Abstract
Cellulose, the important structural compound of cell walls, provides strength and rigidity to cells of numerous organisms. Here, we functionally characterize four cellulose synthase genes (CesA) in the oomycete plant pathogen Phytophthora infestans, the causal agent of potato (Solanum tuberosum) late blight. Three members of this new protein family contain Pleckstrin homology domains and form a distinct phylogenetic group most closely related to the cellulose synthases of cyanobacteria. Expression of all four genes is coordinately upregulated during pre- and early infection stages of potato. Inhibition of cellulose synthesis by 2,6-dichlorobenzonitrile leads to a dramatic reduction in the number of normal germ tubes with appressoria, severe disruption of the cell wall in the preinfection structures, and a complete loss of pathogenicity. Silencing of the entire gene family in P. infestans with RNA interference leads to a similar disruption of the cell wall surrounding appressoria and an inability to form typical functional appressoria. In addition, the cellulose content of the cell walls of the silenced lines is >50% lower than in the walls of the nonsilenced lines. Our data demonstrate that the isolated genes are involved in cellulose biosynthesis and that cellulose synthesis is essential for infection by P. infestans.
INTRODUCTION
The oomycete phylum is a large group of ubiquitous microorganisms that includes both saprophytes and pathogens of plants, insects, fish, nematodes, and vertebrates (Phillips et al., 2008). Plant pathogenic oomycetes infect a wide range of host plants, including crop species, native weeds, ornamental plants, and trees (Erwin and Ribeiro, 1996; van West et al., 2003; Grenville-Briggs and van West, 2005).
One of the model systems to investigate the early stages of the oomycete-plant infection cycle is the Phytophthora infestans–Solanum tuberosum interaction (Birch and Whisson, 2001; Kamoun, 2003; van West and Vleeshouwers, 2004). P. infestans is the causal agent of late blight on both potato (Solanum tuberosum) and tomato (Solanum lycopersicum), costing over $5 billion per annum in control measures and crop losses (Duncan, 1999).
Prior to host plant penetration, P. infestans produces a variety of specialized structures, such as biflagellate zoospores that are able to swim toward host plant cues, walled cysts that germinate upon contact with the host, and appressoria (Walker and van West, 2007). The zoospore lacks a cell wall and maintains cell volume and turgor by means of a water expulsion vacuole (Mitchell and Hardham, 1999). A cell wall is produced during encystment. The cell wall is extended and thickened during germination of the cyst and subsequent production of the appressorium and is therefore important for the establishment of infection. The cell walls of oomycetes consist essentially of (1→3)-β-d-glucans, (1→6)-β-d-glucans, and cellulose (Bartnicki-Garcia, 1968), whereas chitin, which is a major cell wall component of true fungi, occurs in very small amounts in the walls of several oomycetes (Lin and Aronson, 1970; Aronson and Lin, 1978; Campos-Takaki et al., 1982; Bulone et al., 1992). Cellulose has a microfibrillar structure in the walls of oomycetes (Bulone et al., 1992; Helbert et al., 1997) and is thought to contribute in scaffolding in a similar manner as chitin does in the walls of true fungi (Bartnicki-Garcia and Wang, 1983). The other cell wall β-d-glucans constitute most of an amorphous matrix that is laid over and interacts with the inner layer of rather crystalline cellulose microfibrils (Farkas, 1979).
Cellulose is one of the most abundant macromolecules on earth (Delmer, 1999). In addition to oomycetes, it occurs in a vast array of organisms, including all major groups of plants (Brown, 1996), the cellular slime mold Dictyostelium discoideum (Blanton et al., 2000), tunicates (Kimura and Itoh, 1995; Matthysse et al., 2004; Sasakura et al., 2005), and some prokaryotes (reviewed in Ross et al., 1991). Despite the wide distribution of cellulose in nature and its biological and applied importance, its mechanisms of formation are poorly understood. Genes coding for cellulose synthases have nonetheless been isolated in most organisms used as models to study cellulose biosynthesis. The catalytic subunits of the cellulose synthase complexes are processive glycosyltransferases (i.e., enzymes that catalyze the repetitive addition of multiple sugar residues to the growing polysaccharide chains). They belong to glycosyltransferase family 2, which contains other processive enzymes, such as fungal chitin synthases, the chitooligosaccharide synthase Nod C, hyaluronan synthases, and cellulose synthase-like (Csl) proteins (see CAZY database, http://www.cazy.org/). Cellulose synthases are integral membrane proteins that contain multiple transmembrane segments and a cytoplasmic domain bearing the catalytic part of the enzyme (Saxena and Brown, 2000).
As opposed to higher plant and fungal cell wall carbohydrate synthases, which have been very well studied in the last decades, investigations on oomycete polysaccharide synthases are limited to a few enzymes. Examples of the most studied enzymes from oomycete species are the (1→3)-β-d-glucan, cellulose, and chitin synthases from Saprolegnia monoica (Bulone et al., 1990, 1992; Bulone and Fèvre, 1996; Pelosi et al., 2003; Bouzenzana et al., 2006). Despite the economic impact of some Phytophthora species, there is very little biochemical information available on similar enzymes from this genus, and no corresponding gene has been characterized to date. Here, we report the identification of a family of four cellulose synthase genes (CesA) in P. infestans. Structural analysis of all four gene products revealed that they contain multiple transmembrane domains and conserved motifs found in processive glycosyltransferases (Saxena et al., 1995). Having recently developed a double-stranded RNA (dsRNA)-mediated approach to transiently silence genes in P. infestans (Whisson et al., 2005), we used this technique along with a cellulose synthesis inhibitor to functionally characterize these genes and investigate the role of cellulose biosynthesis in the development of P. infestans and its ability to infect potato.
RESULTS
A Family of Four Cellulose Synthase Genes Exists within Phytophthora Species
We identified the sequence of the complete open reading frames of four CesA encoding genes from publicly available EST (Randall et al., 2005) and genomic DNA Phytophthora databases (see Methods for websites) following the discovery of a cellulose synthase fragment through proteomics (see below). Four distinct gene transcripts were identified within P. infestans, and these were designated CesA1, CesA2, CesA3, and CesA4. CesA1, CesA2, and CesA4 share the greatest sequence similarity, with CesA3 being the most divergent member of the family. CesA genes share between 21 and 64% sequence similarity at the DNA level (Table 1). Analysis of the recently partially annotated P. infestans genome sequenced by the Broad Institute (see Methods for website) revealed that CesA1 and CesA2 are located on the same contig (supercontig 17), ∼9.3 Mb apart, and that CesA3 and CesA4 are also located on the same contig (supercontig 47), ∼1.9 Mb apart. Since at least partial genomic sequence information is also publicly available from the Department of Energy (DOE) Joint Genome Institute (http://genome.jgi-psf.org) for two other Phytophthora species, Phytophthora sojae, a soybean pathogen, and Phytophthora ramorum, the cause of sudden oak death, BLAST searches were performed using the P. infestans sequences, and orthologous transcripts to CesA1, CesA2, CesA3, and CesA4 were identified within the genomes of both of these species.
Table 1.
DNA and Amino Acid (in Parentheses) Similarity between Phytophthora CesA Sequences
| Pi CesA1 | Pi CesA2 | Pi CesA3 | Pi CesA4 | Pr CesA1 | Pr CesA2 | Pr CesA3 | Pr CesA4 | Ps CesA1 | Ps CES2 | Ps CesA3 | Ps CesA4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pi CesA1 | 100a | |||||||||||
| Pi CesA2 | 64(71)b | 100a | ||||||||||
| Pi CesA3 | 20(30)d | 34(30)c | 100a | |||||||||
| Pi CesA4 | 57(50)c | 57(53)c | 21(32)d | 100a | ||||||||
| Pr CesA1 | 86(93)a | 71(70)b | 23(29)d | 62(50)b | 100a | |||||||
| Pr CesA2 | 73(70)b | 86(96)a | 38(32)c | 61(54)b | 74(71)b | 100a | ||||||
| Pr CesA3 | 10(20)d | 13(23)c | 87(95)a | 16(23)d | 11(20)d | 14(23)d | 100a | |||||
| Pr CesA4 | 60(49)b | 61(53)b | 39(32)c | 87(95)a | 64(50)b | 64(53)b | 15(24)d | 100a | ||||
| Ps CesA1 | 71(92)b | 86(72)a | 37(30)c | 58(50)c | 89(93)a | 75(71)b | 16(20)d | 63(49)b | 100a | |||
| Ps CesA2 | 68(72)b | 84(94)a | 32(31)c | 57(54)c | 75(71)b | 89(94)a | 16(22)d | 63(53)b | 91(70)a | 100a | ||
| Ps CesA3 | 20(30)d | 34(31)c | 86(94)a | 20(32)d | 23(29)d | 40(32)c | 90(86)a | 37(32)c | 38(30)b | 33(31)c | 100a | |
| Ps CesA4 | 59(50)c | 58(53)c | 36(32)c | 80(95)a | 62(51)b | 63(53)b | 15(23)d | 91(95)a | 61(50)b | 60(54)b | 35(32)c | 100a |
80% or more DNA similarity.
60 to 79% DNA similarity.
30 to 59% DNA similarity.
0 to 29% DNA similarity.
Orthologous Phytophthora CesA genes share a greater similarity to one another than paralogs do. For example, Pi CesA1 and Pr CesA1 share 86% similarity at the DNA level (Table 1). These data strongly suggest that all four CesA genes were present and already diverged from each other in a common ancestor of the Phytophthora lineage.
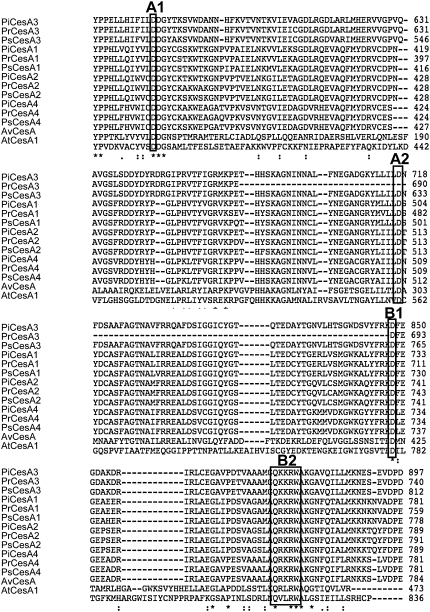
All CesA proteins described to date contain a conserved set of motifs (D,D, D,QXXRW; Figure 1) characterized by the presence of conserved Asp residues separated by a variable number of other amino acids in the globular region of the protein (Saxena et al., 1995; Saxena and Brown, 1997). Domain A (D [motif A1], D [motif A2]) is common to both processive and nonprocessive glycosyltransferases and binds the sugar donor (UDP-glucose), whereas domain B (D [motif B1], QXXRW [motif B2]) forms a putative acceptor binding region. It has been proposed that most family 2 glycosyltransferases, including cellulose synthases, use the A and B domains and the conserved Asp residues to form a single center for glycosyl transfer, but this must be confirmed (Charnock et al., 2001). All 12 putative Phytophthora CesA genes described in this study contain the A and B domains in their predicted amino acid sequences (Figure 1), suggesting that they are able to function as processive glycosyltransferases.
Figure 1.
Sequences of CesAs in Phytophthora sp.
Comparison of the 12 CesA sequences that were identified in Phytophthora sp with other members of the cellulose synthase superfamily. Phytophthora sequences were obtained from publicly available genome sequences published by the Broad Institute and DOE Joint Genome Institute (see Methods for website). ClustalW alignment (segment) of amino acid sequences of CesAs from Phytophthora sp, with a cellulose synthase from Arabidopsis (At CesA1) and Anabaena variabillis (Av CesA). Boxes indicate the conserved Asp residues (A1, A2, and B1) and the QXXRW motif (B2) found in processive glycosyltransferases. Note that conserved residue A2 is absent in Pr CesA3.
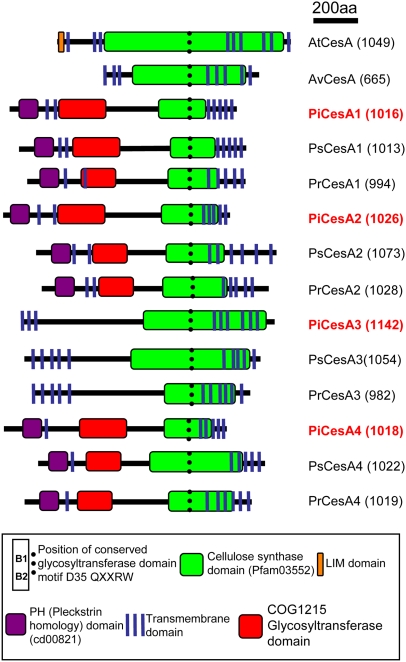
All products of the cellulose synthase genes identified to date are predicted to contain one or more transmembrane domains at the N terminus, followed by a large globular region containing the domain B motifs, and several further transmembrane domains toward the C-terminal end of the protein (Saxena and Brown, 2000). Structural predictions of all twelve Phytophthora proteins were performed using SOSUI (see Methods for website) and supported the hypothesis that these proteins are integral membrane proteins with several transmembrane domains (Figure 2).
Figure 2.
Domain Organization of the Cellulose Synthases from Phytophthora sp.
The predicted gene products share 20 to 94% sequence similarity. Total amino acids (aa) are given in parentheses after the name of each protein. Sequences are aligned according to the position of domain B containing the QXXRW (B2) motif (dotted line). The proteins contain regions similar to cellulose synthases and to other glycosyltransferases. Several transmembrane domains are also predicted. Phytophthora CesA1, CesA2, and CesA4 share the greatest sequence similarity and are predicted to contain PH domains toward the N terminus of each protein. The domain organizations of CesA from Arabidopsis and A. variabillis are given for comparison.
The Phytophthora CesA Proteins Represent a Novel Class of Cellulose Synthases
Conserved domain searches (see Methods for website) were performed using all 12 Phytophthora proteins. The C-terminal end of each amino acid sequence exhibited similarity to cellulose synthase domains found in plants and bacteria. The N-terminal end of each sequence was more divergent, either having similarity to the COG125 glycosyltransferase domain or having no predicted structural similarity at all (Figure 2). In addition, Pi CesA1, Pi CesA2, and Pi CesA4 all contained structural similarity to a Pleckstrin homology (PH) domain toward the N-terminal portion of the protein. This would appear to be a novel feature, as no other cellulose synthase has been reported that contains a PH domain. The zinc finger (or LIM-like) CxxC motif present at the N terminus of all plant cellulose synthases was not present in any of the Phytophthora CesA proteins (Figure 2).
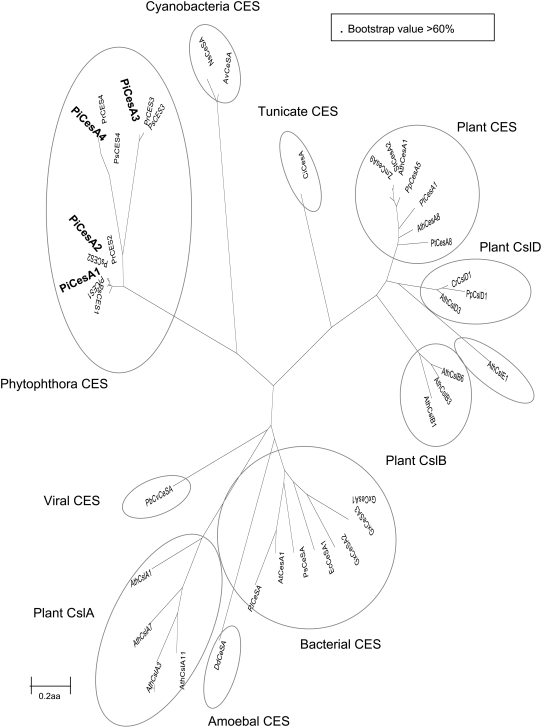
Comparisons with published CesA genes revealed that the Phytophthora genes group as a distinct clade, possibly representing a novel class of cellulose synthases that are more related to published cyanobacterial CesA proteins than to either plant CesA or Csl proteins (Figure 3).
Figure 3.
Dendrograms of Cellulose Synthases Derived from the Oomycete Sequences and Several Sequences from Eukaryotes and Prokaryotes.
Alignments were made using ClustalW and dendrograms produced using Mega 3.1 (see Methods for website) with a maximum evolution algorithm and 1000 bootstrap values. Dendrograms are based on the complete amino acid (aa) sequence (see Supplemental Table 1 online for sequence alignment). Ces(A), cellulose synthase; CslA-F, cellulose synthase-like; Pi, Phytophthora infestans; Pr, Phytophthora ramorum; Ps, Phytophthora sojae; Pt, Populus tremula; Zm, Zea mays; Ath, Arabidopsis thaliana; Cr, Ceratopteris richardii; Pp, Physcomitrella patens; St, Solanum tuberosum; Gx, Gluconacetobacter xylinus; At, Agrobacterium tumefaciens; Ec, Escherichia coli; Ns Nostoc sp; Av, Anabaena variabilis; Dd, Dictyostelium discoideum; Rl, Rhizobium leguminosarum; Ci, Ciona intestinalis; PbCv, Paramecium bursaria Chlorella virus 1; Ps, Pseudomonas syringae.
CesA1 Is More Abundant in Cysts Producing Appressoria Than in Mycelium
During a recent proteomics screen of the preinfection stages of the life cycle of P. infestans, we identified a protein spot on two-dimensional gels as CesA1. ESTs containing the CesA1 sequence were identified using MS-Fit, (see Methods for website) with a score of 184, 10% sequence coverage, and with 5/42 peptides matching between the protein spot and the theoretical digest of the EST encoding CesA1. These matches were toward the C terminus of the protein, and peptides of the following molecular weights were identified: 982.4, 1200.6, 1404.7, 1687.8, and 1831.8. These are consecutive peptides in the theoretical digest, strongly suggesting that the identified protein was indeed CesA1. The CesA1 protein spot was more abundant in germinating cysts producing appressoria than in in vitro–cultured vegetative mycelium (Figure 4). Subsequent analysis revealed that this spot was likely to be a partially degraded fragment of the protein. We reached this conclusion based on the fact that the spot had a much lower molecular weight than predicted and a relatively small percentage of sequence coverage. However, similarity was very high in the matching peptides.
Figure 4.
Identification of a Cellulose Synthase Protein Fragment in Germinating Cysts with Appressoria.
(A) Two-dimensional gel showing proteins isolated from mycelia.
(B) Two-dimensional gel showing proteins isolated from germinating cysts with appressoria.
(C) Enlarged portion of gel in (A) showing location of the spot identified as Pi CesA1 from mycelia.
(D) Enlarged portion of gel in (B) showing location of the spot identified as Pi CesA1 from germinating cysts with appressoria.
Gels were equally loaded with 50 μg total protein and stained with Coomassie blue. Protein pI and mass (kD) are shown in (A) and (B) on the x and y axes, respectively. The spot identified as Pi CesA1 appears more abundant in gel (D) than in gel (C).
The P. infestans Cellulose Synthase Genes Are Transcriptionally Upregulated during Cyst Germination and Appressoria Formation
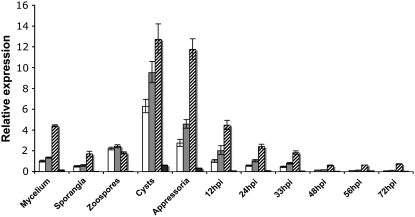
We used real-time RT-PCR to obtain the expression profiles of the P. infestans CesA genes in the preinfection structures and during infection on potato leaves. We compared expression of each gene to the level of CesA1 expression in vegetative nonsporulating mycelium grown in vitro. The expression results confirmed our initial finding with proteomics, that expression of CesA1 was upregulated during the formation of germinating cysts and appressoria. Overall, CesA2 and CesA3 were most highly upregulated, and CesA4 was expressed at comparatively low levels (Figure 5). We found a transcriptional downregulation of all four CesA genes, especially CesA4, in sporangia (∼2-fold, ∼2-fold, ∼2.5-fold, and ∼10-fold respectively). CesA1 and CesA2 were upregulated ∼2-fold within zoospores, and CesA3 and CesA4 appeared to be downregulated at this stage (∼2.5 and ∼10-fold, respectively). All four transcripts were most abundant in germinating cysts (∼6-fold, ∼7-fold, ∼3-fold, and ∼4-fold, respectively) and appressoria (∼3-fold, ∼3-fold, ∼3-fold, and ∼2-fold, respectively). After cyst germination and appressorium formation, the expression levels of all four CesA genes gradually decreased as the infection progressed from biotrophy to necrotrophy (Figure 5). Overall, these results suggest that the CesA genes are of particular importance to the production and germination of cysts and the subsequent formation of the appressorium.
Figure 5.
Expression of CesA Genes throughout the Life Cycle of P. infestans.
Expression profiles of CesA1 (white bars), CesA2 (gray bars), CesA3 (hatched bars), and CesA4 (black bars) in preinfection stages and in planta 12, 24, 33, 48, 56, and 72 h postinoculation (hpi) of susceptible potato cv Bintje. Expression of each sample is relative to the expression of CesA1 in nonsporulating mycelium, which has been assigned the value 1.0. Error bars represent 95% confidence intervals calculated using three biological replicates each containing three technical replicates for each sample within the RT-PCR assay.
The P. infestans Cellulose Synthase Genes Are Required for Normal Appressorium Formation in Vitro
Stable transformation is of low efficiency in many Phytophthora species, and gene knockouts present a significant technical challenge due to the diploid nature of these organisms. Thus, functional characterization of genes in P. infestans is laborious. However, we have recently developed a method of transient gene silencing in P. infestans using dsRNA (Whisson et al., 2005). We exploited this technique to further investigate the role of the cellulose synthases in P. infestans by creating lines in which we transiently silenced all four CesA genes simultaneously. We simultaneously transfected protoplasts with dsRNA molecules homologous to CesA1, CesA2, CesA3, and CesA4 and assessed the resulting regenerated colonies for phenotypic effects and silencing. The dsRNA was designed to target the globular regions of the four CesA genes and was specific for each gene (see Methods). Regions containing the PH domain were avoided to ensure that other genes with similar sequences were not silenced.
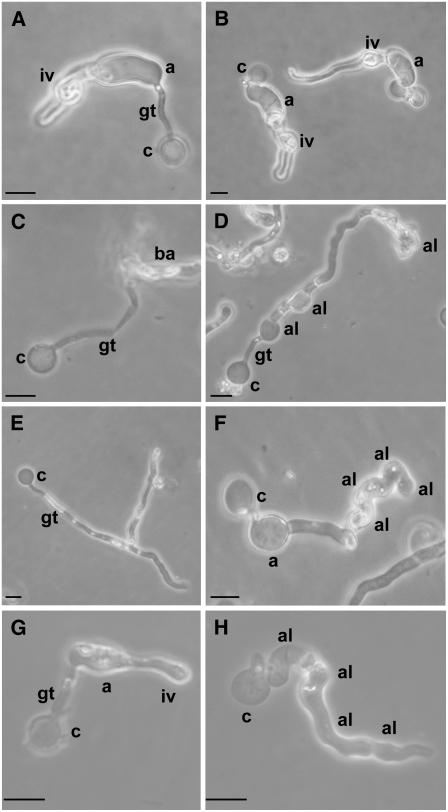
It is possible to induce P. infestans to differentiate through the preinfection stages of the life cycle in vitro; indeed, cells produce very similar characteristics to those induced in planta, with zoospores being released in cold, wet conditions. Encystment of spores can easily be induced with mechanical agitation, and these cysts will then germinate and produce appressoria after incubation in water at 11°C (Kramer et al., 1997; Grenville-Briggs et al., 2005). Cysts typically produce short germ tubes before swelling to produce an appressorium at the growing tip after ∼16 h (Figures 6A and 6B). After continued incubation infection, vesicle-like structures are produced (Kramer et al., 1997; Figures 6A and 6B).
Figure 6.
Morphology of CES1-4–Silenced Lines and DCB-Treated P. infestans after Appressorium Induction in Water at 11°C for 16 h.
(A) Normal appressorium and infection vesicle-like structure produced in control line C1.
(B) Normal appressorium and infection vesicle-like structure produced in control line C6.
(C) Burst appressorium produced in CES1-4–silenced line A1.
(D) Appressoria-like structures produced in CES1-4–silenced line A1.
(E) Cyst with multiple germ tubes in CES1-4–silenced line A9.
(F) Appressoria-like structures produced in CES1-4–silenced line A9.
(G) Normal appressoria and infection vesicle-like structure produced by wild-type P. infestans.
(H) Appressoria-like structures produced by DCB-treated P. infestans. Note the similarity between (D), (F), and (H).
All micrographs were taken in phase contrast. iv, infection vesicle-like structure; a, appressorium; gt, germ-tube; c, cyst; ba, burst appressorium; al, appressoria-like structure. Bars = 10 μm.
At least 60% of wild-type P. infestans cysts produced in vitro go on to produce an appressorium in water at 11°C. A maximum of 40% produce a germ tube but fail to produce an appressorium. We quantified the number of preinfection structures produced in vitro by individual silenced lines. In control lines incubated with the nonendogenous control dsRNA (green fluorescent protein [GFP]), the majority of cysts germinated and at least 60% of these cysts formed normal appressoria. This indicates that control lines behave in an equivalent manner to untreated wild-type P. infestans (Figures 6A, 6B, and 6G). Control lines also went on to produce infection vesicle-like structures, equivalent to those produced by wild-type untreated P. infestans in vitro (Figures 6A and 6B), whereas silenced lines did not (Figures 6C to 6F). In addition, some silenced lines produced either typical or aberrant appressoria, which ruptured at the growing tip where the infection vesicle would normally be produced (Figure 6C). Most of the individual silenced lines showed a marked reduction in the number of normal appressoria that were produced and a corresponding greater number of cysts that germinated but did not go on to produce an appressorium (Figure 6E). Rather, they produced germ tubes containing several abnormal appressorium-like structures, which we presumed were failed attempts to produce an appressorium (Figure 6F). It should be noted that the severity of the reduction in normal appressorium development and the numbers of aberrant appressoria produced directly correlated with the levels of CesA1-4 repression for each individual line (Figures 7A to 7D).
Figure 7.
Expression Levels in CesA1-4–Silenced Lines.
(A) to (D) Relative expression of CesA1, CesA2, CesA3, and CesA4, respectively, in six individual control lines (C1-C6) and 19 individual lines treated with dsRNA from CesA1, CesA2, CesA3, and CesA4 simultaneously (A1-A19). Error bars represent confidence intervals calculated using three technical replicates for each sample within the RT-PCR assay. To allow comparisons of expression levels between lines, expression is shown as a value relative to the mean expression for all control lines for each gene (calibrator value). This calibrator value has been converted to 1.0 to allow comparisons between genes.
(E) Percentages of normal appressoria (black bars), germinating cysts (white bars), and aberrant appressoria-like structures (gray bars) in six individual control lines (C1-C6) and 19 individual CesA1-4–silenced lines. These are the same lines as those shown in (A) to (D).
Real-time RT-PCR analysis of mRNA from germinating cysts from 17 independent lines treated with dsRNA homologous to CesA1, CesA2, CesA3, and CesA4 and six independent lines treated with the control GFP dsRNA revealed that 14 lines showed repression of all four genes (Figures 7A to 7D). Line A8 did not show a clear reduction in CesA1 expression (Figure 7A) but did show reduced expression of CesA2 (Figure 7B), CesA3 (Figure 7C), and CesA4 (Figure 7D). Line A16 showed reduced CesA1 (Figure 7A) and CesA2 (Figure 7B) expression but near wild-type expression of CesA3 (Figure 7C) and CesA4 (Figure 7D). Lines A10 and A18 showed reduced CesA1 expression (Figure 7A) but near wild-type expression of CesA2 (Figure 7B), CesA3 (Figure 7C), and CesA4 (Figure 7D). Lines A17 and A19 showed near wild-type expression for all four CesA genes (Figures 7A to 7D) and a corresponding wild-type phenotype (Figure 7E), suggesting that these lines were not significantly silenced. The high level of variation seen in line A19 may be due to low-level silencing, so that depending on the RNA sampled in each replicate, a proportion of silenced and nonsilenced material may be assayed. An alternative explanation is that this line is actually derived from a colony representing a population that arose from two different protoplasts, with different levels of silencing. If this is the case, the nonsilenced line may mask the phenotype of the silenced line but contribute to significant variation at the RNA level. Line A18 also showed near wild-type expression of CesA1 (Figure 7A), CesA2 (Figure 7B), and CesA4 (Figure 7C) but reduced expression of CesA3 (Figure 7C). However, this line exhibited a near wild-type level of appressorium production (Figure 7E), suggesting that CesA3 is either less important for the production of normal appressoria than the other CesA genes or that it is functionally redundant with other CesA genes compensating for its loss of function. Lines that clearly showed reductions in the transcripts of all four CesA genes showed a clearly altered phenotype in germinating cysts and appressoria formation when compared with the control lines (Figure 7E). Line A6 is the only exception to this, appearing to have produced similar percentages of appressoria and germinating cysts to the control lines (Figure 7E). However, this line produced very few appressoria; thus, it is likely that the percentages recorded do not accurately reflect the real phenotype.
Cellulose Content in Cell Walls Is Affected by Silencing CesA Genes
Twenty-two further lines were produced, and the cellulose content in the appressorial cell walls was determined. Intact appressoria corresponding to six nonsilenced lines (showing an average of 6% appressorium-like structures, 64.5% normal apressoria, and 29.5% germinated cysts that had not produced an appressorium) (nonsilenced pool), eight silenced lines showing an extreme phenotype (composed of an average of 47% appressoria-like structures, 34% normal appressoria, and 19% germinated cysts) (extreme pool), and eight silenced lines showing a more moderate phenotype (composed of an average of 34% appressoria-like structures, 45% normal appressoria, and 21% germinated cysts) (moderate pool) were pooled together to form three samples for cellulose content determination. These pools of whole cells were freeze-dried, and the corresponding cellulose content was determined to investigate the effect of gene silencing on the amount of cell wall cellulose. Cellulose quantitation was performed by extensive enzymatic hydrolysis combined with reducing sugar assay as described in Methods. The specificity of the cellulase mixture used for cellulose hydrolysis and the commercial endo-(1→3)-β-d-glucanase from Aspergillus niger was verified on well-characterized polysaccharides (see main Methods section and Supplemental Methods online), and each glucanase preparation was found to be active only on its specific substrate (see Supplemental Figure 1 online). The use of a mixture of endo- and exocellulases revealed that the amount of cellulose synthesized in the cell walls of appressoria was significantly reduced in the CesA1-4–silenced lines compared with the nonsilenced lines (Figure 8). No reducing sugars were released from the same samples upon digestion with the specific endo-(1→3)-β-d-glucanase from A. niger (data not shown). The amount of released reducing sugars upon cellulase digestion was lower during the first 48 h of hydrolysis for the pool with the most severe phenotype compared with the pool with a moderate phenotype (Figure 8). This observation suggests that the cellulose from the lines showing the moderate phenotype had a lower crystallinity than the cellulose from the lines with the severe phenotype and was thus more readily hydrolyzed. It is unlikely that the observed difference during the first 48 h of hydrolysis is due to the presence of a lower amount of cellulose in the cell walls of appressoria exhibiting the most severe phenotype. Indeed, the kinetics of release of reducing sugars reached a plateau after 72 h of hydrolysis (Figure 8), suggesting that the total amount of cellulose originally present in the samples was fully hydrolyzed and that the lines showing severe and moderate phenotypes originally contained similar amounts of cellulose in their walls. The amount of reducing sugars released from the cell walls of the nonsilenced pool was higher than for the two silenced pools at any time during hydrolysis (Figure 8). As opposed to the data for the silenced lines, it kept increasing after 72 h of digestion, suggesting that the hydrolysis of cellulose in these nonsilenced lines had not reached completion. This indicates the presence of a higher amount of cellulose in the cell walls of the nonsilenced lines. The amount of reducing sugars released from the cell walls of the latter lines was twice as high as that obtained from the silenced lines after 72 h of hydrolysis (Figure 8), indicating that transient gene silencing provoked a reduction of the amount of cellulose in cell walls of at least 50%. Silencing CesA1-4 genes significantly decreases the amount of cellulose in the appressorial cell wall, which in turn leads to a loss of cell wall integrity, leading to the observed phenotypes. Altogether, these data demonstrate that the observed phenotypes are due to a deficiency of cellulose in the walls of the silenced lines and that the CesA1-4 genes are involved in cellulose synthesis.
Figure 8.
The Cellulose Content of Cell Walls Is Affected by Silencing the CesA1-4 Genes.
Cellulose from the cell walls of appressoria corresponding to nonsilenced lines (rectangles, solid line) and to silenced lines (broken lines) with moderate (triangles) and severe (circles) phenotypes was extracted and hydrolyzed by a specific mixture of endo and exocellulases as described in Methods. Aliquots of the hydrolytic reaction mixtures were taken at different time points (0, 24, 48, and 72 h) to assay reducing sugars released from cellulose upon enzymatic hydrolysis. Fresh mixtures of cellulases were added in the rest of the reaction mixtures at the same time points to compensate for cellulase inactivation over time. Reducing sugar equivalents are expressed as micrograms of reducing sugars/milligram of total appressoria.
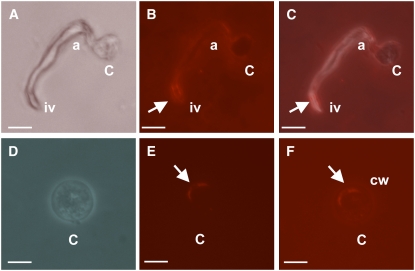
CesA Proteins Localize to the Growing Tip and Infection-Like Vesicles of Appressoria
To gain further insights into the role of the CesA proteins in the biosynthesis of P. infestans cell walls, immunolocalization studies were performed using antibodies produced against a mixture of two synthetic peptides corresponding to segments of a CesA protein from the oomycete S. monoica. The sequences of the peptides are highly similar to the sequences of segments of the four P. infestans CesA proteins (see Supplemental Figure 2 online). For instance, no more than two amino acids were different between the peptides corresponding to Sm CesA and its closest Phytophthora homolog Pi CesA3. The antibodies recognized a single band in plasma membrane preparations from S. monoica and P. infestans, strongly suggesting a specific reactivity with the CesA proteins from both species (see Supplemental Figure 3 online). As a control, we used anti-STR (small tyrosine-rich protein) antibodies from the mycoparasitic oomycete Pythium oligandrum (N.R. Horner and P. van West, unpublished data). The STR sequences used for antibody production are not present in the P. infestans genome, and these antibodies gave no fluorescent signal upon incubation with P. infestans appressoria (data not shown). Wild-type P. infestans appressoria were induced, fixed, permeabilized, and immunolabeled on plastic cover slips and mounted in PBS for visualization. Since the antibodies react exclusively with the membrane-bound CesAs (see Supplemental Figure 3 online), it can be concluded from the immunolocalization data presented in Figure 9 that the CesA proteins specifically localize to the plasma membrane at the growing tip and infection-like vesicles of the appressorium (Figures 9A to 9C) and to the plasma membrane of cysts (Figures 9D to 9F). We were unable to localize these proteins in sporangia, or zoospores (data not shown), reinforcing the concept that cellulose plays a particularly important role in the appressorial cell wall.
Figure 9.
CesA1-4 Proteins Localize to the Growing Tip and Infection-Like Vesicles of Appressoria and Cysts.
Wild-type P. infestans (strain 88069) cysts harvested from 11-d-old solid rye plates were induced to produce appressoria on plastic cover slips, overnight at 11°C. Cells were fixed and labeled with primary anti-CesA antibodies and visualized using Alexa Fluor 555. Phase contrast ([A] and [D]), fluorescent micrographs ([B] and [E]), and combined images ([C] and [F]) of a wild-type appressorium and a cyst. Arrows indicate areas of strong localization of CesA1-4 proteins. c, cyst; a, appressorium; iv, infection vesicle-like structure; cw, cell wall. Note that due to the fixing process used in this procedure, appressoria are dehydrated and therefore appear slightly deflated in (A) to (C). Bars = 10 μm in (A) to (C) and 5 μm in (D) to (F).
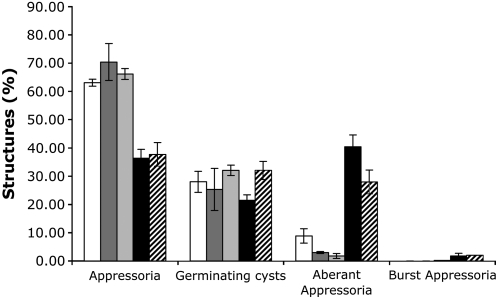
Inhibition of Cellulose Synthesis Using 2,6-Dichlorobenzonitrile Results in Aberrant Appressoria Formation in Vitro
2,6-Dichlorobenzonitrile (DCB) is an inhibitor of cellulose synthesis, acting specifically at micromolar levels (Mayer and Herth, 1978; Montezinos and Delmer, 1980). We next analyzed the effect of this chemical on P. infestans growth and development. An amount of 100 or 40 μM DCB had no significant effects on P. infestans mycelial growth under sporulating (solid plates) or nonsporulating conditions (liquid cultures) (see Supplemental Figure 4 online). Induction of zoosporogenesis, encystment, and incubation under conditions conducive to appressorium formation, in the presence of 100 μM DCB, resulted in a severe reduction in zoospore release and a complete inhibition of cyst germination (see Supplemental Figure 4 online). At 40 μM DCB, rupture of appressoria at the growing tip was observed, suggesting that the cell wall was unable to support sufficient turgor pressure for continued growth. Germ tubes producing aberrant appressoria-like structures, equivalent to those seen in CesA1-4–silenced lines, were also observed (Figures 6G and 6H). A marked reduction in the development of normal appressoria was observed compared with control samples (Figure 10). Comparing these results to the total numbers of structures formed by all silenced lines together shows a similar reduction in appressorium production and increase in appressoria-like structure production (Figure 10). Overall, the cellulose synthesis inhibitor DCB exhibited the same effect on the production of appressoria and appressoria-like structures as silencing of CesA1-4 genes.
Figure 10.
Percentage of Preinfection Structures Produced by DCB-Treated P. infestans and CesA1-4–Silenced Lines.
White bars, water control; mid-gray bars, 1.25% DMSO control; light-gray bars, combined data from 30 RNA interference control lines treated with GFP dsRNA; black bars, treated with 40 μM DCB in 1.25% DMSO; hatched bars, combined data from RNA interference CesA1-4–silenced lines. Mean ± se from three independent experiments.
Thus, we conclude from these data that cellulose production is particularly important for the formation of appressoria in P. infestans.
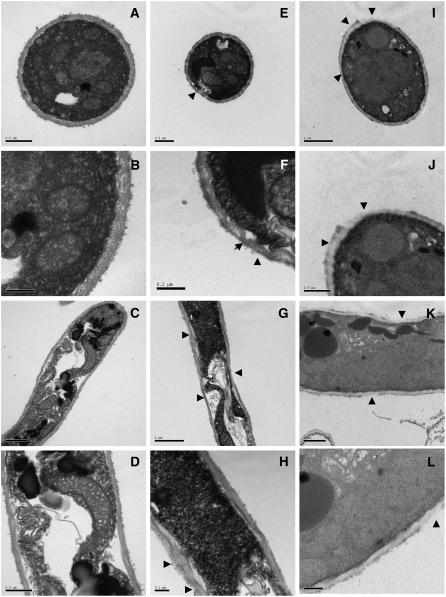
Transmission Electron Microscopy Observations of Cell Walls
To gain further insights into the effects of DCB and of silencing CesA1-4, we examined the cell walls of germ tubes and appressoria using transmission electron microscopy (TEM). Germ tubes in wild-type P. infestans displayed cell walls of uniform thickness (Figures 11A to 11D). Those derived from CesA1-4–silenced lines or from cells treated with DCB showed a nonuniform cell wall structure. The cell wall of germ tubes from CesA1-4–silenced lines showed areas of apparent loss of the outer less well-defined part of the cell wall (Figures 11E and 11F) as well as areas of both thinning and thickening of the wall (Figures 11G and 11H).
Figure 11.
Ultrastructure of Germ Tube Cell Walls of Wild-Type, CesA1-4–Silenced, and DCB-Treated P. infestans in Vitro.
(A) and (B) Cross section of wild-type germ tube cell wall (A) and close-up (B). Note the characteristically uniform cell wall.
(C) and (D) Longitudinal section through a wild-type germ tube (C) and close-up (D). The cell wall surrounding the germ tube is of uniform thickness.
(E) and (F) Cross section of germ tube cell wall from CesA1-4–silenced lines (E) and close-up (F). The wall is less uniform than in the wild type. Note the invagination of the cell membrane, which is caused by an uptake of Epon embedding resin, due to a loss of structure and increased permeability of the cell wall (arrowheads).
(G) and (H) Longitudinal section through a germ tube from a CesA1-4–silenced line (G) and close-up (H). The cell wall is much less uniform and exhibits areas of both thinning and thickening (arrowheads).
(I) and (J) Cross section of cell wall of an appressorium or appressorium-like structure treated with DCB (I) and close-up (J). Note the severe alteration of the cell wall (arrowheads).
(K) and (L) Longitudinal section through a hypha treated with DCB (K) and close-up (L). The cell wall exhibits thinning and apparent disruption, particularly in the outer wall (arrowheads).
Bars = 0.2 μm in (B), (F), and (H), 0.5 μm in (A), (D), (E), (J), and (L), and 1.0 μm in (C), (G), (I), and (K).
Invaginations of the inner cell membrane were observed in silenced lines, often in areas where the cell wall appeared extremely thin or absent (Figure 11F), but not in control or DCB-treated lines. Although invaginations are commonly seen with TEM preparations using Epon resin, this technical observation may reflect biological differences between the CesA1-4–silenced samples and the control and DCB-treated samples. DCB-treated samples showed a more dramatic ultrastructural phenotype, with areas of significant alteration of the outer cell wall structure (Figures 11I and 11J). The cell wall also appeared less electron dense than that of the other samples and frequently contained areas of both thinning and thickening (Figures 11K and 11L).
Cellulose Synthesis Is Essential for Pathogenicity
Under natural conditions, P. infestans zoospores will encyst and germinate upon contact with the host surface. Germ tubes are usually short and produce swollen appressoria at their tips, which characteristically develop to allow penetration at the anticlinal wall between epidermal cells (Figures 12A and 12B), and an infection vesicle is then produced in one of the epidermal cells adjacent to the site of penetration. Hyphae then grow into the mesophyll layer both inter- and intracellularly, producing occasional haustoria and eventually leading to necrosis of host tissues (van West and Vleeshouwers, 2004).
Figure 12.
DCB-Treated P. infestans Is Nonpathogenic.
(A) and (B) Scanning electron micrograph of wild-type cysts that have produced short germ tubes and appressoria and have penetrated a potato leaflet, 16 hpi.
(C) and (D) Cysts treated with DCB and inoculated in the presence of DCB have produced long germ tubes and appresorium-like structures and are unable to penetrate the potato leaflet, 16 hpi.
(E) Zoospores produced in the presence of DCB are completely nonpathogenic, while control zoospores produce large water-soaked necrotic lesions. All observations were performed 6 d after inoculation. (i) and (iii) Zoospores treated with 40 μM DCB, in 1.25% DMSO, and inoculated in the presence of 40 μM DCB. (ii) and (iv) Control zoospores treated with 1.25% DMSO control and inoculated in the presence of 1.25% DMSO. (v) Zoospores produced in the presence of 40 μM DCB and washed and inoculated in water are able to recover their pathogenicity. (vi) Leaflets inoculated with 40 μM DCB only show no symptoms.
To investigate the need for cellulose synthesis during plant infection, we inoculated susceptible potato cv Craigs Royal leaflets with zoospores produced in the presence of 40 μM DCB and recorded disease progress (host plant necrosis) 6 d after inoculation. We found that cells treated with 40 μM DCB were completely nonpathogenic. Severe necrosis and water-soaked lesions were observed on all leaflets inoculated with non-DCB-treated zoospores (Figure 12E). To gain a further understanding of this loss of pathogenicity, we examined infected material 16 h after inoculation using scanning electron microscopy. By this time, wild-type P. infestans zoospores had encysted, germinated, and produced many appressoria on the surface of leaflets and appeared to have penetrated the host (Figures 12A and 12B). Zoospores treated with DCB had encysted, producing long germ tubes and aberrant appressoria-like structures, which were similar to those observed in vitro (Figures 12C and 12D). These structures resembled failed appressoria on the surface of the leaf. That is, it appeared that cysts had germinated and repeatedly attempted to produce appressoria but had failed to penetrate the leaf (Figures 12C and 12D). We therefore conclude that a lack of cellulose in the cell wall leads to an inability to form normal appressoria. Appressoria-like structures were formed, but these were unable to penetrate host cells presumably due to a loss of cell wall strength associated with reduced levels of cellulose. Therefore, we can conclude that a cellulosic cell wall is required for pathogenicity of P. infestans.
DISCUSSION
A Family of Four P. infestans Cellulose Synthase Genes Represent a Novel Class of CesA Genes
Since components of the cell wall are involved in adherence, recognition of a suitable host, and the protection of the cell from defense products, changes in the structure or composition of the cell wall may affect virulence. Oomycete cell walls are predominantly composed of cellulose (Bartnicki-Garcia, 1968). Here, we used the recently published P. infestans, P. ramorum, and P. sojae genome sequences to identify a novel family of oomycete cellulose synthases.
Each genome contained four CesA genes, with similarity between orthologs much greater than similarity between paralogs, suggesting that these genes were ancient and present in a common Phytophthora ancestor. The Phytophthora CesA1, CesA2, and CesA4 proteins contain a PH domain toward the N-terminal region of the protein. PH domains are conserved domains first identified in Homo sapiens Pleckstrin1, the major protein kinase C substrate of platelets (Tyers et al., 1988). PH domains have since been found in many different eukaryotic proteins involved in signal transduction and cytoskeletal organization that require association with the cell membrane (Mayer et al., 1993; Lemmon et al., 1996). The presence of PH domains in cellulose synthases represents a novel adaptation specific to the oomycetes, though their function remains unclear.
In our phylogenetic analysis, we found that Phytophthora proteins grouped together as a distinct clade, separate from either bacterial or plant cellulose synthases and more closely related to cyanobacterial cellulose synthases than to those from plants.
Cellulose Synthesis Is Required for Normal Appressorium Formation and Pathogenicity
Expression profiling and gene silencing of the CesA gene family in P. infestans revealed that the CesA genes are upregulated during the germination of cysts and the subsequent production of appressoria. Cellulose confers stability to the cell wall, and it is therefore not surprising that at the point in the life cycle when a new cell wall needs to be produced and expanded, cells either lacking in cellulose synthase expression, or chemically inhibited in the synthesis of cellulose, show altered cell morphology. Here, we have shown that cellulose production is important for the morphology of the cell wall in germinating cysts and appressoria prior to penetration of the host plant cuticle.
To determine the function of the CesA gene family in more detail, we transiently silenced the entire family using homologous dsRNA, designed specifically to each transcript. We achieved significant silencing of all four genes (up to 90% for CesA1, CesA2, and CesA4 in some lines). Levels of CesA3 were also significantly reduced, but the maximum knockdown seen was ∼60%. The severity of the loss of appressorium production and the levels of appressoria-like structures produced by individual lines directly correlated to the observed transcript levels. In addition, transient silencing of CesA1-4 provoked a decrease of >50% of the cellulose content in the walls of appressoria, thereby confirming the involvement of the Phytophthora CesA genes in cellulose biosynthesis.
The CesA proteins localize to the growing tip wall and infection vesicles of appressoria and to the plasma membrane of cysts, which is in good agreement with the expected active cellulose production during wall expansion and is consistent with our RT-PCR expression profiles, which indicate that all four genes are upregulated in cyst germination and appressorium production. This is also consistent with the localization of chitin synthase (CHS) proteins, which perform an equivalent function in fungi (Munro et al., 2001; Madrid et al., 2003; Weber et al., 2006). We were unable to localize the CesA proteins in the plasma membranes of sporangia or in the membranes of zoospores, confirming that cellulose biosynthesis is particularly important during the production of germ tubes and appressoria and does not play a significant role in the preceding preinfection stages. Since the antibodies produced against S. monoica CesA peptides likely react with all four Pi CesA proteins indistinguishably, we were unable to localize these proteins individually using this method. Future experiments using GFP or monomeric red fluorescent protein fusions would allow us to assess the role and localization of the individual proteins in more detail.
The cellulose synthesis inhibitor DCB had no effect on mycelial growth in the oomycete Achlya bisexualis, presumably reflecting the fact that cellulose is not observed in the elongating tip of A. bisexualis hyphae (Shapiro and Mullins, 2002). However, the effects of DCB have not previously been assessed in oomycete appressoria. Our results indicate that a functional appressorium with its characteristic cellulosic cell wall is essential for P. infestans pathogenicity. Both in vitro and on the surface of potato leaves, P. infestans inhibited from cellulose synthesis produced aberrant appressoria-like structures. In vitro, these structures were not dissimilar to the infection vesicles seen in wild-type P. infestans and produced from an appressorium. However, since single germinating cysts produced several of these structures in linear succession, it is more likely that they represent repeated attempts by the germinated cysts to produce a functional appressorium. This is also confirmed by the observation that on the leaf surface, several of these structures are often produced from a single germinating cyst, with no sign of penetration of the host plant cuticle. This is presumably due to the cell wall being unable to maintain sufficient integrity required during host penetration.
P. infestans Was Rendered Completely Nonpathogenic in the Presence of DCB
It is possible that this is due not only to the inability of P. infestans to produce functional appressoria but may also be due, in part, to subtle effects of DCB on the potato leaves themselves, since DCB is a known inhibitor of cellulose synthesis in plants. However, if DCB were to decrease cellulose synthesis within host cells, we would expect to see an increase in susceptibility to P. infestans, which was not observed in our experiments. It may be possible that DCB has triggered a general defense response, resulting in an increase in plant resistance. However, we did not observe any of the normal defense responses associated with P. infestans resistance, such as a hypersensitive response. Rather, we found aberrant P. infestans appressoria, such as those found in vitro, suggesting that DCB was acting primarily on P. infestans rather than the plant.
Oomycetes such as P. infestans produce appressoria from swollen hyphal tips presumably to provide a focal point for the concentration of resources to breach the host plant cuticle either by mechanical force, hydrolysis of host cell barriers, or a combination of both processes. However, unlike the fungus Magnaporthe grisea, breach of the host cuticle is likely not achieved through mechanical force alone, since oomycetes also possess genes for cell wall hydrolysis that are upregulated in germinating cysts (Jarvis et al., 1981; McLeod et al., 2003). Cellulases are likely to be part of the repertoire of cell wall hydrolyzing enzymes secreted by plant pathogenic oomycetes, since cellulose is an important part of the host plant wall (Masaaki et al., 1973; Picard et al., 2000). However, these enzymes are likely to be under very tight control to avoid immediate host cell death and highly specific for plant cellulosic compounds to avoid degradation of their own outer cell wall (Pieterse et al., 1992).
Our results suggest that cell wall stability and integrity is required for appressorium development. Therefore, appressorial integrity is likely to be an essential element in P. infestans pathogenesis, since without it, P. infestans is nonpathogenic. Chitin is the major component of the fungal cell wall, and a similar role for CHSs has been reported in fungal pathogens of both plants and humans (Munro et al., 2001; Madrid et al., 2003; Liu et al., 2004; Weber et al., 2006). In those studies, disruption of the fungal CHS genes led to hypersensitivity to host oxidative defense responses, osmotic sensitivity, or changes in the composition of the cell wall. These findings confirm that changes in the structure and composition of the cell wall in many pathogenic fungi and, now shown in oomycetes, can lead to significantly reduced pathogenicity.
In lines silenced for CesA1-4, the cell wall showed a less uniform structure, with areas of thinning or of thickening and a higher degree of disorganization than in wild-type cell walls. This phenotype was even more pronounced following treatment with DCB. However, the cell walls of germ tubes derived from silenced lines sometimes showed invaginations of the membrane and cell wall–like material, but this was not observed after DCB treatment. Since the CesA proteins are predicted to be integral membrane proteins and that they localize to the growing appressorial cell wall, it is possible that these invaginations are associated with a reduction in cellulose synthase proteins in the membrane due to transient gene silencing, something that would not occur with DCB treatment. In comparison, DCB inhibition assays involve normal levels of CesA proteins, which are inhibited at the site of action. This therefore suggests a structural role for the CesA proteins in stabilizing the plasma membrane.
Glycoproteins with cellulose binding activity, termed CBEL proteins (cellulose binding, elicitor of defense, lectin-like) have been identified in a number of Phytophthora and Pythium species, including P. infestans, P. sojae, P. parasitica, P. citricola, and Pythium irregulare (Gaulin et al., 2002). In P. parasitica var nicotianae, CBEL is required for the proper deposition of cell wall polymers, and silencing CBEL results in distinct areas of cell wall thickening (Gaulin et al., 2002), a phenotype not dissimilar to that seen in our CesA1-4–silenced lines. Since CBEL contains two cellulose binding domains that do not show enzymatic activity, they may serve to locally cross-link two cellulose microfibrills within the cell wall and therefore play a role in the organization of the cell wall. CBELs also play a role in adherence on cellulosic surfaces and may also bind host cellulose in the establishment of infection.
METHODS
Growth of Phytophthora infestans Life Cycle Stages, Appressoria Formation, Potato Plants, and Plant Inoculation
P. infestans strain 88069 (A1 mating type, race 1.3.4.7) was maintained on Rye medium supplemented with 2% sucrose as described by Caten and Jinks (1968). The P. infestans strains used were grown under the Scottish Executive Environment and Rural Affairs license number PH/29/2006. Mycelia for protein, DNA, or RNA isolation was grown for 6 d in liquid rye broth, culture filtrate was removed using a vacuum manifold, and the resulting mycelial tissue stored at −70°C until use. P. infestans life cycle stages of sporangia, zoospores, germinating cysts, and germinating cysts developing appressoria were prepared as described by Grenville-Briggs et al. (2005).
Potato (Solanum tuberosum) infection for real-time RT-PCR analysis was performed as described elsewhere (Avrova et al., 2003). Leaves were harvested before inoculation (B0) and at 12 hpi (B12), 24 hpi (B24), 33 hpi (B33), 48 hpi (B48), 56 hpi (B56), and 72 hpi (B72) as described by Grenville-Briggs et al. (2005). These time points correspond to the biotrophic interaction (B12-B33), a transition phase between biotrophy and necrotrophy (B33-B48), and hyphal ramification and sporulation in host tissue during the necrotrophic phase (B56-B72). Collected leaf samples were frozen in liquid nitrogen and then stored at −70°C prior to RNA extraction. Additional plants (inoculated and uninoculated) kept for 5 d after inoculation showed high levels of infection and no infection, respectively (data not shown).
Extraction of Proteins, Two-Dimensional Gel Electrophoresis, and Protein Identification
Protein extraction and identification was performed as described previously (Grenville-Briggs et al., 2005). In brief, solubilized proteins were subjected to two-dimensional electrophoresis in an immobilized pH 4 to 7 gradient. The protein spot corresponding to CesA1 was excised from a two-dimensional gel, and peptide fingerprints were obtained using matrix-assisted laser-desorption ionization time of flight mass spectrometry. MS-FIT (http://prospector.ucsf.edu) was used with the Syngenta Phytophthora Consortium database (https://xgi.ncgr.org/spc/) to identify corresponding ESTs.
Sequence Analysis and Phylogeny
CesA sequences were identified from the P. infestans genome DNA sequence recently made available by the Broad Institute (http://www.broad.mit.edu/annotation/genome/phytophthora_infestans/Home.html) and from an EST collection maintained by the Syngenta Phytophthora Consortium (https://xgi.ncgr.org/spc/) (Randall et al., 2005). P. ramorum and P. sojae CES sequences were obtained from the DOE Joint Genome Institute (http://genome.jgi-psf.org/Phyra1_1/Phyra1_1.home.html and http://genome.jgi-psf.org/Physo1_1/Physo1_1.home.html). Other cellulose synthase sequences used for phylogenetic analysis were obtained from the protein databases at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/). Alignments were made with ClustalW (Higgins et al., 1994). Phylogenetic dendrograms were constructed using MEGA 2.1 (www.megasoftware.net) (Kumar et al., 2004), with the minimum evolution algorithms using 1000 bootstrap replications. Prediction of transmembrane domains was done using SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/) (Hirokawa et al., 1998). Conserved domain searches were performed using the NCBI CD search program (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer and Bryant, 2004).
RNA Extraction
Total RNA from individual stages of the life cycle of P. infestans was extracted from frozen samples ground in liquid nitrogen using a Qiagen RNeasy plant mini kit, following the manufacturer's protocol. Total RNA was extracted from frozen material deriving from single individual colonies generated from regenerated dsRNA-treated protoplasts using a Qiagen RNeasy plant mini kit following the manufacturer's protocol but with the following modification. Pellets were vortex-disrupted using 500-μL glass beads per sample in extraction buffer prior to RNA extraction. Prior to cDNA synthesis, all RNA samples were DNaseI treated using the Qiagen on-column DNase kit, following the manufacturer's protocol. Integrity of the RNA was tested by agarose gel electrophoresis. First-strand cDNA was synthesized from 1 to 20 μg total RNA by oligo(dT) priming using the first-strand cDNA synthesis kit (Amersham Pharmacia Biotech) following the manufacturer's protocol.
SYBR Green Real-Time RT-PCR Assays
Primer pairs (Table 2) were designed to anneal specifically to each of the four transcripts from P. infestans for real-time RT-PCR analysis. Template cDNA was derived from mycelium grown in rye broth, sporangia, zoospores, germinating cysts, and germinating cysts with appressoria, as well as potato leaves before inoculation (B0) and at 12 hpi (B12), 24 hpi (B24), 33 hpi (B33), 48 hpi (B48), 56 hpi (B56), and 72 hpi (B72). The actA gene from P. infestans was used as a constitutively expressed endogenous control, and the expression of each gene in mycelium was determined relative to actA as described by Avrova et al. (2003). Expression of all genes in different life cycle stages was compared with the level of their expression in a calibrator sample, which was cDNA from mycelium. The expression of CesA1 in the mycelium cDNA sample was assigned the value of 1.0. For expression of genes in potentially silenced lines, the calibrator sample was the mean expression value from all the control lines for each gene. This mean number was assigned the value of 1.0 to allow comparisons to be made between genes. RT-PCR assays for the expression of genes throughout the life cycle were performed using three biological replicates each containing three technical replicates. RT-PCR assays for the expression of genes in silenced lines were performed only using three technical replicates, as the transient nature of the RNA interference inhibition and the cell growth in this system limited available tissue. Instead, individual lines serve as biological replicates for the experiment as a whole.
Table 2.
Oligonucleotide Primer Pairs and Amplicon Sizes Used in Template Preparation for dsRNA Synthesis and Real-Time RT-PCR
| Gene | Primer Name | Primer Sequence (5′–3′) | Application | Amplicon Size (bp) |
|---|---|---|---|---|
| Pi CesA1 | CesA1T7F | GTAATACGACTCACTATAGGGCGTGCCGAAGCTTATCGT | dsRNA synthesis | 240 |
| CesA1T7R | GTAATACGACTCACTATAGGGACAGCGCCAGAGAACACC | dsRNA synthesis | ||
| CesA1TaqF | CTCCATTGCTGTGGGTCTCAT | Real-time RT-PCR | 90 | |
| CesA1TaqR | GGCTCAACCTAGATACACCTCCTCTA | Real-time RT-PCR | ||
| Pi CesA2 | CesA2T7F | GTAATACGACTCACTATAGGGCTACCGCAGAGCTACCGAAC | dsRNA synthesis | 195 |
| CesA2T7R | GTAATACGACTCACTATAGGGATTCCTGGATGCTCATACGC | dsRNA synthesis | ||
| CesA2TaqF | TGTCGGCGCTGAGCAA | Real-time RT-PCR | 77 | |
| CesA2TaqR | GAACGCGTAGGCGAACCA | Real-time RT-PCR | ||
| Pi CesA3 | CesA3T7F | GTAATACGACTCACTATAGGGAGATGCCTACACGGGTAACG | dsRNA synthesis | 206 |
| CesA3T7R | GTAATACGACTCACTATAGGGGGTCGACTTCGCTCTCATTC | dsRNA synthesis | ||
| CesA3TaqF | TTCACGCTGCTCTTCAGTCAA | Real-time RT-PCR | 80 | |
| CesA3TaqR | ACGTAGTTCCACGGGTTCGT | Real-time RT-PCR | ||
| Pi CesA4 | CesA4T7F | GTAATACGACTCACTATAGGGGAGTCTGTGGCTGCTGCTCT | dsRNA synthesis | 189 |
| CesA4T7R | GTAATACGACTCACTATAGGGGATCGGGTACACCGTCAAGT | dsRNA synthesis | ||
| CesA4TaqF | TGGCGCTGGTGCCTAAG | Real-time RT-PCR | 82 | |
| CesA4TaqR | CGCAGCACGTCATCGTTATC | Real-time RT-PCR | ||
| GFP | GFPT7F | GTAATACGACTCACTATAGGGGCAGATTGCGTGGACAGGT | dsRNA synthesis | 199 |
| GFPT7R | GTAATACGACTCACTATAGGGCTGGAGTACAACTACAACT | dsRNA synthesis | ||
| ActA | ActAF2 | CATCAAGGAGAAGCTGACGTACA | Real-time RT-PCR | 69 |
| ActAR2 | GACGACTCGGCGGCAG | Real-time RT-PCR |
RNA Interference
Oligonucleotide primers were designed to amplify 200- to 250-bp amplicons from CesA1, CesA2, CesA3, and CesA4 (Table 2), and the T7 promotor sequence was added to the 5′ end of both the forward and reverse primer for each gene. Amplicons were designed to be specific to each gene and were not located within the PH domain or within the core of the putative cellulose synthase catalytic domain. BLASTN searches within the Syngenta Phytophthora Consortium EST database and P. infestans genome sequence revealed that the sequences of these amplicons were specific to each gene. The gene encoding gfp from vector p34GFN (Si-Ammour et al., 2003) was used as a nonendogenous negative control as described by Whisson et al. (2005). Test primers for real-time RT-PCR quantification of gene expression after exposure to dsRNA products were also designed. These primers were located outside the amplicon used for dsRNA synthesis. Sufficient PCR reactions were performed to yield 5 μg of PCR product for use in dsRNA synthesis. Synthesis of dsRNA was performed using the Megascript RNA interference kit (Ambion) following the manufacturer's protocol. Protoplasting and transfection were performed as described previously (Whisson et al., 2005). Fifteen days after transfection, agar plates containing single colonies were flooded with 10 mL of sterile distilled water, and zoospores were harvested, encysted, and left to germinate for 1 to 2 h at room temperature as described by Grenville-Briggs et al. (2005). A portion of each sample (2 mL) was reserved for phenotypic analysis of germinating cysts producing appressoria as described by Grenville-Briggs et al. (2005), and the remainder was collected by centrifugation and used for RNA extraction.
Quantitation of Cellulose in the Cell Walls of Silenced and Nonsilenced Lines
Total appressoria corresponding to six nonsilenced lines (showing an average of 6% appressorium-like structures, 64.5% normal appressoria, and 29.5% germinating cysts that had not produced an appressorium) (nonsilenced pool), eight silenced lines showing an extreme phenotype (composed of an average of 47% appressoria-like structures, 34% normal appressoria, and 19% germinated cysts with no appressorium) (extreme pool), and eight silenced lines showing a more moderate phenotype (composed of an average of 34% appressoria-like structures, 45% normal appressoria, and 21% germinated cysts with no appressorium) (moderate pool) were pooled together to form three samples for cellulose content determination.
The amount of cellulose in these three pools was determined by enzymatic hydrolysis to confirm the involvement of the CesA gene family in cellulose biosynthesis. An equal amount of freeze-dried appressoria corresponding to the different pooled lines was washed with 5 mL of water and collected by centrifugation at 4000g for 15 min. The samples were resuspended in 5 mL of 80% methanol containing 5% KOH, and the suspension was heated for 15 min at 100°C. The extraction of alkali-soluble cell wall components was repeated on the insoluble material recovered by centrifugation at 1000g for 5 min. The last pellets containing the cellulose extracted from the appressoria walls were washed three times with 0.5 M acetic acid and water until the suspension reached a final pH of 5. They were finally resuspended in 1.5 mL of water and subjected to the action of a mixture of recombinant cellulases from Thermobifida fusca expressed in Escherichia coli (generous gift from D.B. Wilson, Cornell University). Aliquots (200 μL) corresponding to each pool were transferred to Eppendorf tubes. The total volume of each sample was adjusted to 1 mL with 50 mM sodium acetate, pH 5.0, and a mixture of endocellulases Cel 6A and Cel 9A (2.0 μg/mL each, final concentration), and exocellulase Cel 6B (9.5 μg/mL, final concentration) was added to each tube. Triplicates were run in parallel on each pool. The samples were then incubated for 72 h at 50°C. Aliquots were taken out at 0, 24, 48, and 72 h while the rest of the reaction mixture was simultaneously supplemented with a fresh mixture of cellulases at the same concentrations as indicated above. The amount of (1→3)-β-d-glucan eventually present in the cellulose samples was determined in the same conditions as described for the cellulase digestion by replacing the cellulase mixture by an endo-(1→3)-β-d-glucanase from Aspergillus niger (Megazyme; dilution 1/1000). In the latter case, the samples were incubated at 40°C in 4.5 mM sodium acetate buffer, pH 4.5. Negative controls were performed by replacing the hydrolytic enzymes with water. The specificity of the cellulase mixture and the endo-(1→3)-β-d-glucanase were verified using different sources of cellulose (Avicel; carboxymethylcellulose 4 M from Megazyme) and (1→3)-β-d-glucan (laminarin and curdlan) substrates (see Supplemental Figure 1 online). The amount of reducing sugars released upon enzymatic hydrolysis was measured by the Nelson-Somogyi assay (Nelson, 1944) using a standard curve made with glucose.
Production of Anti-CesA Antibodies
The sequence of a cellulose synthase from Arabidopsis thaliana (At CesA1; GenBank accession number NP_194967) was used to search the Oomycete Genomics Database (http://www.oomycete.org; Gajendran et al., 2006) using the online TBLASTN tool (Altschul et al., 1997). An EST sequence from Saprolegnia parasitica (GenBank accession number DN616186) that exhibited significantly high similarity with a segment of the plant cellulose synthase was used to design the following primers: 5′-CCACGCGTCCGCACGATT-3′, 5′-CCAGTACGGTACCCAGACGGAAGAT-3′, and 5′-GCTGGGTCAACGTAAGCGTTGG-3′. The latter were used for the amplification of the cDNA coding for the putative catalytic subunit of the cellulose synthase from Saprolegnia monoica. The Total RNA extraction kit from Qiagen was used for purifying total RNA from a 3-d-old mycelium culture of S. monoica grown on Machlis medium (Machlis, 1953), following the manufacturer's protocol for plant cells and tissues and filamentous fungi. cDNA amplification was performed using the FirstChoice RLM RACE kit (Ambion). The amino acid sequences deduced from the amplified cDNA fragments were used to design the two following peptides: ANRSVDSNDVWRAQ and GWDSIYFRKDFERR. The latter were synthesized (Fmoc chemistry) and conjugated to keyhole limpet hemocyanin, and a mixture of both conjugates was used for antibody production in rabbits by the company CovalAb using standard protocols. These peptides were similar to peptides found in P. infestans (see Supplemental Figure 2 online), and their specificity toward CesA proteins from S. monoica and P. infestans was shown by protein gel blot analyses of plasma membrane proteins (see Supplemental Figure 3 and Supplemental Methods online).
In Situ Immunolabeling of the P. infestans Cellulose Synthase
Wild-type P. infestans (strain 88069) cysts were induced to produce appressoria on plastic cover slips, overnight at 11°C. Cells were fixed in 4% paraformaldehyde and permeabilized using 0.1% Triton X-100. Incubation with primary anti-CesA antibodies or control anti-STR antibodies (N.R. Horner and P. van West, unpublished data) was performed at 37°C for 1 h. Cells were labeled with goat, anti-rabbit Alexa Fluor 555 for 45 min at room temperature in the dark. Phase contrast and fluorescence microscopy were performed using an Axioplan 2 microscope (Zeiss) with standard rhodamine and UV filter sets. Images were captured using a Hamamatsu CCD camera and analyzed using Openlab 5.0.3 (Improvision).
Effects of the Cellulose Biosynthesis Inhibitor DCB on P. infestans Development and Pathogenicity
To test DCB for inhibitory effects on P. infestans cellulose biosynthesis, 11-d-old sporulating agar cultures were flooded with either a 40 μM DCB solution (prepared in 1.25% [v/v] DMSO), a 1.25% (v/v) DMSO control, or a deionized water control. After 4 h at 4°C, zoospores were aspirated from plates, encysted, transferred to empty Petri dishes, and left to germinate at 11°C. Plates were assessed for germination and appressoria development after 16 to 18 h.
To test whether P. infestans cultures were altered in pathogenicity when induced to develop in the presence of DCB, 11-d-old sporulating agar cultures were treated as described for in vitro samples. After 4 h at 4°C, zoospores produced under the conditions described in the previous paragraph were aspirated from plates and encysted, and the concentration of each sample was adjusted to 7 × 103 zoospores/mL. Zoospores were either inoculated directly onto potato leaves or were first washed twice with sterile water and resuspended in 40 μM DCB. Approximately forty 10-μL droplets (containing 70 zoospores each) were drop inoculated onto the abaxial side of detached leaves of potato cultivar Craigs Royal (compatible interaction). To allow for variation in infection, each sample was inoculated onto 12 separate leaflets. Leaves were also mock-inoculated with water only, a 1.25% (v/v) DMSO solution, and a 40 μM solution of DCB. No signs of disease or other symptoms were observed on mock-inoculated leaves. Leaves were incubated at 20°C for 7 d at 100% relative humidity and were assessed daily for disease development.
Electron Microscopy
Appressoria from P. infestans treated with DCB or from CesA1-4–silenced lines were pelletted and fixed in 2.5% glutaraldehyde and processed conventionally as described by Wastling et al. (1992) through to Epon embedding. Sectioning was performed as described previously (Bishop et al., 1999), and samples were viewed at an accelerated voltage of 80 kV using a Philips CM10 TEM fitted with a Gatan bioscan CCD camera. For scanning electron microscopy, potato leaves were inoculated with zoospores produced in the presence of DCB as described above and were incubated at 11°C for 16 h, after which appressoria are normally formed. Samples were fixed in 2.5% glutaraldhyde and processed conventionally as described by Wastling et al. (1992) through to drying. Dried specimens were attached to aluminum stubs and sputter-coated with gold using an Emitech K550 sputter coater. Coated stubs were examined at 10 kV using a Jeol 35CF scanning electron microscope.
Accession Numbers
Sequence data from this article can be found in the Genbank/EMBL data libraries under accession numbers ABP96902 (Pi CesA1), ABP96903 (Pi CesA2), ABP96904 (Pi CesA3), ABP96905 (Pi CesA4), ABP96906, (Ps CesA1) ABP96907 (Ps CesA2), ABP96908 (Ps CesA3), ABP96909 (Ps CesA4), ABP96910 (Pr CesA1), ABP96911 (Pr CesA2), ABP96912 (Pr CesA3), and APB96913 (Pr CesA4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Specificity of the Hydrolytic Enzymes Used for the Quantitative Analysis of the Cellulose and (1→3)-β-Glucan in the Walls of Wild-Type and Silenced Appressoria.
Supplemental Figure 2. Alignment of the Four CesA Sequences from P. infestans (Pi CesA1-4) with a Segment of the CesA Protein from S. monoica (Sm CesA).
Supplemental Figure 3. Protein Gel Blot Analysis of the Anti-CesA Antibodies.
Supplemental Figure 4. Effects of DCB on Radial Growth, Zoospore Release, and Cyst Germination in P. infestans.
Supplemental Data Set 1. Alignment of 42 CES Family Members for Phylogenetic Analysis.
Supplemental Methods. Isolation of Plasma Membranes, Protein Gel Blot Analysis, Effects of DCB on Radial growth, Zoospore Release, and Cyst Germination.
Supplementary Material
Acknowledgments
We thank Liz Stewart, Ian Davidson, Alistair Mckinnon, Kevin MacKenzie, and Debbie Marshall for technical help. Carol Munro is acknowledged for helpful comments on the manuscript. We thank D.B. Wilson (Cornell University, Ithaca, NY) for the generous gift of the Cel 6A, Cel 9A, and Cel 6B cellulases. Funding for P.v.W., L.J.G.-B., V.L.A., and A.W. was provided by the British Biotechnology and Biological Sciences Research Council, the Royal Society, and the University of Aberdeen. Funding for A.O.A., S.C.W., and P.R.J.B. was provided by the Scottish Executive Environment and Rural Affairs Department. Financial support to V.B. and J.F. was from the Swedish Centre for Biomimetic Fiber Engineering.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Pieter van West (p.vanwest@abdn.ac.uk).
Online version contains Web-only data.
References
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson, J.M., and Lin, C.C. (1978). Hyphal cell wall chemistry of Leptomitus lacteus. Mycologia 70 363–369. [Google Scholar]
- Avrova, A.O., Venter, E., Birch, P.J., and Whisson, S.C. (2003). Profiling and quantifying differential gene transcription in Phytophthora infestans prior to and during the early stages of potato infection. Fungal Genet. Biol. 40 4–14. [DOI] [PubMed] [Google Scholar]
- Bartnicki-Garcia, S. (1968). Cell wall chemistry, morphogenesis and taxonomy of fungi. Annu. Rev. Microbiol. 22 87–108. [DOI] [PubMed] [Google Scholar]
- Bartnicki-Garcia, S., and Wang, M.C. (1983). Biochemical aspects of morphogenesis in Phytophthora. In Phytophthora, Its Biology, Taxonomy, Ecology and Pathology, D.C. Erwin, S. Bartnicki-Garcia, and P.H. Tsao, eds (St. Paul, MN: American Phytopathological Society Press), pp. 121–137.
- Birch, P.R.J., and Whisson, S.C. (2001). Phytophthora infestans enters the genomics era. Mol. Plant Pathol. 2 257–263. [DOI] [PubMed] [Google Scholar]
- Bishop, E.T., Bell, G.T., Bloor, S., Broom, I.J., Hendry, N.F.K., and Wheatley, D.N. (1999). An in vitro model of angiogenesis: Basic features. Angiogenesis 3 335–344. [DOI] [PubMed] [Google Scholar]
- Blanton, R.L., Fuller, D., Iranfar, N., Grimson, M.J., and Loomis, W.F. (2000). The cellulose synthase gene of Dictyostelium. Proc. Natl. Acad. Sci. USA 97 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzenzana, J., Pelosi, L., Briolay, A., Briolay, J., and Bulone, V. (2006). Identification of the first Oomycete annexin as a (1-3)-β-D-glucan synthase activator. Mol. Microbiol. 62 552–565. [DOI] [PubMed] [Google Scholar]
- Brown, R.M., Jr. (1996). The biosynthesis of cellulose. J. Macromol. Sci. 10 1345–1373. [Google Scholar]
- Bulone, V., Chanzy, H., Gay, L., Girard, V., and Fèvre, M. (1992). Characterisation of chitin and chitin synthase from the cellulosic cell wall fungus Saprolegnia monoica. Exp. Mycol. 16 8–21. [Google Scholar]
- Bulone, V., and Fèvre, M. (1996). A 34-kDa polypeptide is associated with the (1→3)-β-glucan synthase activity from the fungus Saprolegnia monoica. FEMS Microbiol. Lett. 140 145–150. [DOI] [PubMed] [Google Scholar]
- Bulone, V., Girard, V., and Fèvre, M. (1990). Separation and partial purification of 1,3-β-glucan and 1,4-β-glucan synthases from Saprolegnia. Plant Physiol. 94 1748–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Takaki, G.M., Dietrich, S.M.C., and Mascarenhas, Y. (1982). Isolation and characterization of chitin from the cell walls of Achlya radiosa. J. Gen. Microbiol. 128 207–209. [Google Scholar]
- Caten, C.E., and Jinks, L. (1968). Spontaneous variability of single isolates of Phytophthora infestans I, cultural variation. Can. J. Bot. 46 329–347. [Google Scholar]
- Charnock, S.J., Henrissat, B., and Davies, G.J. (2001). Three-dimensional structures of UDP-sugar glycosyltransferases illuminate the biosynthesis of plant polysaccharides. Plant Physiol. 125 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer, D.P. (1999). Cellulose biosynthesis: Exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 245–276. [DOI] [PubMed] [Google Scholar]
- Duncan, J.M. (1999). Phytophthora - An abiding threat to our crops. Microbiol. Today 26 114–116. [Google Scholar]
- Erwin, D.C., and Ribeiro, O.K. (1996). Phytophthora Diseases Worldwide. (St. Paul, MN: American Phytopathological Society Press).
- Farkas, V. (1979). Biosynthesis of cell walls of fungi. Microbiol. Rev. 43 117–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajendran, K., Gonzales, M.D., Farmer, A., Archuleta, E., Win, J., Waugh, M.E., and Kamoun, S. (2006). Phytophthora functional genomics database (PFGD): Functional genomics of Phytophthora-plant interactions. Nucleic Acids Res. 34 D465–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulin, E., Jauneau, A., Villaba, F., Rickauer, M., Esquerre-Tugaye, M.-T., and Bottin, A. (2002). The CBEL glycoprotein of Phytophthora parasitica var nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J. Cell Sci. 115 4565–4575. [DOI] [PubMed] [Google Scholar]
- Grenville-Briggs, L.J., Avrova, A., Bruce, C.R., Williams, A., Birch, P.R.J., and van West, P. (2005). Elevated amino acid biosynthesis in Phytophthora infestans during appressorium formation and potato infection. Fungal Genet. Biol. 42 244–256. [DOI] [PubMed] [Google Scholar]
- Grenville-Briggs, L.J., and van West, P. (2005). The biotrophic stages of oomycete-plant interactions. In Advances in Applied Microbiology, Vol. 57, A.I. Laskin, J.W. Bennett, and G.M. Gadd, eds (San Diego, CA: Academic Press), pp. 217–243. [DOI] [PubMed]
- Helbert, W., Sugiyama, J., Ishihara, M., and Yamanaka, S. (1997). Characterization of native crystalline cellulose in the cell walls of oomycota. J. Biotechnol. 57 29–37. [Google Scholar]
- Higgins, D., Thompson, J., Gibson, T., Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998). SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14 378–379. [DOI] [PubMed] [Google Scholar]
- Jarvis, M.C., Threlfall, D.R., and Friend, J. (1981). Potato cell wall polysaccharides: Degradation with enzymes from Phytophthora infestans. J. Exp. Bot. 32 1309–1319. [Google Scholar]
- Kamoun, S. (2003). Molecular genetics of pathogenic oomycetes. Eukaryot. Cell 2 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, S., and Itoh, T. (1995). Evidence for the role of glomerulocyte in cellulose synthesis in the tunicate Metandrocarpa uedia. Protoplasma 186 24–33. [Google Scholar]
- Kramer, R., Freytag, S., and Schmelzer, E. (1997). In vitro formation of infection structures of Phytophthora infestans is associated with synthesis of stage specific polypeptides. Eur. J. Plant Pathol. 103 43–53. [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lemmon, M.A., Ferguson, K.M., and Schlessinger, J. (1996). PH domains: Diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell 85 621–624. [DOI] [PubMed] [Google Scholar]
- Lin, C.C., and Aronson, J.M. (1970). Chitin and cellulose in the cell walls of the Oomycetes Apodachlya. Arch. Mikrobiol. 72 111–114. [DOI] [PubMed] [Google Scholar]
- Liu, H., Kauffmann, S., Becker, J.M., and Szaniszlo, P.J. (2004). Wangiella (Exophiala) dermitidis WdChs5p, a class V chitin synthase, is essential for sustained cell growth at temperature of infection. Eukaryot. Cell 3 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlis, L. (1953). Growth and nutrition of watermolds in the subgenus Euallomyces II - Optimal composition of the minimal medium. Am. J. Bot. 40 449–460. [Google Scholar]
- Madrid, M.P., Di Pietro, A., and Roncero, M.I. (2003). Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defense compounds. Mol. Microbiol. 47 257–266. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., and Bryant, S.H. (2004). CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 32 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaaki, Y., Hajime, M., and Kiichi, K. (1973). Macerating enzyme of Phytophthora. l. Macerating activity of crude enzyme secreted by Phytophthora capsici. Ann. Phytopathological Soc. Jpn. 39 389–395. [Google Scholar]
- Matthysse, A.G., Deschet, K., Williams, M., Marry, M., White, A.R., and Smith, W.C. (2004). A functional cellulose synthase from ascidian epidermis. Proc. Natl. Acad. Sci. USA 101 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, B.J., Ren, R., and Clark, R.L. (1993). A putative modular domain present in diverse signalling molecules. Cell 73 629–630. [DOI] [PubMed] [Google Scholar]
- Mayer, Y., and Herth, W. (1978). Chemical inhibition of cell wall formation and cytokinesis but not of nuclear division in protoplasts of Nicotiana tabacum L. cultured in vitro. Planta 142 253–262. [DOI] [PubMed] [Google Scholar]
- McLeod, A., Smart, C.D., and Fry, W.E. (2003). Characterization of 1,3-β-glucanase and 1,3;1,4-β glucanase genes from Phytophthora infestans. Fungal Genet. Biol. 38 250–263. [DOI] [PubMed] [Google Scholar]
- Mitchell, H.J., and Hardham, A.R. (1999). Characterization of the water expulsion vacuole in Phytophthora nicotianae zoospores. Protoplasma 206 118–130. [Google Scholar]
- Montezinos, D., and Delmer, D.P. (1980). Characterization of inhibitors of cellulose synthesis in cotton fibres. Planta 148 305–311. [DOI] [PubMed] [Google Scholar]
- Munro, C.A., Winter, K., Buchan, A., Henry, K., Becker, J.M., Brown, A.J.P., Bulawa, C.E., and Gow, N.A.R. (2001). Chs1 of Candida albicans is an essential chitin synthase required for synthesis of the septum and cell integrity. Mol. Microbiol. 39 1414–1426. [DOI] [PubMed] [Google Scholar]
- Nelson, N. (1944). A photometric adaptation of the Somogyi method for the determination of glucose. J. Biol. Chem. 153 375–381. [Google Scholar]
- Pelosi, L., Imai, T., Chanzy, H., Heux, L., Buhler, E., and Bulone, V. (2003). Structural and morphological diversity of (1→3)-β-d-glucans synthesized in vitro by enzymes from Saprolegnia monoica. Comparison with a corresponding in vitro product from blackberry (Rubus fruticosus). Biochemistry 42 6264–6274. [DOI] [PubMed] [Google Scholar]
- Phillips, A.J., Anderson, V.L., Robertson, E.J., Secombes, C.J., and van West, P. (2008). New insights into animal pathogenic oomycetes. Trends Microbiol. 16 13–19. [DOI] [PubMed] [Google Scholar]
- Picard, K., Tirilly, Y., and Benhamou, N. (2000). Cytological effects of cellulases in the parasitism of Phytophthora parasitica by Pythium oligandrum. Appl. Environ. Microbiol. 66 4305–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J., DeWit, P.J.G.M., and Govers, F. (1992). Molecular aspects of the potato-Phytophthora infestans interaction. Neth. J. Plant Pathol. 98 85–92. [Google Scholar]
- Randall, T.A., et al. (2005). Large-scale gene discovery in the oomycete Phytophthora infestans reveals likely components of phytopathogenicity shared with true fungi. Mol. Plant Microbe Interact. 18 229–243. [DOI] [PubMed] [Google Scholar]
- Ross, P., Mayer, R., and Benziman, M. (1991). Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55 35–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura, Y., Nakashima, K., Awazu, S., Matsuoka, T., Nakayama, A., Azuma, J., and Satoh, N. (2005). Transposon-mediated insertional mutagenesis revealed the functions of animal cellulose synthase in the ascidian Ciona intestinalis. Proc. Natl. Acad. Sci. USA 102 15134–15139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, I., and Brown, R. (2000). Cellulose synthases and related enzymes. Curr. Opin. Plant Biol. 3 523–531. [DOI] [PubMed] [Google Scholar]
- Saxena, I.M., and Brown, R.M. (1997). Identification of cellulose synthase(s) in higher plants: Sequence analysis of processive β-glycosyl-transferases with the common motif ‘D,D,D35Q(R,Q)XRW.’ Cellulose 4 33–49. [Google Scholar]
- Saxena, I.M., Brown, R.M., Fèvre, M., Geremia, R.A., and Henrissat, B. (1995). Multidomain architecture of β-glycosyl transferases: Implications for mechanism of action. J. Bacteriol. 177 1419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, A., and Mullins, J.T. (2002). Hyphal tip growth in Achlya bisexualis. II. Distribution of cellulose in elongating and non-elongating regions of the wall. Mycologia 94 273–279. [PubMed] [Google Scholar]
- Si-Ammour, A., Mauch-Mani, B., and Mauch, F. (2003). Quantification and induced resistance against Phytophthora species expressing GFP as a vital marker; β-aminobutyric acid but not BTH protects potato and Arabidopsis from infection. Mol. Plant Pathol. 4 237–248. [DOI] [PubMed] [Google Scholar]
- Tyers, M., Rachubinski, R.A., Stewart, M.I., Varrichio, A.M., Shorr, R.G., Haslam, R.J., and Herley, C.B. (1988). Molecular cloning and expression of the major protein kinase C substrate of platelets. Nature 333 470–473. [DOI] [PubMed] [Google Scholar]
- van West, P., Appiah, A.A., and Gow, N.A.R. (2003). Advances in research on root pathogenic oomycetes. Physiol. Mol. Plant Pathol. 62 99–113. [Google Scholar]
- van West, P., and Vleeshouwers, V.G.A.A. (2004). The Phytophthora infestans-host interaction. In Plant Pathogen Interactions. Annual Pathogen Reviews, Vol. 11, N.J. Talbot, ed (Oxford, UK: Blackwell Scientific Publishers), pp. 217–242.
- Walker, C.A., and van West, P. (2007). Zoospore development in the Oomycetes. Fungal Biol. Rev. 21 10–18. [Google Scholar]
- Weber, I., Aßmann, D., Thines, E., and Steinberg, G. (2006). Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wastling, J.M., McKenzie, K., and Chappell, L.H. (1992). Effects of cyclosporin A on the morphology and tegumentary ultrastructure of Hymenolepis microstoma in vivo. Parasitology 104 531–538. [DOI] [PubMed] [Google Scholar]
- Whisson, S.C., Avrova, A.O., van West, P., and Jones, J.T. (2005). A method for dsRNA mediated transient gene silencing in Phytophthora infestans. Mol. Plant Pathol. 6 153–163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.