in tobacco leaf extracts (from ∼1 g of fresh tissue) were determined by a method adapted from a procedure described elsewhere (Raskin et al., 1989). HPLC separation was achieved using a 3-μm C18 reverse-phase column (50 mm × 2 mm; Luna) operated at 23°C, with a 400 μL/min flow rate using solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in methanol) running a gradient of 20% solvent B to 80% solvent B in 6 min. SA concentrations were measured using a single quadrupole mass spectrometer (Agilent 1100) coupled directly to the HPLC column, operated in the negative mode, running single-ion mode on ions 137.0, 208.0, and 263.2 m/z. Each sample was run twice, once spiked with an internal control containing a known amount of SA.
Microarray Analysis
Microarray data from experiments on the effects of the plant hormones from Affymetrix ATH1 GeneChips were downloaded from the TIGR database. Complete descriptions of experimental details and the data files are available at http://Arabidopsis.org/info/expression/ATGenExpress.jsp. Data analysis was performed using the R statistical language (http://www.r-project.org/) with the Bioconductor packages limma and affy (Gautier et al., 2004; Gentleman et al., 2004; Smyth, 2005). Background-corrected, normalized, and log-transformed perfect match expression values were obtained using the robust multiarray average summary method (Irizarry et al., 2003). Differentially expressed genes were identified using the rank products method with a false discovery rate of ⩽0.05 (Breitling et al., 2004). The overlap of genes differentially expressed in the hormonal treatment with those in vfb knockdown plants were analyzed using custom in-house perl scripts (available on request). The Multi-Experiment Viewer software from the TIGR website (Saeed et al., 2003) was used to cluster similarly expressed genes using K-means clustering (Soukas et al., 2000) and figure of merit (Yeung et al., 2001). Promoter analysis was performed using Athena (O'Connor et al., 2005).
Accession Numbers
Sequence data from this article can be found in the GenBank data libraries under the following accession numbers: tobacco ACIF1/ACRE189, AY220479; solanaceous ACIF1 orthologs (Sl ACIF1, SGN-U218766/TIGR-TC164112 and Sl-SGN-U237904; St ACIF1, SGN-U273676/SGN-U273677/SGN-U282566); and ACIF1 paralogs (Sl SGN-U239302, St SGN-U292620, and Nb TC9508). Arabidopsis locus numbers are as follows: SKIP2/VFB4/At5g67250, Fbl16/VFB2/At3g50080, Fbl8/Fbl24/VFB3/At4g07400, and VFB1/At1g47056. Populus trichocarpa ACIF1 homolog identifiers are as follows: 347627, 591999, 655088, and 651111. The rice ACIF1 homolog number is Os4g42670. The microarray data for the vfb knockdown plant has accession number GSM3863 (http://www.ncbi.nlm.nih.gov/geo). The effects of the plant hormones at different time points were surveyed by the AtGenExpress Consortium and deposited as NASC Array 172 to 184 (http://affymetrix.Arabidopsis.info/narrays/).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of the 14 Putative LRRs of ACIF1.
Supplemental Figure 2. The F-Box Motif Is Required for the Interactions with ASKs in Y2H.
Supplemental Figure 3. TRV-Based VIGS of ACIF1 Compromises N-Mediated HR in N. benthamiana but Not Resistance to Infection by TMV:GFP or PVX:GFP.
Supplemental Figure 4. Reduced Growth Inhibition of RNAi-ACF1 Line J Seedlings When Grown on Medium Containing IF+Avr9.
Supplemental Figure 5. Bacterial Growth of Pst (+/−AvrPto) Is Not Affected by Silencing ACIF1 or Expression of Mutant ACIF1 in Tobacco.
Supplemental Figure 6. Silencing of ACIF1 Reduced Speck Disease Symptom (Lesions) on Tomato but Did Not Affect the Proliferation of P. s. pv tomato DC3000 or T1 (+AvrPto).
Supplemental Figure 7. TRV-Based VIGS of ACIF1 Compromises Avr9-Dependent HR in Cf9 Tomato.
Supplemental Figure 8. Knockdown of Arabidopsis ACIF1 Homologs (VFBs) Positively Regulates a Set of MeJA- and ABA-Responsive Genes.
Supplemental Figure 9. Tomato RNAi-ACIF1 Line D Are Slightly Stunted Compared with Control Plants.
Supplemental Table 1. Alignment Used to Create the Phylogenetic Tree of ACIF1 Homologs (Fasta Format).
Supplemental Table 2. MeJA-Responsive Marker Genes (Including Genes Encoding the Biosynthetic Pathway of MeJA) and PR Genes That Show Differential Gene Expression in the vfb Knockdown Mutant.
Supplemental Table 3. Lists of Genes Differentially Regulated in the Triple Knockout vfb1-1 vfb2-1 vfb3-1/RNAi-VFB4 That Are Also Differentially Regulated by MeJA.
Supplemental Table 4. List of Genes Differentially Regulated in the Triple Knockout vfb1-1 vfb2-1 vfb3-1/RNAi-VFB4 That Are Also Differentially Regulated by ABA.
Supplemental Table 5. Overlap between Differential Gene Expression Observed after Different Hormone Treatments (30, 60, and 180 min Combined; for SA, only 180 min) and Genes Differentially Expressed in the vfb Knockdown Mutant Background.
Supplemental Table 6. Gene Clusters Representing the Differential Gene Expression in the vfb Knockdown Mutants and Their Regulation in Wild-Type Arabidopsis (Col-0) after Different Hormone Treatments (30, 60, and 180 min).
Supplemental Table 7. DNA Sequences of the PCR Primers Used.
Supplementary Material
Acknowledgments
We thank M. Smoker for generating transgenic plants. We thank T. Kretsch for providing the pVT102-U vector carrying the ASK constructs, the CUL1-AD construct, and the yeast strains PJ69-4A and PJ69-4α; A. Devoto and J.G. Turner for providing the ASK1 and ASK2 constructs; and J. Peart and J. Rathjen for providing the various binary vectors for expressing elicitors, R proteins, TMV-U1, and the Pseudomonas strains used. Microarray data files for the vfb1-1 vfb2-1 vfb3-1 triple knockout and the RNAi-VFB4 mutant were kindly provided by C. Schwechheimer. H.A.v.d.B. was supported by a Marie Curie fellowship (HPMMF-CT-2002-01882) and by the Netherlands Organization for Scientific Research (NWO/VENI Grant 863.04.018). D.I.T. was supported by European Union FP6 STREP Grant 511983-TransDeath. The Jones laboratory is supported by the Gatsby Charitable Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jonathan D.G. Jones (jonathan.jones@sainsbury-laboratory.ac.uk).
Online version contains Web-only data.
References
- Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.P., Badruzsaufari, E., Schenk, P.M., Manners, J.M., Desmond, O.J., Ehlert, C., Maclean, D.J., Ebert, P.R., and Kazan, K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel, J.L., Nomura, K., Bandyopadhyay, S., Shimizu, R., Collmer, A., and He, S.Y. (2003). Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL-ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol. Microbiol. 49 1239–1251. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., Kanyuka, K., and Baulcombe, D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt, M.R., Grabov, A., Brearley, J., Hammond, K.K., and Jones, J.D.G. (1999). K+ channels of Cf-9 transgenic tobacco guard cells as targets for Cladosporium fulvum Avr9 elicitor-dependent signal transduction. Plant J. 19 453–462. [DOI] [PubMed] [Google Scholar]
- Boter, M., Ruiz-Rivero, O., Abdeen, A., and Prat, S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brading, P.A., Hammond-Kosack, K.E., Parr, A., and Jones, J.D.G. (2000). Salicylic acid is not required for Cf-2- and Cf-9-dependent resistance of tomato to Cladosporium fulvum. Plant J. 23 305–318. [DOI] [PubMed] [Google Scholar]
- Breitling, R., Armengaud, P., Amtmann, A., and Herzyk, P. (2004). Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573 83–92. [DOI] [PubMed] [Google Scholar]
- Brigneti, G., Martin-Hernandez, A.M., Jin, H., Chen, J., Baulcombe, D.C., Baker, B., and Jones, J.D.G. (2004). Virus-induced gene silencing in Solanum species. Plant J. 39 264–272. [DOI] [PubMed] [Google Scholar]
- Büche, C., Poppea, C., Schäfera, E., and Kretsch, T. (2000). eid1: A new Arabidopsis mutant hypersensitive in phytochrome A–dependent high-irradiance responses. Plant Cell 12 547–558. [PMC free article] [PubMed] [Google Scholar]
- Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T.J., Higgins, D.G., and Thompson, J.D. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J.M., Lorenzo, O., Garcia-Casado, G., Lopez-Vidriero, I., Lozano, F.M., Ponce, M.R., Micol, J.L., and Solano, R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 666–671. [DOI] [PubMed] [Google Scholar]
- Cope, G.A., and Deshaies, R.J. (2006). Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 7 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., Boniotti, M.B., and Gutierrez, C. (2002). Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14 3057–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto, A., Nieto-Rostro, M., Xie, D., Ellis, C., Harmston, R., Patrick, E., Davis, J., Sherratt, L., Coleman, M., and Turner, J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32 457–466. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005. a). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., Weijers, D., Lechner, E., Yamada, M., Hobbie, L., Ehrismann, J.S., Jurgens, G., and Estelle, M. (2005. b). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9 109–119. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E., Rowland, O., Piedras, P., Hammond-Kosack, K.E., and Jones, J.D.G. (2000). cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farras, R., Ferrando, A., Jasik, J., Kleinow, T., Okresz, L., Tiburcio, A., Salchert, K., del Pozo, C., Schell, J., and Koncz, C. (2001). SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 20 2742–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin, L.K., Krishnamurthy, N., Tor, M., Sjolander, K.V., and Jones, J.D.G. (2005). Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 138 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X., Richards, D.E., Fleck, B., Xie, D., Burton, N., and Harberd, N.P. (2004). The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCF(SLY1) E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261 754–756. [DOI] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S.H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Smalle, J., Gingerich, D.J., Walker, J.M., Yoo, S.D., Yanagisawa, S., and Vierstra, R.D. (2004). Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA 101 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, L., Cope, L., Bolstad, B.M., and Irizarry, R.A. (2004). Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20 307–315. [DOI] [PubMed] [Google Scholar]
- Gentleman, R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe, R., Tsitsigiannis, D.I., Ludwig, A.A., Panicot, M., Shirasu, K., and Jones, J.D.G. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18 1067–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan, S., Wei, W., and He, S.Y. (1996). hrp gene-dependent induction of hin1: A plant gene activated rapidly by both harpins and the avrPto gene-mediated signal. Plant J. 10 591–600. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414 271–276. [DOI] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115 667–677. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1994). Incomplete dominance of tomato Cf genes for resistance to Cladosporium fulvum. Mol. Plant Microbe Interact. 7 58–70. [Google Scholar]
- Hammond-Kosack, K.E., Tang, S., Harrison, K., and Jones, J.D.G. (1998). The tomato Cf-9 disease resistance gene functions in tobacco and potato to confer responsiveness to the fungal avirulence gene product Avr9. Plant Cell 10 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese, A., Ludwig, A.A., and Jones, J.D.G. (2005). Rapid phosphorylation of a syntaxin during the Avr9/Cf-9-race-specific signaling pathway. Plant Physiol. 138 2406–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Hiraga, S., Ito, H., Yamakawa, H., Ohtsubo, N., Seo, S., Mitsuhara, I., Matsui, H., Honma, M., and Ohashi, Y. (2000). An HR-induced tobacco peroxidase gene is responsive to spermine, but not to salicylate, methyl jasmonate, and ethephon. Mol. Plant Microbe Interact. 13 210–216. [DOI] [PubMed] [Google Scholar]
- Horsch, R.B., Rogers, S.G., and Fraley, R.T. (1985). Transgenic plants. Cold Spring Harb. Symp. Quant. Biol. 50 433–437. [DOI] [PubMed] [Google Scholar]
- Hoth, S., Morgante, M., Sanchez, J.P., Hanafey, M.K., Tingey, S.V., and Chua, N.H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115 4891–4900. [DOI] [PubMed] [Google Scholar]
- Huitema, E., Vleeshouwers, V.G., Cakir, C., Kamoun, S., and Govers, F. (2005). Differences in intensity and specificity of hypersensitive response induction in Nicotiana spp. by INF1, INF2A, and INF2B of Phytophthora infestans. Mol. Plant Microbe Interact. 18 183–193. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Jain, M., Nijhawan, A., Arora, R., Agarwal, P., Ray, S., Sharma, P., Kapoor, S., Tyagi, A.K., and Khurana, J.P. (2007). F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 106–111. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Kaminaka, H., Nake, C., Epple, P., Dittgen, J., Schutze, K., Chaban, C., Holt, B.F., III, Merkle, T., Schafer, E., Harter, K., and Dangl, J.L. (2006). bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 25 4400–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.S., and Delaney, T.P. (2002). Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 14 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, E.O., Ward, M.K., and Raney, D.E. (1954). Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44 301–307. [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Kuroda, H., Takahashi, N., Shimada, H., Seki, M., Shinozaki, K., and Matsui, M. (2002). Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol. 43 1073–1085. [DOI] [PubMed] [Google Scholar]
- Lechner, E., Achard, P., Vansiri, A., Potuschak, T., and Genschik, P. (2006). F-box proteins everywhere. Curr. Opin. Plant Biol. 9 631–638. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., and Dinesh-Kumar, S.P. (2004). Involvement of MEK1 MAPKK, NTF6 MPAK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J. 38 800–809. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Serino, G., Deng, X.W., and Dinesh-Kumar, S.P. (2002. a). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.L., Schiff, M., and Dines-Kumar, S.P. (2002. b). Virus-induced gene silencing in tomato. Plant J. 31 777–786. [DOI] [PubMed] [Google Scholar]
- Lopez-Solanilla, E., Bronstein, P.A., Schneider, A.R., and Collmer, A. (2004). HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol. Microbiol. 54 353–365. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O., Chico, J.M., Sanchez-Serrano, J.J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, A.A., Saitoh, H., Felix, G., Freymark, G., Miersch, O., Wasternack, C., Boller, T., Jones, J.D.G., and Romeis, T. (2005). Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc. Natl. Acad. Sci. USA 102 10736–10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, S., and Bohnert, H.J. (2007). Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol. 8 R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmeisse, R., Van den Ackerveken, G.F.J.M., Goosen, T., De Wit, P.J.G.M., and Van den Broeck, H.W.J. (1993). Disruption of the avirulence gene Avr9 in two races of the tomato pathogen Cladosporium fulvum causes virulence on tomato genotypes with the complementary resistance gene Cf9. Mol. Plant Microbe Interact. 6 412–417. [Google Scholar]
- Marrocco, K., Zhou, Y., Bury, E., Dieterle, M., Funk, M., Genschik, P., Krenz, M., Stolpe, T., and Kretsch, T. (2006). Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant J. 45 423–438. [DOI] [PubMed] [Google Scholar]
- Mestre, P., and Baulcombe, D.C. (2006). Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 18 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur, L.A., Bi, Y.M., Darby, R.M., Firek, S., and Draper, J. (1997). Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV-infected tobacco. Plant J. 12 1113–1126. [DOI] [PubMed] [Google Scholar]
- Murphy, A.M., Chivasa, S., Siungh, D.P., and Carr, J.P. (1999). Salicylic acid-induced resistance to viruses and other pathogens: A parting of ways? Trends Plant Sci. 4 155–160. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Zipfel, C., Rowland, O., Keller, I., Robatzek, S., Boller, T., and Jones, J.D.G. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov, V., Ludwig, A.A., and Jones, J.D.G. (2006). CITRX thioredoxin is a putative adaptor protein connecting Cf-9 and the ACIK1 protein kinase during the Cf-9/Avr9-induced defence response. FEBS Lett. 580 4236–4241. [DOI] [PubMed] [Google Scholar]
- Nemhauser, J.L., Hong, F., and Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475. [DOI] [PubMed] [Google Scholar]
- Nicholson, R., and Hammerschmidt, R. (1992). Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30 369–389. [Google Scholar]
- Niki, T., Mitsuhara, I., Seo, S., Ohtsubo, N., and Ohashi, T. (1998). Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol. 39 500–507. [Google Scholar]
- O'Connor, T.R., Dyreson, C., and Wyrick, J.J. (2005). Athena: A resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21 4411–4413. [DOI] [PubMed] [Google Scholar]
- Panter, S.N., Hammond-Kosack, K.E., Harrison, K., Jones, J.D.G., and Jones, D.A. (2002). Developmental control of promoter activity is not responsible for mature onset of Cf-9B-mediated resistance to leaf mold in tomato. Mol. Plant Microbe Interact. 15 1099–1107. [DOI] [PubMed] [Google Scholar]
- Peart, J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski, M.D., and Deshaies, R.J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6 9–20. [DOI] [PubMed] [Google Scholar]
- Piedras, P., Hammond-Kosack, K.E., Harrison, K., and Jones, J.D.G. (1998). Rapid, Cf-9- and Avr9-dependent production of active oxygen species to tobacco suspension cultures. Mol. Plant Microbe Interact. 11 1155–1166. [Google Scholar]
- Potuschak, T., Lechner, E., Parmentier, Y., Yanagisawa, S., Grava, S., Koncz, C., and Genschik, P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115 679–689. [DOI] [PubMed] [Google Scholar]
- Raskin, I., Turner, I.M., and Melander, W.R. (1989). Regulation of heat production in the inflorescences of an Arum lily by endogenous salicylic acid. Proc. Natl. Acad. Sci. USA 86 2214–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rivas, S., Romeis, T., and Jones, J.D.G. (2002). The Cf-9 disease resistance protein is present in an approximately 420-kilodalton heteromultimeric membrane-associated complex at one molecule per complex. Plant Cell 14 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rivas, S., Rougon-Cardoso, A., Smoker, M., Schauser, L., Yoshioka, H., and Jones, J.D.G. (2004). CITRX thioredoxin interacts with the tomato Cf-9 resistance protein and negatively regulates defence. EMBO J. 23 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rivas, S., and Thomas, C.M. (2005). Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu. Rev. Phytopathol. 43 395–436. [DOI] [PubMed] [Google Scholar]
- Rohila, J.S., Chen, M., Cerny, R., and Fromm, M.E. (2004). Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38 172–181. [DOI] [PubMed] [Google Scholar]
- Romeis, T., Ludwig, A.A., Martin, R., and Jones, J.D.G. (2001). Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 20 5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, T., Piedras, P., and Jones, J.D.G. (2000). Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell 12 803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, T., Piedras, P., Zhang, S.Q., Klessig, D.F., Hirt, H., and Jones, J.D.G. (1999). Rapid AVR9- and Cf-9-dependent activation of MAP kinases in tobacco cell cultures and leaves: Convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald, P.C., Salmeron, J.M., Carland, F.M., and Staskawicz, B.J. (1992). The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J. Bacteriol. 174 1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland, O., Ludwig, A.A., Merrick, C.J., Baillieul, F., Tracy, F.E., Durrant, W.E., Fritz-Laylin, L., Nekrasov, V., Sjolander, K., Yoshioka, H., and Jones, J.D.G. (2005). Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell 17 295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed, A.L., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34 374–378. [DOI] [PubMed] [Google Scholar]
- Schulman, B.A., Carrano, A.C., Jeffrey, P.D., Bowen, Z., Kinnucan, E.R., Finnin, M.S., Elledge, S.J., Harper, J.W., Pagano, M., and Pavletich, N.P. (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408 381–386. [DOI] [PubMed] [Google Scholar]
- Schwager, K.M., Calderon-Villalobos, L.I., Dohmann, E.M., Willige, B.C., Knierer, S., Nill, C., and Schwechheimer, C. (2007). Characterization of the VIER F-BOX PROTEINE genes from Arabidopsis reveals their importance for plant growth and development. Plant Cell 19 1163–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G.K. (2005). Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber, eds (New York: Springer), pp. 397–420.
- Soukas, A., Cohen, P., Socci, N.D., and Friedman, J.M. (2000). Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 14 963–980. [PMC free article] [PubMed] [Google Scholar]
- Stirnberg, P., Furner, I.J., and Ottoline Leyser, H.M. (2007). MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50 80–94. [DOI] [PubMed] [Google Scholar]
- Stolpe, T., Susslin, C., Marrocco, K., Nick, P., Kretsch, T., and Kircher, S. (2005). In planta analysis of protein-protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma 226 137–146. [DOI] [PubMed] [Google Scholar]
- Strader, L.C., Ritchie, S., Soule, J.D., McGinnis, K.M., and Steber, C.M. (2004). Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc. Natl. Acad. Sci. USA 101 12771–12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, X., Calderon-Villalobos, L.I., Sharon, M., Zheng, C., Robinson, C.V., Estelle, M., and Zheng, N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645. [DOI] [PubMed] [Google Scholar]
- Thilmony, R.L., Chen, Z., Bressan, R.A., and Martin, G.B. (1995). Expression of the tomato Pto gene in tobacco enhances resistance to Pseudomonas syringae pv tabaci expressing avrPto. Plant Cell 7 1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., Nomura, K., He, S.Y., Howe, G.A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448 661–665. [DOI] [PubMed] [Google Scholar]
- Thomas, C.M., Tang, S., Hammond-Kosack, K., and Jones, J.D.G. (2000). Comparison of the hypersensitive response induced by the tomato Cf-4 and Cf-9 genes in Nicotiana spp. Mol. Plant Microbe Interact. 13 465–469. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., van Esse, H.P., Crous, P.W., and de Wit, P.J.G.M. (2005). Cladosporium fulvum (syn Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol. Plant Pathol. 6 379–393. [DOI] [PubMed] [Google Scholar]
- Thomson, J.H. (2006). Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 16 1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz, G., Huq, E., and Quail, P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei, K., Naito, Y., Takahashi, F., Haraguchi, T., Ohki-Hamazaki, H., Juni, A., Ueda, R., and Saigo, K. (2004). Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 32 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn, R.A., Wulff, B.B., Rivas, S., Durrant, M.C., van der Ploeg, A., de Wit, P.J.G.M., and Jones, J.D.G. (2005). Structure-function analysis of Cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 17 1000–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen, G., Van den Burg, H.A., Cornelissen, B.J.C., and Takken, F.L.W. (2007). Structure and function of resistance proteins in solanaceous plants. Annu. Rev. Phytopathol. 45 43–72. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Willems, A.R., Schwab, M., and Tyers, M. (2004). A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695 133–170. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W.L., Ma, H., Peng, W., Huang, D., and Xie, D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P., Zhang, Y., Kang, L., Roossinck, M.J., and Mysore, K.S. (2006). Computational estimation and experimental verification of off-target silencing during post-transcriptional gene silencing in plants. Plant Physiol. 142 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, V., Mallappa, C., Gangappa, S.N., Bhatia, S., and Chattopadhyay, S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17 1953–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.W., Gonzalez-Lamothe, R., Ewan, R.A., Rowland, O., Yoshioka, H., Shenton, M., Ye, H., O'Donnell, E., Jones, J.D.G., and Sadanandom, A. (2006). The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18 1084–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, K.Y., Haynor, D.R., and Ruzzo, W.L. (2001). Validating clustering for gene expression data. Bioinformatics 17 309–318. [DOI] [PubMed] [Google Scholar]
- Zheng, N., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416 703–709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.