Abstract
To form the building blocks of isoprenoids, isopentenyl diphosphate (IPP) isomerase activity, which converts IPP to dimethylallyl diphosphate (DMAPP), appears to be necessary in cytosol, plastids, and mitochondria. Arabidopsis thaliana contains only two IPP isomerases (Isopentenyl Diphosphate Isomerase1 [IDI1] and IDI2). Both encode proteins with N-terminal extensions similar to transit peptides and are expressed in all organs, with IDI1 less abundant than IDI2. Examination of enhanced green fluorescent protein fusions established that IDI1 is mainly in the plastid, whereas IDI2 is mainly in the mitochondria. Both proteins are also in the cytosol as a result of their translation from naturally occurring shorter transcripts lacking transit peptides, as demonstrated by 5′ rapid amplification of cDNA ends cloning. IPP isomerase activity in the cytosol was confirmed by uniform labeling of IPP- and DMAPP-derived units of the cytoplasmic isoprenoid product, sitosterol, when labeled mevalonate was administered. Analysis of mutant lines showed that double mutants were nonviable, while homozygous single mutants had no major morphological or chemical differences from the wild type except for flowers with fused sepals and underdeveloped petals on idi2 mutants. Thus, each of the two Arabidopsis IPP isomerases is found in multiple but partially overlapping subcellular locations, and each can compensate for the loss of the other through partial redundancy in the cytosol.
INTRODUCTION
Isoprenoids are essential compounds in all organisms, but their largest structural variety occurs in plants. There, they serve not only universal roles as photosynthetic pigments, redox cofactors, and hormones but also specialized ecological roles as feeding deterrents, pollination vector attractants, and phytoalexins. Animals and fungi produce the precursors for isoprenoid biosynthesis via the mevalonic acid pathway, and many bacteria synthesize the same building blocks through the methylerythritol phosphate (MEP) pathway. Plants, by contrast, possess both pathways, with the mevalonic acid pathway operating in the cytosol (McGarvey and Croteau, 1995) and the MEP pathway operating in plastids (Rodríguez-Concepción and Boronat, 2002). Although proceeding via chemically distinct routes, the two pathways converge at the formation of isopentenyl diphosphate (IPP). Isoprenoid biosynthesis also requires the formation of dimethylallyl diphosphate (DMAPP), a double bond isomer of IPP, as both of these C5 diphosphate compounds are used by prenyltransferases to form the C10, C15, C20, and larger prenyl diphosphates that serve as precursors in the monoterpene, sesquiterpene, and diterpene biosynthetic pathways, respectively. In each reaction series, DMAPP is the initial allylic diphosphate to which successive units of IPP are added via head-to-tail condensations (Burke et al., 1999).
While the plastidic MEP pathway results in the synthesis of both IPP and DMAPP (Hoeffler et al., 2002), the cytosol-localized mevalonate pathway produces only IPP. IPP can be isomerized to DMAPP by isopentenyl diphosphate isomerase (IDI), a divalent metal ion–requiring enzyme found in all living organisms (Gershenzon and Kreis, 1999). IDI can also catalyze the reverse reaction from DMAPP to IPP, but the equilibrium favors the forward reaction. Animals, fungi, and some bacteria contain an enzyme designated type I IPP isomerase (Muehlbacher and Poulter, 1988; Anderson et al., 1989; Carrigan and Poulter, 2003); other bacteria have a nonhomologous type II IPP isomerase (Kaneda et al., 2001). In plants, only DNA sequences encoding proteins homologous with type I IPP isomerase have been identified to date (Blanc and Pichersky, 1995; Campbell et al., 1997; Cunningham and Gantt, 2000; Nakamura et al., 2001; Page et al., 2004).
Based on the pathways of isoprenoid biosynthesis found in plants, the molar ratio of IPP to DMAPP needed to make various terpenoid classes varies from 1:1 for monoterpenes to 2:1 for sesquiterpenes and sterols, 3:1 for diterpenes, carotenoids, and phytol, and much higher for long-chain polyprenols and polyterpenes (Gershenzon and Kreis, 1999) (Figure 1). Thus, by controlling the conversion of IPP to DMAPP, IPP isomerase may have an important role in regulating the biosynthesis of the major types of isoprenoid products formed. In principle, DMAPP for isoprenoid formation in nonplastidic compartments could arise not only from IPP isomerase but also directly from the plastidial MEP pathway via transport from plastids to other sites of isoprenoid biosynthesis. However, since the IPP:DMAPP ratio produced in vitro by 1-hydroxy-2-methylbutenyl 4-diphosphate reductase, the last step in the MEP pathway, is 6:1 (Rohdich et al., 2003; Eisenreich et al., 2004), IPP isomerase activity might still be necessary to optimize the relative amounts of these two C5 intermediates for efficient isoprenoid biosynthesis both in the plastids and elsewhere in the cell.
Figure 1.
Overview of Isoprenoid Biosynthesis by Compartment in a Higher Plant Cell.
Indicated are the IPP and DMAPP requirements for various classes of isoprenoids. AcCoA, acetyl-CoA; FPP, farnesyl diphosphate; GPP, geranyl diphosphate; GGPP, geranylgeranyl diphosphate; G3P, glyceraldehyde 3-phosphate; MEP, methylerythritol phosphate pathway; MVA, mevalonate pathway; OPP, diphosphate moiety. The block arrows denote multiple steps, and the asymmetric arrows reflect the expected equilibrium of the reversible IPP isomerase (IDI) reaction.
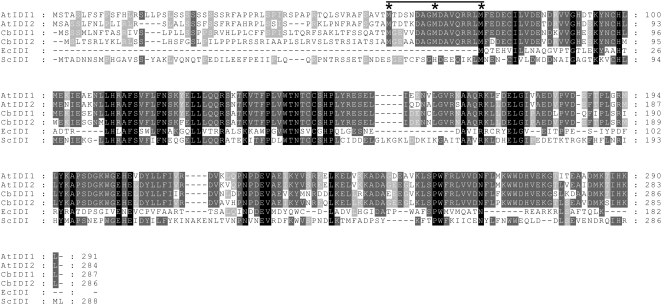
With the distribution of the various branches of isoprenoid biosynthesis in different subcellular locations, the compartmentation of IPP isomerase could have a significant influence on the flux to different end products. Intriguingly, most known plant genes encoding type I IPP isomerases appear to encode proteins with N-terminal transit peptides, suggesting organellar localization (Figure 2). For example, Clarkia breweri IDI1 and IDI2 have N-terminal extensions of 70 and 69 amino acids, respectively, compared with Escherichia coli IDI (Blanc and Pichersky, 1995), and Arabidopsis thaliana has been shown to contain only two IPP isomerase genes (At5g16440, designated IDI1, and At3g02780, designated IDI2), both of which encode a protein with an N-terminal extension (compared with the E. coli IPP isomerase) that has been shown to not be necessary for catalytic activity (Campbell et al., 1997) (Figure 2). An exception was found in tobacco (Nicotiana tabacum), in which two cDNAs, designated IDI1 and IDI2, were obtained, one encoding a protein with an apparent transit peptide, while the other did not. When these proteins were expressed as GFP fusions in this plant, the tobacco IDI1 protein was found to be targeted to the plastid, while the IDI2 protein was found in the cytosol (Nakamura et al., 2001). However, it was not conclusively demonstrated that the tobacco IDI2 cDNA was in fact a copy of a full-length transcript (e.g., by comparison with a genomic sequence or by primer extension experiments).
Figure 2.
Amino Acid Sequence Alignment of Plant, Yeast, and E. coli Type I IPP Isomerase Proteins.
At, Arabidopsis thaliana IDI1 (At5g16440) and IDI2 (At3g02780); Cb, Clarkia breweri IPP isomerase 1 (X82627) and IPP isomerase 2 (U48963); Ec, Escherichia coli IPP isomerase (NP_417365); Sc, Saccharomyces cerevisiae IPP isomerase (P15496). Residues in black are absolutely conserved in all sequences shown, while those in dark gray and light gray are conserved among 70 and 50%, respectively. Alignments were performed using Blosum62 (AlignX, VectorNTI 10) with a gap-opening penalty of 10 and a gap-extension penalty of 0.1. Asterisks indicate Met residues conserved in nearly all known plant species. The horizontal bar represents the conserved 16-residue region of the N-terminal extension (relative to E. coli IDI).
The presence of N-terminal extensions on the IPP isomerases found in Arabidopsis and other species suggests that they might be directed to plastids or mitochondria, both sites of isoprenoid formation in plants. Plastids, in addition to the MEP pathway, also contain biosynthetic machinery for monoterpene, diterpene, gibberellin, carotenoid, and phytol production. Mitochondria, on the other hand, are sites of ubiquinone biosynthesis (Lütke-Brinkhaus et al., 1984; Ohara et al., 2006) and form other isoprenoids in organisms outside the plant kingdom (Felkai et al., 1999; Marbois et al., 2005). Curiously, in both C. breweri and Arabidopsis, none of the IDI proteins appear to be targeted to the cytosol (since they all appear to have transit peptides), even though this compartment produces sesquiterpenes, sterols, and dolichols and the cytosolic mevalonate pathway supplies only IPP and not DMAPP. Conceivably, alternative splicing or alternative transcription start sites of the IDI genes could lead to cytosolic protein targeting, as is known for another Arabidopsis protein involved in isoprenoid biosynthesis, farnesyl diphosphate synthase1, which can be located in both the cytosol and the mitochondria (Cunillera et al., 1997). Thus, it is not clear at present which plant compartments possess IPP isomerase activity and whether such activity is essential for the synthesis of isoprenoid compounds in that compartment.
Here, we present evidence that in Arabidopsis, one IPP isomerase (IDI1) is targeted to plastids, the other (IDI2) is targeted to mitochondria, and additional IPP isomerase activity encoded by both genes is present in the cytoplasm. While even one functional IDI gene is sufficient to support growth and development, we demonstrate that IDI1 and IDI2 are not completely redundant and play distinct roles in isoprenoid metabolism.
RESULTS
Transcripts of Arabidopsis IDI1 and IDI2 Encode Proteins with an N-Terminal Extension
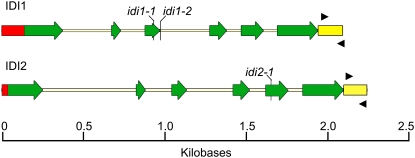
The present annotation in The Arabidopsis Information Resource (TAIR; http://www.plantgdb.org/AtGDB/) shows that the IDI1 gene contains six exons and five introns, as does IDI2 (Figure 3). These two genes encode proteins of 291 and 284 amino acids, respectively, which contain N-terminal extensions of 75 and 68 residues, respectively, compared with the sequence of E. coli IDI (Figure 2). This extension, present in most plant IDI sequences (Cunningham and Gantt, 2000), is quite variable, with the exception of the 16 residues just upstream of the Met residue in the equivalent position to the starting Met in the E. coli sequence (Figure 2). This 16-residue sequence starts with a Met that is conserved in almost all known plant IDI sequences and contains a second conserved Met residue halfway through (Cunningham and Gantt, 2000) (Figure 2). Interestingly, yeast IDI also contains a large N-terminal extension relative to E. coli IDI, but this sequence as a whole shows little similarity to the corresponding plant sequences, with the exception of the above-mentioned 16-residue region. The yeast sequence does not contain any Met residues in this region (Figure 2), but since some enzymes of the mevalonate pathway have been associated with mitochondria in yeast (Shimizu et al., 1971), it may indeed serve some noncatalytic function. When the N-terminal extension upstream of this 16-residue region, which is also highly variable between At IDI1 and At IDI2, is discounted, At IDI1 and At IDI2 are 91% identical to each other, 84 and 83% identical, respectively, to C. breweri IDI1, 45 and 46% identical, respectively, to yeast IDI, and 27 and 26% identical, respectively, to E. coli IDI. Interestingly, the first exon of At IDI1 and At IDI2 (236 and 215 nucleotides, respectively, starting from the initiating ATG codon) encodes the entire N-terminal part of the protein that has no equivalent in the E. coli IDI protein plus a few additional amino acids.
Figure 3.
Genomic Structures of Arabidopsis IDI1 (At5g16440) and IDI2 (At3g02780).
Green arrows indicate exons, while the thin yellow lines in between indicate introns. The red boxes show the 5′ untranslated regions. The 3′ untranslated regions are shown as yellow boxes. qRT-PCR forward and reverse primers are represented as black arrowheads. The insertion sites of the T-DNA mutants used in this study are indicated with black vertical lines. They are idi1-1 (GABI insertion line 217HO4, exon 3, 799 nucleotides downstream of the predicted start codon), idi1-2 (SALK insertion line 006330, intron 3, 841 nucleotides downstream of the predicted start codon), and idi2-1 (SALK insertion line 014625, exon 4, 1651 nucleotides downstream of the predicted start codon).
The prediction of N-terminal extensions for both IDI1 and IDI2, as outlined above, is consistent with other available sequence information for these genes. For example, the starting codon of IDI1 is likely to be as indicated in Figure 2 due to the presence of an in-frame stop codon in the 5′ untranslated region (UTR) of IDI1 at position −9 relative to the predicted initiating ATG in several available cDNAs. For IDI2, available cDNAs include a maximum of 26 nucleotides of 5′ UTR sequence upstream of the predicted ATG initiating codon, and this region does not include any additional start codons or stop codons in-frame with the proposed start codon. However, an in-frame stop codon is present at −57 with no additional start codons in between, making it extremely likely that the proposed start codon for IDI2 is also the correct one. Examination of 36 cDNAs for IDI1 and 41 cDNAs for IDI2 in the databases also supported the identification of all introns as annotated in TAIR (except when the cDNAs were incomplete). The sequences of RT-PCR products that we obtained for IDI1 and IDI2 using oligonucleotides based on the beginning and end of the open reading frames as annotated in TAIR (idi1-attB1 and idi1-attB2 [for IDI1] or idi2-attB1 and idi2-attB2 [for IDI2] [see Supplemental Table 1 online]) also confirmed the intron–exon demarcation.
In order to gain insight into the possible subcellular localizations of the two IDI enzymes as directed by the N-terminal extensions, the full-length deduced amino acid sequences for the two proteins were used to query several public servers, including ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) (Emanuelsson et al., 1999), TargetP (http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al., 2000), Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html), MitoProt (http://mips.gsf.de/cgi-bin/proj/medgen/mitofilter) (Claros and Vincens, 1996), and PSORT (http://wolfpsort.org/) (Nakai and Horton, 1999). Most algorithms suggested plastid localization with a high level of certainty for IDI1, although MitoProt also suggested mitochondrial localization for this protein. There was less agreement regarding IDI2, with targeting predictions roughly split between the mitochondria and plastid. Interestingly, ChloroP predicted plastid localization for both proteins, while MitoProt predicted mitochondrial localization for both.
IDI1 Is Directed to the Plastid and IDI2 Is Directed to the Mitochondrion
To determine the subcellular localization of both IDI proteins, we transformed Arabidopsis with constructs in which the genomic sequence for either IDI1 or IDI2 had been fused to enhanced green fluorescent protein (eGFP) under the control of the cauliflower mosaic virus 35S promoter and visualized targeting by confocal and fluorescence microscopy. Two other transformed lines were used as controls. First, we cloned the eGFP gene into the same eGFP-ready Gateway plant transformation vector (pB7FWG2) (Karimi et al., 2002), producing an eGFP dimer lacking any transit peptide, which served as a control for cytosolic localization. Second, we used Arabidopsis plants transformed with a signal peptide fused to eGFP, for which plastid localization of the eGFP fusion protein had already been confirmed (Marques et al., 2004).
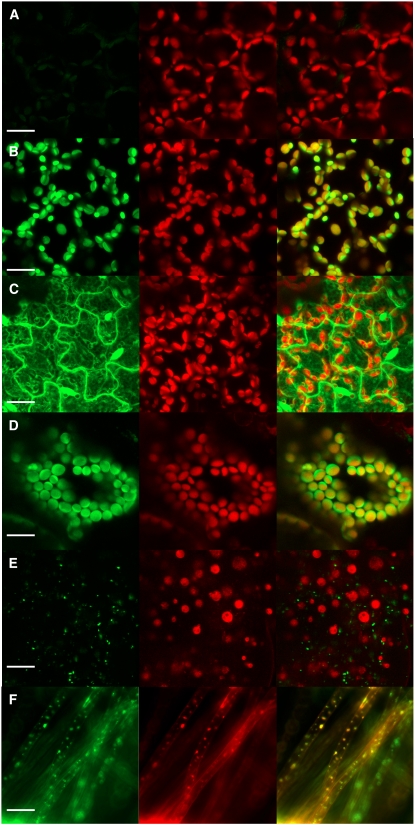
Comparisons of confocal micrographs of the plastid and cytosolic controls (Figures 4B and 4C, respectively) and nontransformed controls (Figure 4A) viewed under similar instrument settings indicated that virtually no green fluorescence was detectable in nontransformed controls, whereas red autofluorescence derived from chlorophyll was readily visible (Figure 4A). In the plastid control, intense green fluorescence colocalized with chloroplast autofluorescence (Figure 4B). However, in the cytosolic localization control, green fluorescence was visible in the cytosol, the nucleus, and at the cell wall but not in the red autofluorescent plastids (Figure 4C). Thus, under these experimental conditions, plastid targeting could readily be distinguished from cytosolic localization.
Figure 4.
Subcellular Localization of IDI-eGFP Fusion Proteins in Transformed or Control Arabidopsis Leaf Tissue.
The first column shows green fluorescence from eGFP, the second shows red autofluorescence from chlorophyll, and the third shows an overlay of the two channels. All confocal images were scanned using similar laser gain and offset settings. Confocal images are as follows: nontransformed Arabidopsis (A), plastid localization–positive control ferredoxin NADP(H) oxidoreductase (Marques et al., 2004) (B), cytosolic eGFP expression (empty vector control) (C), genomic IDI1-eGFP fusion (D), and genomic IDI2-eGFP fusion (E). (F) shows an epifluorescence micrograph of MitoTracker Orange–stained root hairs of Arabidopsis stably transformed with the IDI2-eGFP construct. eGFP fluorescence is shown in the left column, MitoTracker Orange fluorescence in the central column, and the overlay of the two in the right column. Bars = 20 μm.
Plant lines transformed with IDI1 were visualized in a similar way and revealed eGFP fluorescence that colocalized with plastids in roots, stems, and leaves (Figure 4D; localization in stems and roots not shown). However, plants transformed with IDI2 exhibited green fluorescence in discrete subcellular organelles that did not colocalize with plastid autofluorescence (Figure 4E). When seedling roots from these same transformed lines were stained with a mitochondria-specific orange fluorescent dye, eGFP fluorescence was observed to colocalize with the MitoTracker Orange–derived fluorescence (Figure 4F), confirming targeting to mitochondria.
The same transformation vectors containing the cDNAs instead of the full-length genomic sequences were also tested in this way. Cloning and sequencing of the transcripts using transgene-specific primers showed that plants transformed with the genomic sequence and cDNAs produced identical transcripts. Although the cDNAs were found to have generally the same localization patterns as the genomic sequences, some expression of the IDI1 cDNA was occasionally detected in both the cytosol and plastid simultaneously (Figure 5A). A third set of eGFP fusion constructs for IDI1 and IDI2 bearing the native promoter was analyzed, but expression levels were too low to visualize eGFP fluorescence.
Figure 5.
Confocal Micrographs Showing the Simultaneous Plastid and Cytosolic Localization of IDI1c-eGFP in the Stem of a Wild-Type Plant and of the Truncated cytIDI-eGFP in the Cytoplasm of Leaf Tissue in the idi2-1 Mutant Background.
(A) IDI1c-eGFP expression in the stem (wild type).
(B) cytIDI-eGFP expression in leaf (idi2-1).
The first column shows green fluorescence from eGFP, the second shows red autofluorescence from chlorophyll, and the third shows an overlay of the two channels. All confocal images were scanned using similar laser gain and offset settings. Cytosolic expression of IDI did not restore the fused-sepal phenotype of the mutant plant but made possible the isolation of plants homozygous for the idi2 allele and heterozygous for the idi1 allele. Confocal conditions were the same as those described for Figure 4. Bars = 20 μm.
At Least One IPP Isomerase Gene Is Required for Arabidopsis Viability, and IDI2 Is Necessary for Normal Flower Development
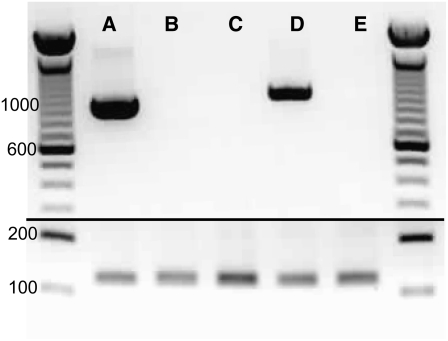
Lines of Arabidopsis with mutations at each IPP isomerase locus were investigated to learn more about the roles of these genes in planta. Using publicly available T-DNA insertion line collections, we obtained two lines with insertions in the IDI1 gene (idi1-1, GABI 217_ H04, and idi1-2, SALK_006330, N506330) and one line with an insertion in the IDI2 gene (idi2-1, SALK_014625, N514625). The insertion in idi1-1 is in exon 3, the insertion in idi1-2 is after the fourth nucleotide in intron 3, and the insertion in idi2-1 is in exon 5 (Figure 3). RT-PCR revealed that none of these mutant lines produced a functional full-length transcript from the affected gene (Figure 6). All single idi mutants (Figure 3) were confirmed as homozygous for their respective insertions in the T3 generation by DNA gel blot analysis, T-DNA–specific PCR, and left border sequencing of the resulting PCR products. The segregation of T-DNA insertions and associated herbicide resistance markers was Mendelian in all cases.
Figure 6.
RT-PCR Analysis of Wild-Type and idi Mutant Lines.
Total RNA was converted to cDNA using a poly(dT) primer and then used in a PCR either with primers that amplify the full-length transcript of IDI1 (columns A to C) or IDI2 (columns D and E) (top panel) or a normalizer gene (RP2LS; bottom panel) to confirm the conversion of RNA into cDNA template. cDNA templates are as follows: wild type (A), idi1-1 (B), idi1-2 (C), wild type (D), and idi2-1 (E). IDI1 was amplified with primers flanking the start and stop codons and is predicted to produce a product of 930 bp, whereas IDI2 was amplified with primers that included a small portion of the 3′ UTR and is thus predicted to amplify a slightly larger product of 988 bp.
Plants homozygous for idi1-1 or idi1-2 alleles showed no obvious phenotype. However, plants homozygous for the idi2-1 allele formed flowers in which the sepals were fused and the petals were highly underdeveloped (Figure 7A). The pollen from these mutants was viable, and backcrossing of this mutant for three generations to ecotype Columbia (Col-0) wild-type plants followed by self-fertilization confirmed that the phenotype was linked to the idi2-1 mutant allele.
Figure 7.
Phenotype of idi2-1 Mutant Flowers and Genetic Complementation.
Flowers from the idi2-1 mutant ([A], left) demonstrate a fused-sepal phenotype with highly underdeveloped petals but produce fertile pollen and can self-fertilize. A Col-0 wild-type flower ([A], right) is shown for comparison. Two homozygous idi2 lines (out of 36) expressing cytosolic IDI showed partial reversion to the wild-type flower morphology (B). The black arrow in (B) indicates a mutant flower, whereas the white arrows show wild-type flowers. The fused-sepal phenotype of the idi2-1 mutant was not affected by transformation with the IDI1 cDNA (C), while this phenotype was fully restored to that of the wild type by transformation of this mutant with a cDNA encoding IDI2 (D). Shown are three independent lines transformed with the IDI1 cDNA (C) and three lines transformed with the IDI2 cDNA (D); a Col-0 wild-type flower is pictured at left in (D).
We were unsuccessful in obtaining double homozygous mutants by crossing either idi1 mutant with idi2-1. Following screening of >400 progeny from each of these crosses (>800 plants total), no double homozygous plant was observed (1 in 16 was expected). In addition, we were unable to detect any plants that were homozygous at one locus and heterozygous at another. The expected frequency of this genotype, assuming strictly Mendelian segregation, would be one in four plants.
In an attempt to rescue the double homozygous mutant containing both the idi1-1 and idi2-1 alleles, we transformed double heterozygous lines with a truncated version of the IDI1 cDNA lacking the putative signal peptide but fused to eGFP in an effort to provide IPP isomerase activity to the cytosol (see kinetic characterization for a description of the active, truncated forms of IDI1 and IDI2). The retention of the truncated fusion protein (cytIDI-eGFP) in the cytoplasm was confirmed by confocal microscopy (Figure 5B). Transformed plants were screened by PCR for the presence of the idi1-1 or idi2-1 allele. We determined the genotypes of 40 plants by PCR and further monitored the expression of the transgene in these plants by measuring GFP fluorescence in crude protein extracts of seedlings (data not shown). Based on this screening strategy, we selected six independent lines from the T0 generation that were heterozygous for both the idi1-1 and idi2-1 mutant alleles and had high levels of expression of the truncated IDI1 (as determined by GFP fluorescence) and allowed them to self-fertilize. Screening of 240 individuals in the T1 generation again failed to yield a double homozygous mutant. However, we were able to find plants that were heterozygous at the IDI1 locus and homozygous for idi2-1. Of 240 plants, 36 had such a genotype. In nearly every case, these plants displayed the same flower morphology defect as the idi2-1 single mutant, although two lines (cytIDI-11 and cytIDI-14) displayed partial restoration to the wild type (Figure 7B). No plants homozygous for the idi1-1 mutant allele and heterozygous at the IDI2 locus were isolated in this screen.
Transformation of the idi2-1 Mutant with the Full-Length IDI2 cDNA but Not the Full-Length IDI1 cDNA Restores the Wild-Type Floral Phenotype
Since the fused sepal phenotype was observed to segregate with the idi2-1 allele, we attempted to restore the wild-type phenotype in this mutant by transformation with vectors encoding either the IDI1 or IDI2 full-length cDNA. T1 glufosinate ammonium–resistant plants were allowed to self-fertilize, and seeds were collected from individual transformed lines. T2 plants were grown on sterile Murashige and Skoog (MS)–glufosinate ammonium plates, and transformed lines with apparent single inserts (as judged by a 3:1 resistant-to-susceptible ratio) were transferred to soil and allowed to flower under long-day conditions. Plants transformed with the IDI1 cDNA retained the mutant phenotype (Figure 7C), while the flowers of those transformed with the IDI2 cDNA were completely restored to the wild type (Figure 7D). The subcellular localization of IDI2 in the rescued mutant background was confirmed with confocal microscopy and determined to be identical to that shown in Figure 4E.
Loss of Either IDI1 or IDI2 Individually Has a Minimal Effect on Isoprenoid Content in Leaf Tissue or in Flowers
The single homozygous mutants were subjected to chemical analyses to quantify the major isoprenoid metabolites produced in the chloroplasts and mitochondria. Chlorophyll and carotenoids are probably the most abundant isoprenoid products of the chloroplasts, while ubiquinone is the major isoprenoid produced in mitochondria. Given the role of carotenoids as photoprotectants of the photosynthetic apparatus, we anticipated that if either IDI protein played a limiting role in carotenoid production, their absence would be most clearly noted under high light conditions. Therefore, we compared carotenoid and chlorophyll levels in acetone extracts of leaves from wild-type and mutant plants grown under high light levels (600 μmol·m−2·s−1) as measured by HPLC. A simple ion-trap liquid chromatography–mass spectrometry (LC-MS) method was implemented to selectively detect ubiquinone levels in the same extracts. We detected no differences between wild-type plants and idi1-1 and idi1-2 mutant lines for any of the isoprenoids analyzed, except that a slight decrease (20%) in both the carotenoids and chlorophylls was noted for idi2-1 (Figure 8). A sample of the plants used for HPLC analysis was also used to again confirm the absence of functional transcripts in the mutant lines.
Figure 8.
Carotenoid, Chlorophyll, and Ubiquinone Contents of Wild-Type and idi Mutant Leaf Tissue Exposed to Continuous High Light (600 μmol·m−2·s−1).
Frozen, ground leaf tissue (500 mg) from six pooled individual plants was extracted with 2.5 mL of acetone:water (9:1) and analyzed by HPLC (for carotenoids and chlorophyll) or ion-trap LC-MS (for ubiquinone). Values shown are averages ± sd of eight replicates. Quantification was based on external standard curves. Ubiquinone-9 levels are shown on the right vertical axis.
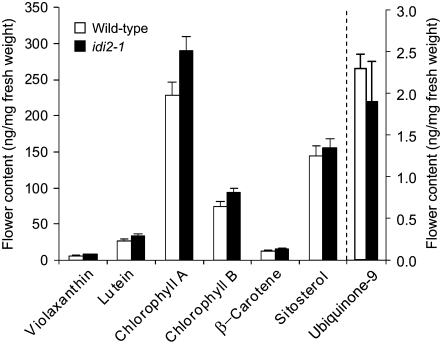
To examine whether the idi2-1 mutant flower phenotype was due to a deficiency in a specific class of essential isoprenoids, the above chemical analysis was repeated using wild-type and idi2-1 flowers grown under normal conditions. No significant differences were noted between the levels of carotenoids, chlorophylls, phytosterols, or ubiquinone in flowers from the idi2-1 mutant versus those of wild-type flowers (Figure 9).
Figure 9.
Analysis of Isoprenoid Levels in idi2-1 Mutant Flowers.
Carotenoid and chlorophyll levels were determined by HPLC analysis (monitoring at 445 and 650 nm). An aliquot of the same sample was used for the analysis of ubiquinone-9 levels by ion-trap LC-MS as described in Methods. Quantification was based on external standard curves. Quantification of sitosterol was based on the relative peak area of stigmastanol added to flower tissue prior to extraction as an internal standard (10 μg), while identification was based on comparison with a derivatized authentic sitosterol standard. Error bars indicate sd from seven replicates. Ubiquinone-9 levels are shown on the right vertical axis.
IDI2 Transcripts Are More Abundant Than Those of IDI1 Overall, but Both Genes Have Similar Organ-Specific Expression Profiles
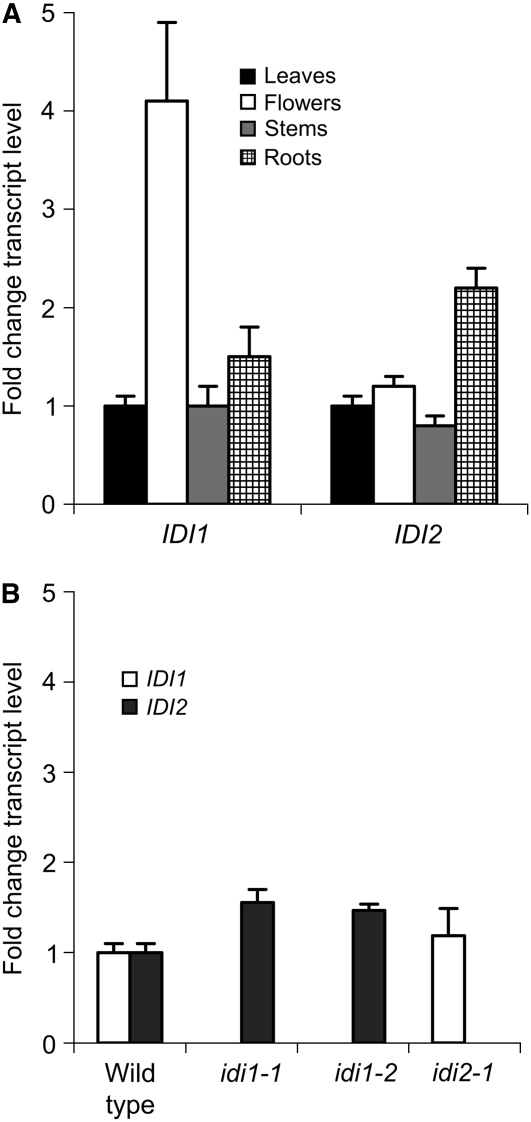
The organ-specific expression of IDI1 and IDI2 was monitored by quantitative real-time PCR (qRT-PCR) using primers designed to recognize a portion of the 3′ UTR of each gene. Steady state transcript levels of each gene in four organs (roots, stems, leaves, and flowers) were measured and normalized to those of leaves. Within a given organ, overall expression between IDI1 and IDI2 was also compared based on the Ct values of the common normalizer gene. Qualitatively, expression of both genes was detectable in all organs surveyed. Quantitatively, expression of IDI1 was 4.1-fold higher in flowers than in leaves, 1.5-fold higher in roots than leaves, and about the same in stems as in leaves (Figure 10A). Expression of IDI2 was about the same in flowers and stems as in leaves but ∼2.5-fold higher in roots, in agreement with Campbell et al. (1997) (Figure 10A). When the expression of the two IDI genes was compared within a given organ, IDI2 was always more abundant, with IDI2 transcripts outnumbering IDI1 by ∼10-fold in leaves, 22-fold in roots, 4.9-fold in flowers, and 4.6-fold in stems.
Figure 10.
Steady State Transcript Levels of IDI1 and IDI2 in Arabidopsis as Measured by qRT-PCR.
Relative quantification was performed according to the Pfaffl method (Pfaffl, 2001). Error bars indicate sd.
(A) Organ-specific expression. Fold change values compare expression levels of each gene with that in the leaf. When IDI1 and IDI2 were compared in the same organ, IDI2 was found to be more abundant in all organs surveyed (described in text). cDNA template loading was normalized to the Ct value of the Arabidopsis RP2LS gene (At4g35800).
(B) Expression of the remaining IDI gene in idi mutant lines. Total RNA was extracted from whole seedlings, converted to cDNA, and used in relative quantification experiments with wild-type seedling extracts.
The steady state transcript level of the remaining functional IDI gene in each mutant line was similarly measured. In this case, whole mutant seedlings were used and compared with samples obtained from wild-type seedlings. No mutant showed a significant increase in the remaining IDI gene relative to the wild type (≤1.5 fold increase; Figure 10B).
The IDI1 and IDI2 Proteins Have Similar Kinetic Properties
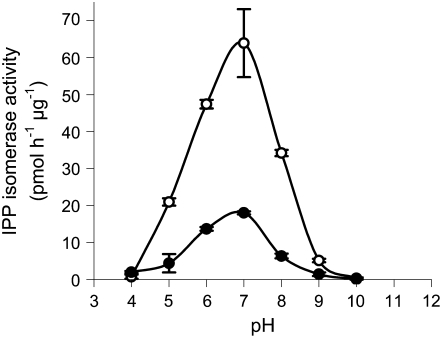
To look for further differences in the roles of the two genes, we compared the catalytic properties of the encoded proteins expressed in vitro. It was previously shown that the N-terminal extensions of IDI1 and IDI2 were not required for catalytic activity (Campbell et al., 1997). Therefore, we expressed both proteins in truncated form based on their alignment to the start codon of E. coli IDI (Figure 2). The purification of the recombinant proteins was aided by expression with an N-terminal His tag. However, due to the catalytic role of divalent metal ions in the IPP isomerase reaction (Carrigan and Poulter, 2003), the presence of the His tag posed problems for accurately measuring the affinity of each enzyme for IPP. Therefore, after introducing a thrombin cleavage site, the His tags were removed by thrombolytic cleavage following purification on a charged Ni affinity column. The Km values of IDI1 and IDI2 for IPP were determined to be 20 ± 3.8 and 16 ± 2.6 μM, respectively, while their kcat values were 2.24 ± 0.1 and 0.89 ± 0.04 min−1, consistent with previous literature reports for this class of IDI enzymes (Muehlbacher and Poulter, 1988; Anderson et al., 1989; Street and Poulter, 1990; Carrigan and Poulter, 2003). A pH optimum curve of each protein revealed that both had a similar preference for a neutral pH (Figure 11).
Figure 11.
pH Optima of the Two Mature IDI Proteins from Arabidopsis.
Activity is based on the conversion of [1-14C]IPP to its acid-labile isomer, DMAPP, and scintillation counting of an ether extract of the reaction following acid hydrolysis. Error bars indicate sd of four replicates at each pH. Linear conditions were established previously by assaying a range of enzyme concentrations and incubation times. IDI1 and IDI2 are represented by open and closed circles, respectively.
Incorporation of [2-13C]Mevalonate into Sitosterol Demonstrates IPP Isomerase Activity in the Cytosol
Except for the floral phenotype of the idi2 mutant, there were no significant morphological or chemical alterations in the individual idi mutants, making it hard to draw any conclusions about the specific roles of these genes in isoprenoid metabolism. The negative impact of a single idi mutation may be ameliorated by DMAPP supplied from the isomerase encoded by the other IDI gene. To investigate this possibility, we compared the biosynthesis of sitosterol, the most abundant plant sterol, in the wild type versus all three idi mutant lines. Sitosterol is known to be biosynthesized in the cytosol. Thus, to assess the source of the IPP and DMAPP for sitosterol biosynthesis, we labeled the cytosolic mevalonate pathway by supplying [2-13C]mevalonate to plants in a MS growth medium while simultaneously administering mevastatin, an inhibitor of an essential step in the mevalonate pathway, HMG-CoA reductase, to block the endogenous formation of unlabeled mevalonate (Rodríguez-Concepción et al., 2004). The biosynthesis of a sitosterol molecule requires four IPP units and two DMAPP units (see Supplemental Figure 1 online). If there were a cytosolic IPP isomerase, all IPP and DMAPP units should be labeled by [2-13C]mevalonate, resulting in an incorporation of five 13C atoms (one 13C atom is lost during demethylation of the cycloartenol intermediate) (Schaeffer et al., 2001). However, if there is no IDI activity in the cytosol, only the IPP units would be labeled directly from [2-13C]mevalonate, resulting in the incorporation of four 13C atoms. The DMAPP would have to come either from IPP generated in the cytosol and isomerized to DMAPP in the plastids or mitochondria or from IPP generated in the plastid that was isomerized there. In either case, the incorporation of label would be expected to be lower than for a cytosol-generated DMAPP unit.
We detected high levels of [2-13C]mevalonate incorporation into sitosterol in these experiments using GC-MS and 13C NMR. The mass spectra for the trisilyl-sitosterol derivatives from the wild type and idi mutant lines were identical (Figure 12). In extracts from labeled plants (Figure 12B), a [M + 5]+· molecular peak dominated the molecular ion cluster in the wild type and in all three mutant lines, indicating the existence of a cytosolic IPP isomerase activity in all of these plants. When spectra from plants supplemented with [2-13C]mevalonate were compared with those derived from unlabeled plants (Figures 12B versus 12A), many differences in the fragmentation patterns were evident. Aside from the dominant mass ions (m/z 491 versus 486), sitosterol fragments from labeled plants bearing one (m/z 130 versus 129), two (m/z 161 versus 159), three (m/z 258 versus 255), four (m/z 361 versus 357), and five (m/z 401 vs. 396) 13C labels were detected, corresponding to sitosterol fragments containing one to five 13C-labeled isoprene units. The strongest mass spectral evidence for a cytosolic isomerase comes from the detection of 13C labeling in the aliphatic side chain of the sitosterol skeleton, most of whose carbon atoms originate from DMAPP. Specific fragments corresponding to this side chain showed a significant enrichment of 1 mass unit (m/z 44 versus 43 and m/z 86 versus 85) in all samples supplemented with [2-13C]mevalonate (Figure 12B) but not in the unlabeled controls (Figure 12A), consistent with 13C labeling of the C26 position of the aliphatic chain of sitosterol via a DMAPP unit derived from [2-13C]mevalonic acid (see Supplemental Figure 1 online). The absolute levels of sitosterol in the plants used in these experiments were similar in mutant and wild-type lines grown on MS medium alone (132 to 141 ng/mg fresh weight, based on comparison with the internal stigmastanol standard). Meanwhile, plants grown in the presence of [2-13C]mevalonate and mevastatin displayed a decrease of ∼30% in the levels of sitosterol (99 to 109 ng/mg fresh weight).
Figure 12.
Sitosterol Mass Spectra from Arabidopsis Plants Grown in the Absence or Presence of [2-13C]Mevalonate and Mevastatin.
(A) Plants grown on MS medium.
(B) Plants grown on MS medium containing [2-13C]mevalonate mevastatin.
In each case, no difference was detected between spectra from wild-type or idi mutant plants (spectra from a wild-type plant are shown; see also Supplemental Figure 2 online). The inset in (A) shows the mass spectrum of authentic sitosterol standard; the inset in (B) shows the incorporation of [2-13C]mevalonate into sitosterol in wild-type and idi lines grown in the presence of mevastatin, as determined by the relative proportions of isotopomers in the mass ion cluster representing [M]+ (m/z 486) through [M+5]+ (m/z 491) species after subtraction of the natural abundance of 13C in the control samples. Error bars indicate sd of four replicates. Sterols were extracted from 2-week-old seedlings with methanol:CH2Cl2 (2:1, v/v) using stigmastanol as an internal standard, derivatized with N-methyl-N- trimethylsilyltrifluoroacetamide, and analyzed by GC-MS.
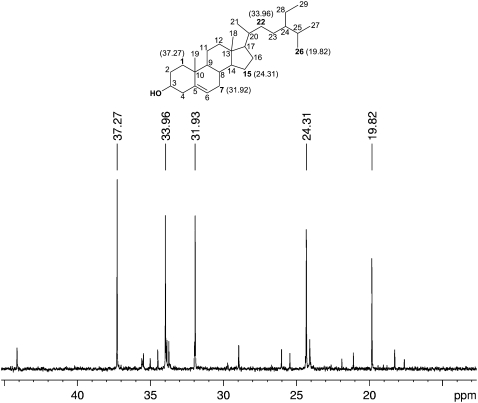
To confirm that the labeling of sitosterol mass spectral fragments was due to DMAPP originating from an isomerase in the cytosol, labeled sitosterol from wild-type and mutant plants was purified by preparative HPLC and then measured by 13C NMR. The incorporation of [2-13C]mevalonic acid would be expected to label five specific carbon atoms in the sitosterol skeleton (see Supplemental Figure 1 online). For sitosterol isolated from both mutant and wild-type plants, each of these five atoms was indeed found to be enriched (Figure 13). Most notable was the strong enrichment of a signal at 19.82 ppm corresponding to C26, a carbon atom in the side chain derived from DMAPP. This furnished strong evidence that the DMAPP used in the formation of sterols is derived from the cytosolic mevalonate pathway via a cytosolic IPP isomerase.
Figure 13.
Upfield Portion of the 13C NMR Spectra of Purified Sitosterol Extracted from Wild-Type and idi Mutant Arabidopsis Seedlings Cultivated in the Presence of [2-13C]Mevalonate and Mevastatin.
Sitosterol from wild-type and mutant plants gave virtually identical spectra (see Supplemental Figure 3 online). The wild-type spectrum is shown above. These signals were identified by previously published chemical shift values for sitosterol (Goad and Akihisa, 1997). These allowed unambiguous assignment of all carbon atoms in the molecule. Labeling was found to occur at the predicted positions based on the scheme for sitosterol biosynthesis from IPP and DMAPP shown in Supplemental Figure 1 online. The peaks at 24.31 (C15), 31.93 (C7), 33.96 (C22), and 37.27 (C1) ppm were enriched by labeling via IPP. The peak at 19.82 ppm (C26) was enriched by labeling via DMAPP. Since the extent of IPP and DMAPP labeling of sitosterol, a cytosolic isoprenoid product, is comparable, both substrates must come from the cytosol, indicating the presence of a cytosolic IPP isomerase. The structure and carbon atom numbering of sitosterol are shown above the spectrum, with enriched carbons shown in boldface; numbers in parentheses indicate the reported chemical shifts in parts per million.
Shortened Transcripts of Both IDI1 and IDI2 Are Expressed That Lack the N-Terminal Transit Peptide for Organellar Targeting
The presence of the IDI protein in the cytosol might be due to the translation of a shorter transcript lacking the N-terminal signaling peptide. To determine whether such alternative transcription occurred at a significant rate, 5′ rapid amplification of cDNA ends (RACE) was employed to assess the lengths of the IDI1 and IDI2 transcripts. Using reverse primers for each gene optimized for 5′ RACE (see Supplemental Table 1 online), we found two distinct size classes of 5′ RACE products for IDI1 and IDI2. Because the reverse primers were designed to be complementary to exon 2 (IDI1) and exon 4 (IDI2), we were able to rule out genomic DNA contamination as a possible source for these clones. Sequence analysis of these fragments indicated that the longer 5′ RACE products from transcripts of both genes from both leaf and flower tissues came from full-length transcripts that included the proposed start codons (Figure 14; fragments 1 and 3 representing IDI1 and IDI2, respectively). For IDI1, even longer transcripts than those represented by fragment 1 were observed in both tissues. Importantly, 5′ RACE analysis and sequencing detected shorter transcripts for both IDI1 and IDI2 (represented by fragments 2 and 5 for IDI1 in leaf and flowers, respectively, and by fragment 4 for IDI2 in both tissues). Fragment 2 represents an IDI1 transcript that begins 180 bp downstream from the predicted start codon, while fragment 5 represents an IDI1 transcript beginning 48 bp downstream from the same position. Fragment 4 represents an IDI2 transcript that begins 52 bp from its start codon. The shorter transcripts include the region encoding the Met-rich, conserved portion of the N-terminal extension corresponding to Met codons at positions −16 and −8 (IDI2 fragment 4 and IDI1 fragment 5) or only at position −8 (IDI1 fragment 4) relative to the E. coli IDI start codon (Figure 2). A highly truncated IDI1 RACE product was also observed from leaf templates, which is unlikely to produce functional proteins.
Figure 14.
The 5′ RACE Products of IDI1 and IDI2 in Leaves and Flowers of Wild-Type Arabidopsis Plants.
Nested RACE products were separated on a 4% agarose gel, excised, cloned, and sequenced. A nucleotide alignment of five representative RACE products is shown for bands representing the IDI1 full-length transcript (1), the IDI1 short transcript (2 and 5), the IDI2 full-length transcript (3), and the IDI2 short transcript (4). Alignments were performed using Blosum62 (VectorNTI 10) with a gap-opening penalty of 10 and a gap-extension penalty of 0.1. The presumed start codons for the full-length IDI1 and IDI2 proteins are boxed, while the dotted lines represent possible alternative start codons for shortened transcripts, including that corresponding to the E. coli start codon (defined as Met +1) and conserved Met codons at amino acid positions −8 and −16 relative to the E. coli start position. The sizes of representative marker bands are indicated in base pairs.
DISCUSSION
The Arabidopsis IDI1 and IDI2 Genes Encode Similar Proteins with Divergent N Termini That Are Predicted to Serve as Transit Peptides
Our studies of the two Arabidopsis IPP isomerase genes, IDI1 and IDI2, indicated no great differences in organ expression patterns between them, and the catalytic properties of the encoded proteins were also similar. However, there are some distinctions in their metabolic roles caused by differences in subcellular localization resulting from divergence in their N-terminal sequences.
An analysis of the gene structure of IDI1 and IDI2, based on available cDNAs and our own RT-PCR products, indicated that the annotated versions available in public databases correctly show the intron and exon structure of these genes and that alternative splicing of primary transcripts, if it occurs, must be rare. Furthermore, the presence of upstream in-frame stop codons in the 5′ UTR (or promoter region) of both genes strongly suggested that the start codons shown in Figure 2 are the true translational start signals used in planta for the complete transcript. The predicted amino acid sequences include the presence of a 65- to 75-residue N-terminal extension in both proteins compared with the E. coli homolog. These extensions are highly divergent from one another (and from the corresponding sequences in other plant IDI proteins) with the exception of the last 16 residues. This observation, together with the results of targeting prediction software and the observation that both proteins are catalytically active when expressed as truncations without the extension (Campbell et al., 1997; this study), all indicated likely organellar targeting of IDI1 and IDI2 when translated from full-length complete transcripts.
The IDI1 Protein Participates in Plastid Isoprenoid Formation, but Knocking Out the IDI1 Gene Has No Morphological or Chemical Effects
Expression of the IDI1 gene fused to the eGFP open reading frame in plants resulted in clear plastid localization of this protein (Figure 4). While the terminal step of the MEP pathway produces both IPP and DMAPP (Eisenreich et al., 2004), previous work has suggested that IDI activity in the plastid is necessary for maximal rates of synthesis of isoprenoids such as carotenoids (Albrecht and Sandmann, 1994; Page et al., 2004). Furthermore, it has also been shown that in E. coli cells that were engineered to synthesize carotenoids from DMAPP and IPP produced via the MEP pathway, IDI activity was a limiting factor in carotenoid biosynthesis (Kajiwara et al., 1997; Cunningham and Gantt, 2000). That exogenously supplied mevalonate can complement a block in 1-deoxyxylulose 5-phosphate synthase activity and chlorophyll biosynthesis (presumably following the transport of IPP to the plastid) also suggests the presence of IDI activity in this compartment (Estévez et al., 2000; Nagata et al., 2002). Thus, plastid localization for a plant IDI is not surprising. However, plants homozygous for the idi1 null alleles are viable and contain wild-type levels of carotenoids and chlorophylls even under high light conditions (Figure 8), suggesting that IDI2 may somehow compensate for the lack of IDI1.
The IDI2 Protein Participates in Mitochondrial Isoprenoid Formation, and idi2 Mutants Have Abnormal Floral Morphology
The expression in plants of IDI2 fused to eGFP unambiguously demonstrated localization to a nonplastid organelle (Figure 4E). Using a mitochondria-specific fluorescent dye, we were able to show the site of IDI2 targeting as the mitochondria (Figure 4F). A mitochondria-specific isoform of IPP isomerase is not surprising given the previously reported role of mitochondria in the biosynthesis of some essential isoprenoids, such as ubiquinone (Lütke-Brinkhaus et al., 1984), and the targeting of a specific isoform of FPP synthase to this compartment (Cunillera et al., 1997). While IPP has been shown to be taken up by mitochondria (Lütke-Brinkhaus et al., 1984), if DMAPP cannot similarly be absorbed, then there would be an absolute requirement for an IPP isomerase in the mitochondrion to provide DMAPP for the construction of isoprenoids. Direct import of FPP into mitochondria, inferred from the observation that the administration of [3H]farnesol to cultured tobacco BY-2 cells resulted in labeled ubiquinone (Hartmann and Bach, 2001), would obviate the need for an IPP isomerase there. But it is possible that the label from farnesol in this study was imported in a form other than FPP, such as ubiquinone itself, which has been shown to be imported into mitochondria in tobacco after assembly in the endoplasmic reticulum (Swiezewska et al., 1993; Ohara et al., 2004).
Compared with IDI1, IDI2 is more highly expressed overall. At the organ level, we observed that IDI2 expression is highest in roots but that IDI1 expression is highest in flowers. These results partially agree with those of Campbell et al. (1997), who found that IDI1 and IDI2 were both most highly expressed in roots. This discrepancy may be due to cross-hybridization of the cDNA probes used with RNA gel blots in the previous study, since the portions of IDI1 and IDI2 that encode the mature protein share 89% identity at the nucleotide level. In our case, the design of primers in the 3′ UTR, where IDI1 and IDI2 share no similarity, and the sequencing of multiple qRT-PCR products ensured the specificity of IDI transcript measurements.
The mitochondrial localization of IDI2 and the fused-sepal phenotype of the idi2-1 mutant (Figure 7) led us to speculate that specific isoprenoid metabolites derived from the mitochondria are necessary for normal floral development. In addition to the synthesis of ubiquinone, some protein prenylation is thought to take place in the mitochondria of spinach (Spinacia oleracea) (Shipton et al., 1995). However, in Arabidopsis, no protein prenyltransferase activity is known to occur in this organelle (Crowell, 2000). While mitochondria have previously been shown to be involved in floral organ development in Nicotiana spp (Bonnett et al., 1991), it is not currently clear why a lack of IDI2 activity in Arabidopsis would produce the observed floral phenotype.
Representative isoprenoids biosynthesized in the plastid, cytosol, and mitochondrion were analyzed in idi2-1 flowers in an attempt to identify the cause of the mutant phenotype. However, these analyses failed to show any specific isoprenoid deficiency in mutant flowers. Furthermore, transformation experiments in which a truncated isomerase expressed in the idi2-1 mutant background failed to fully restore the wild-type phenotype suggest that the basis of the idi2-1 phenotype is not related to any of the compounds we have examined here or to the lack of cytosolic isomerase activity.
Single Mutants in IDI1 or IDI2 Are Rescued by the Remaining IDI
Characterization of mutant lines in which the IDI genes had been disrupted showed that the simultaneous loss of both genes results in a lethal phenotype. However, there were no major morphological or chemical abnormalities associated with the loss of either gene singly, with the exception of the loss of IDI2 in flowers. What is the source of DMAPP in these single idi mutants? We can rule out the MEP pathway as a significant source of DMAPP in the single mutant lines, because then the double idi mutant (which should have a functioning MEP pathway) would also be viable. By the same reasoning, we can exclude the existence of a heretofore unrecognized type II IPP isomerase in Arabidopsis or the presence of any other protein catalyzing the conversion of IPP to DMAPP in planta. Other explanations for the viability of the single idi mutant lines require the remaining IDI protein to be the source of the DMAPP. Although it was impossible to obtain viable plants with three idi null alleles in crosses between the mutants, overexpressing IDI1 as a cytosolic protein gave viable plants that were homozygous for the idi2 null allele and heterozygous for an idi1 null allele. To be sure, there are limits to the ability of one IDI activity to complement another, in that expression of IDI1 as a plastidic protein or a cytosolic protein completely failed to restore the idi2-1 flower phenotype to that of the wild type (Figure 7). Nevertheless, it seems clear that a single IDI gene is sufficient for plant viability, whether it encodes IDI1, which we observed in the plastids, or IDI2, which was found in the mitochondria. In both cases, the transport of DMAPP or later intermediates between organelles would seem essential for normal isoprenoid formation.
Scattered evidence suggests that isoprenoid biosynthetic intermediates can be transported among subcellular components. For example, IPP import into mitochondria (Lütke-Brinkhaus et al., 1984) and exchange of IPP between the cytosol and plastid have been documented by inhibitor studies in Arabidopsis (Laule et al., 2003) and in vivo feeding studies in several plant systems, including tobacco BY-2 cultures (Hemmerlin et al., 2003), snapdragon (Antirrhinum majus) (Dudareva et al., 2005), chamomile (Chamaemelum nobile) (Adam et al., 1999), and lima bean (Phaseolus lunatus) (Piel et al., 1998). In vitro studies with isolated membranes also support the transport of IPP from plastids to the cytosol (and possibly DMAPP, albeit at very low levels) (Bick and Lange, 2003) and from the cytosol to plastids (Kreuz and Kleinig, 1984; Soler et al., 1993; Flügge and Gao, 2005). Among earlier isoprenoid intermediates, exogenously supplied mevalonate, an intermediate in the cytosolic IPP pathway, has been shown to partially rescue a block in the plastidic MEP pathway (Estévez et al., 2000; Nagata et al., 2002; Gutiérrez-Nava et al., 2004; Rodríguez-Concepción et al., 2004). The transport of bona fide MEP pathway intermediates such as 1-deoxy d-xylulose 5-phosphate (DXP) from the cytosol into plastids has been shown to proceed via an inorganic phosphate antiport transporter (Flügge and Gao, 2005). In fact, a cytosolic xylulose kinase was recently identified in Arabidopsis that converts 1-deoxy d-xylulose to DXP (Hemmerlin et al., 2006), although a role for DXP in the cytosol is unclear at present.
To find the source of DMAPP in cytosolic isoprenoid formation, we investigated the origin of the C5 units of a major cytosol-produced isoprenoid, sitosterol, in both wild-type and single idi mutant Arabidopsis plants. When [2-13C]mevalonate, an intermediate of the cytosolic pathway, was supplied, all C5 units in all idi1 and idi2 mutant lines were uniformly labeled whether derived from IPP or DMAPP. These findings point to the existence of a cytosolic IPP isomerase activity arising from both IDI1 and IDI2 and provide no support for the incorporation of DMAPP units originating in another compartment (which would be unlabeled) or for the transport of cytosolic IPP to the plastids or mitochondria for the isomerization to DMAPP followed by incorporation upon return to the cytosol (which would give lower specific labeling).
Arabidopsis Has IPP Isomerase Activity in the Cytosol Arising from Shorter Transcripts of IDI1 and IDI2
While the GFP studies indicated the localization of IPP isomerase protein only in plastids (IDI1) or mitochondria (IDI2), inspection of the gene sequences indicated additional downstream Met codons of IDI1 and IDI2 that could serve as initiators of translation of shortened proteins without signal peptides that might be localized to the cytosol. Using 5′ RACE, shorter transcripts were isolated for both genes; these lacked the 5′ UTR and 48 bp or more of the coding sequence of the full-length transcripts. These short transcripts can be expected to give shortened proteins without transit peptides, providing further evidence of cytosol-localized IPP isomerase. The fact that the in vitro activity of both proteins has an optimum at pH 7.0 is also consistent with a cytosolic location. However, in relation to mitochondrial and plastidial IPP isomerase forms, such truncated cytosolic proteins probably represent only a small proportion of total IPP isomerase proteins. While 5′ RACE is not a quantitative technique, it is expected to favor the amplification of shorter fragments. Hence, the observation that RACE products corresponding to full-length transcripts are more abundant than RACE products representing shorter ones (Figure 14) suggests that full-length transcripts predominate in planta. The greater proportion of longer plastid-targeted (IDI1) or mitochondria-targeted (IDI2) transcripts versus cytosolic transcripts (from both IDI1 and IDI2) may also explain the general lack of cytosolic eGFP expression observed in IDI-eGFP–transformed lines. Fluorescence from a small amount of cytosolically localized fusion protein dispersed over the large space of the cytosol is likely much less visible by confocal microscopy than large amounts of plastidial and mitochondrial fusion proteins accumulated in the smaller volumes of these organelles. However, since our IDI-eGFP constructs did not include the native 5′ UTR sequences, the analysis of 5′ RACE products is probably a more physiologically relevant indicator of protein targeting.
The presence of IPP isomerase activity in the cytosol is not only consistent with the sitosterol feeding studies and the existence of shorter transcripts but also rationalizes the lack of gross phenotypes in the idi mutants. It also provides a logical way to support the formation of sesquiterpenes, sterols, dolichols, and a range of other isoprenoids in this compartment, since the basic isoprenoid pathway in the cytosol produces only IPP and not DMAPP, the C5 starter unit for terpenoid construction.
The Properties and Localization of IDI May Vary among Plant Species
While the IPP isomerase of E. coli is dispensable for this bacterium (Hahn et al., 1999), plants require this activity to supply the C5 starter units for isoprenoid chain elongation; indeed, most plant species studied, like Arabidopsis, have at least two different IDI genes (Cunningham and Gantt, 2000; Nakamura et al., 2001). The products of multiple IDI genes may be targeted to different subcellular locations, as shown here for Arabidopsis, because isoprenoid formation in plants is typically distributed among different subcellular compartments. Previous studies with tobacco showed that two IDI cDNAs encode proteins that are localized to the cytoplasm and the plastid, respectively (Nakamura et al., 2001), although it remains to be determined whether these cDNAs in fact represent actual transcripts of IDI forms present in this species.
Interestingly, sequence analyses of IDI genes have demonstrated that proteins from the same species are more similar to each other than to IDI sequences of other species (Cunningham and Gantt, 2000). This is also true in Arabidopsis, in which IDI1 and IDI2 share 92% identity when the N-terminal region is excluded from comparison but are only 84 to 90% identical to other known plant IPP isomerase sequences. Hence, IDI gene duplication is relatively recent on the scale of higher plant evolution, and there may be differences in IPP isomerase properties or localization among different plant lineages (Ramos-Valdivia et al., 1997a, 1998). For example, in Cinchona robusta cell cultures that produce anthraquinones requiring a DMAPP unit, two IDI proteins are known with different kinetic characteristics and patterns of occurrence (Ramos-Valdivia et al., 1997b). These results are in contrast with this study, in which it was shown that the two Arabidopsis proteins have no significant differences in kinetic properties but have different patterns of subcellular localization.
There are vast differences among plant species in the amounts and types of terpenoid secondary metabolites they produce. Given that particular terpene types differ in their relative requirements for IPP and DMAPP, and in the subcellular compartments in which they are produced, various species may require distinct types of IPP isomerases operating in separate compartments to efficiently regulate their IPP and DMAPP pools. Arabidopsis, with its relatively low output of terpenoid secondary metabolites (Chen et al., 2003; D'Auria and Gershenzon, 2005) and two biochemically similar IPP isomerases operating in various compartments, may provide a basic model for IPP isomerase diversification and distribution in a plant species.
METHODS
Plant Materials and Treatments
Arabidopsis thaliana (ecotype Col-0) plants were grown on soil in a climate-controlled growth chamber (22°C, 55% RH, and 100 μmol·m−2·s−1 photosynthetically active radiation) under either short days (12 h of light/12 h of dark) or long days (16 h of light/8 h of dark) for 4 to 6 weeks. The three independent T-DNA insertion mutants used in this study were idi1-1 (SALK_006330, N506330), idi1-2 (GABI 217_H04), and idi2-1 (SALK_014625, N514625). Seed stocks were obtained either from the European Arabidopsis Stock Center or from the GABI-KAT consortium (Bielefeld University). Initial segregation analysis was performed by seeding plants on MS medium plates supplemented with either 50 μg/mL kanamycin (SALK insertion lines) or 7.5 μg/mL sulfadiazine (GABI insertion line) as described previously (Alonso et al., 2003; Rosso et al., 2003). In addition, a PCR-based assay was designed to detect the presence of the T-DNA insertion. The primers used for this assay consisted of T-DNA left border–specific primers (SALK_LBb1or GABI_LBb1) in combination with the gene-specific primers IDI1L, IDI1R, IDI2L, and IDI2R (see Supplemental Table 1 online). Plants segregating as homozygous as confirmed by herbicide resistance and PCR were further analyzed by DNA gel blot analysis performed as described previously (D'Auria et al., 2007). Hybridization probes included either a HindIII fragment from the genomic clone of IDI1 or the XbaI–NcoI fragment obtained from the genomic clone of IDI2.
Homozygous idi2-1 plants were backcrossed for three generations to Col-0 plants for confirmation of phenotype segregation. In every generation, 30 progeny were screened for the presence of the T-DNA insert, and a minimum of five positive plants based on the PCR assay were used as the pollen donors for backcrossing. Plants positive for a T-DNA insert after the third generation of backcrossing were allowed to self-fertilize, and their progeny were genotyped by DNA gel blot. In order to obtain plant lines containing T-DNA insertion alleles for both IDI1 and IDI2, homozygous mutants of each of the four insertion lines were crossed in a reciprocal manner and analyzed in the self-fertilized F2 for a double mutant genotype.
Isotopically Labeled Precursor Feeding and Sterol Analysis
Seeds of the four homozygous T-DNA insertion lines, along with Col-0 seeds, were sterilized and plated on MS medium supplemented with a combination of 10 μM mevastatin (Sigma-Aldrich) and 4.5 mM [2-13C]mevalonolactone (Sigma-Aldrich) or with either compound alone as controls. Plants were grown for 2 weeks. Sterols were then extracted as follows: 100 mg of plant material (fresh weight) was ground in liquid N2 and extracted with 1.5 mL of methanol:CH2Cl2 (2:1) containing 10 μg of stigmastanol (Sigma-Aldrich) as an internal standard. Samples were incubated at 40°C for 30 min and centrifuged at 4200g for 10 min. The supernatant was removed and the pellet was reextracted twice with an additional 1.5 mL of methanol:CH2Cl2, followed by pooling of the supernatants. These were then back-extracted with 2 mL of 0.8% aqueous NaCl, and the lower organic phase was removed with a glass pipette and transferred to a new glass vial. The aqueous phase was extracted two additional times with 2 mL of CH2Cl2, and phases were separated as before. The pooled organic fractions were dried under a nitrogen stream and resuspended in 200 μL of CH2Cl2. After transfer to a glass GC vial insert, the samples were dried again, redissolved with 70 μL of tetrahydrofuran, incubated with 30 μL of N-methyl-N-trimethylsilyltrifluoroacetamide, and immediately analyzed.
The samples were analyzed on a Hewlett-Packard 6890 gas chromatograph coupled to a Hewlett-Packard 5973 quadrupole mass selective detector. Separation was performed on a DB5 column (5% phenylmethylpolysiloxane; J&W Scientific) measuring 30 m × 0.25 mm i.d. with a 0.25-μm film thickness. Helium was utilized as the carrier gas at a constant flow rate of 2 mL/min. A 1-μL aliquot of each sample was injected in a splitless mode at 250°C. The temperature program was as follows: 2-min hold at 200°C, then 15°C/min to 280°C, followed by a second gradient of 2°C/min to 300°C, with a final 3-min hold. The mass detector was run in scan mode from m/z 33 to 555 at 70 eV. Sitosterol, the dominant phytosterol in Arabidopsis, eluted at 14.21 min (data not shown). Isotopic labeling patterns and incorporation efficiencies were determined for sitosterol following identification by retention time and mass spectra compared with those of an authentic standard.
For NMR analysis, the methanol:CH2Cl2 plant extracts were concentrated and sitosterol was purified by preparative HPLC using a Supelcosil LC-18 DB semipreparative HPLC column (Supelco) and a gradient beginning at 10% water (solvent A) and 90% acetone (solvent B), reaching 100% B at 11 min, followed by a hold at 100% B for 5 min before a return to initial conditions at 20 min. Analytes were monitored at 210 nm, and sitosterol eluted as a minor peak at 10.65 min based on comparison with an authentic standard. Sitosterol fractions from consecutive runs were pooled, evaporated under a nitrogen stream, and resuspended in CDCl3. A Bruker DRX 500 NMR spectrometer was used for recording of 1H and 13C NMR spectra of sitosterol. Operating frequencies were 500.13 MHz for the acquisition of 1H spectra and 125.75 MHz for 13C spectra. CDCl3 was used as the solvent, and chemical shifts were referenced to trimethylsilane added as an internal standard.
Vector Construction and Plant Transformation
Gateway technology (Invitrogen) was used for the generation of all transformation constructs, which generally consisted of a target gene bearing a C-terminal fusion to eGFP under the control of the cauliflower mosaic virus 35S promoter for plant transformations or a target gene bearing an N-terminal fusion to a His tag under the control of the T7 promoter for bacterial expression. attB adaptor–bearing PCR primers (see Supplemental Table 1 online) were designed for the generation of attB PCR products for recombination with the donor vector pDONR207 via BP Clonase reactions (Invitrogen). Primer and template combinations used to generate the full-length cDNA or genomic clones for IDI1 and IDI2 are shown in Supplemental Table 1 online. Primers used to subclone IDI1 and IDI2 into the pB7FWG2 transformation vector (Karimi et al., 2002) bearing a C-terminal eGFP fusion included the presumed start codon (but none of the 5′ UTR), encompassed the entire genomic sequence, including all introns, and up to and including the last amino acid codon of exon 6. However, the stop codons were left out to produce a continuous open reading frame with eGFP fused to the C terminus of either protein. The region connecting eGFP to IDI1 or IDI2 encoded a short tether sequence (HPAFLYKVVIS) which is predicted to form a random coil. Fully sequenced entry clones were recombined in LR Clonase reactions with the pB7FWG2 vector (Karimi et al., 2002). After each 10-μL recombination reaction, 3 μL was used to transform TOP10 chemically competent cells (Invitrogen), and the resulting transformed cells were then used to inoculate 5-mL overnight cultures grown at 37°C in the presence of antibiotic (50 μg/mL gentamycin [Duchefa] for pDONR entry clones or 100 μg/mL spectinomycin [Duchefa] for plant transformation expression vectors). Plant transformation vectors were introduced into Agrobacterium tumefaciens strain GV3101 and used in vacuum infiltration transformations with Arabidopsis (ecotype Col-0) (Bechtold and Pelletier, 1998).
T0 seeds were sown in soil, stratified for 3 d at 4°C, and sprayed with 50 μg/mL glufosinate ammonium two to three times daily starting at day 10. Resistant plant lines were allowed to self-pollinate. Resulting T1 seeds were characterized as follows. Single insert lines were identified by segregation analysis based on resistance to glufosinate ammonium when grown on sterile MS phytagar plates. Transgene expression levels of independent lines were estimated by measuring the levels of green fluorescence in seedling total protein extracts with a fluorescence plate reader and a SYBR Green fluorescence filter. For this measurement, 100 mg of seedlings from each independent line was homogenized in triplicate in a cold, 2-mL glass Tenbroek homogenizer containing 500 μL of extraction buffer (100 mM Tris, pH 7.5, 10% glycerol, and 1 mM DTT). The homogenate was spun at 4°C in a benchtop microcentrifuge at maximum speed for 10 min, and a 50-μL aliquot of each supernatant was transferred to a plate well for fluorescence measurement and compared with wild-type (nontransformed) extracts. Apparent single insert lines (based on 3:1 glufosinate ammonium resistance ratios) having the highest green fluorescence values were carried on to the next generation and used in confocal microscopy analysis. To confirm the presence of the full-length transgene in each independent transformed line, total RNA was extracted from each line used for microscopy and converted to cDNA as described below in Gene Expression Measurements. This was then used as a template in RT-PCR with the 35SF and eGFPR primers (see Supplemental Table 1 online). Amplicons were cloned using the TOPO-TA-pCR4 cloning kit (Invitrogen) and sequenced with M13, M13 reverse, and internal primers to confirm that all plants used in microscopy expressed a full-length transcript, including a complete transit peptide, that was in-frame with the N terminus of eGFP.
Microscopy
Stably transformed Arabidopsis seedlings from the T2 or T3 generation representing four transformation constructs (35S:IDI1c-eGFP, 35S:IDI2c-eGFP, 35S:IDI1g-eGFP, and 35S:IDI2g-eGFP) were grown on sterile MS agarose plates containing 25 μg/mL glufosinate ammonium (Sigma-Aldrich) for 10 to 15 d. To test for chloroplast localization of eGFP fusion proteins, true leaves, stems, and roots from each group were dissected and mounted on a microscope slide with water and used in confocal microscopy. Scans were performed on plants from three independent lines per construct using a Zeiss LSM 510 Meta system, lasers specific for eGFP (488-nm excitation, detection from 500 to 530 nm) and for chloroplast autofluorescence (561-nm excitation, 575-nm long-pass detection), and a 40× LD C-Apochromat objective (water-corrected). To test for mitochondrial localization, transformed Arabidopsis seedlings were stained overnight in PBS, pH 7.2, containing 2% DMSO and 500 nM MitoTracker Orange CMTMRos (Invitrogen) at 4°C in light-protected vessels with gentle shaking. Stained seedlings were washed two times with PBS at 4°C for 4 h with gentle shaking. Roots and stems were mounted in PBS and visualized using a Zeiss Axiovert 200 inverted fluorescence microscope, eGFP (filterset 38) and orange (filterset 20) fluorescent filter blocks, an attached MRc5 Axiocam color digital camera, and an LD Achroplan 40× objective.
Gene Expression Measurements
The expression of IDI1 and IDI2 in wild-type plant organs and homozygous mutant seedlings was monitored by qRT-PCR and SYBR Green assays. Primers were designed for each gene using Beacon Designer (version 5; PremierBiosoft) in the 3′ UTR (IDI1utrF, IDI1utrR, IDI2utrF, and IDI2utrR) (see Supplemental Table 1 online). The efficiencies of the primers were determined by the standard curve method (Pfaffl, 2001) and according to their individual amplification curves using the LinRegPCR program (Ramakers et al., 2003). For comparative quantification, six normalizer gene candidates were screened for similarity of overall expression levels among different organs using purified cDNA from wild-type Arabidopsis roots, stems, flowers, and leaves. For comparison of organ-specific expression of IDI1 and IDI2, 2.0 μg of total RNA from each of those tissues was reverse-transcribed using SuperScript III (Invitrogen) in a 20-μL reaction according to the manufacturer's instructions using 50 pmol of an anchored poly(dT) primer [d(T18V)]. For each tissue, four independent biological replicates derived from individual soil-grown plants were reverse-transcribed into cDNA. After 1 h at 50°C, reactions were diluted 1:10 with pure water. A 1-μL aliquot of this diluted template containing ∼0.2 ng of cDNA was analyzed in triplicate in 25-μL SYBR Green assays in which leaves were used as a calibrator against each of the other organs. Fold change differences for IDI1 and IDI2 expression in each of the organs surveyed were calculated according to the efficiency-corrected model (Pfaffl, 2001) using RP2LS (At4g35800) as a reference gene. To measure IDI gene expression in homozygous knockout mutants, whole plate-grown seedlings were used for total RNA extractions as described above (four biological replicates and three technical replicates of each), then analyzed by qRT-PCR as described above using wild-type seedling transcript levels as a calibrator for fold change calculations.
Biochemical Characterization
For a comparison of the kinetic properties of the two IDI proteins, a truncated version of each cDNA lacking the signal peptide but containing a thrombin cleavage site was generated using the cytIDI1clv-attB1 and cytIDI1stp-attB2 primers for idi1 and cytIDI2clv-attB1 and cytIDI2stp-attB2 for idi2 (see Supplemental Table 1 online). Entry clones were generated from these attB PCR products using pDONR207 as described above. Fully sequenced entry clones were then used in LR Clonase reactions with a Gateway-compatible form of the bacterial expression vector pET28 (Yu and Liu, 2006) bearing nine N-terminal His residues to yield cytIDI1-pH28 and cytIDI2-pH28. The truncation site was based on the alignment of IDI1 and IDI2 to the start Met of the Escherichia coli IDI protein (GI: 1789255). Following transformation of E. coli BL21(DE3) competent cells with purified plasmids encoding cytIDI1-pET28 and cytIDI2-pET28, a single colony of each was used to inoculate a 200-mL culture of OverNight Express autoinduction medium (Novagen) supplemented with 1% (v/v) glycerol and 50 μg/mL kanamycin. Cultures were grown at room temperature for 48 h (220 rpm), then centrifuged for 20 min at 4400g and 4°C. The pellet was resuspended in 10 mL of ice-cold 50 mM Tris, pH 7.5, 10% (v/v) glycerol, 500 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 10 mM MgCl2, 10 mM imidazole, and 50 μg/mL lysozyme and sonicated three times for 1 min each using a Bandelin HD2070 Sonoplus sonicator at 70% power. Lysates were centrifuged for 20 min at 31,000g and 4°C. Crude extracts were purified on a Pharmacia HisTrapHQ Ni affinity column using a fast protein liquid chromatography protein purification system (Pharmacia-Amersham) by washing the bound sample with resuspension buffer lacking phenylmethylsulfonyl fluoride and lysozyme and eluting with the same buffer containing 125 mM imidazole.
Following affinity purification, the His tags were removed with a thrombin cleavage capture kit (Novagen). The biotinylated thrombin was removed with a strepavidin–agarose spin column, and the digested proteins, judged to be >97% pure by SDS-PAGE, were subsequently desalted into IDI assay buffer (50 mM Tris, pH 7.0, 10% [v/v] glycerol, 5 mM MgCl2, 200 mM KCl, 1 mM DTT, 0.1 mM MnCl2, and 0.1 mM ZnCl2) using DG-10 desalting columns (Bio-Rad) and quantified using the Bradford reagent (Bio-Rad). Proteins were diluted to 0.25 mg/mL and assayed by the acid lability method (Satterwhite, 1985). For Km and kcat measurements, reactions were performed in 100-μL volumes at six different concentrations of [1-14C]IPP (American Radiochemicals; 50 mCi/mmol). For pH optima studies, a broad-range pH buffer consisting of 50 mM acetate, MES, and bis-Tris-propane was adjusted at whole pH intervals from 4 to 10, and reactions were performed under linear conditions at each pH in triplicate. Each reaction included 2.5 μg of protein and took place at room temperature. After 15 min, reactions were stopped by the addition of 1 volume of 1 n HCl and 1 mL of diethyl ether. Hydrolysis of the acid-labile DMAPP isomer was performed at 37°C for 30 min, followed by extraction two times with 1 mL of diethyl ether. The organic phases were pooled, purified over a Pasteur pipette column containing glass wool, silica gel, and anhydrous MgSO4, and finally quantified by liquid scintillation counting. Km and kcat calculations were determined using SigmaPlot version 7.0 and the Enzyme Kinetics module version 1.1.
Isoprenoid Analysis
Five hundred milligrams of leaf tissue from wild-type and idi mutant plants grown in 250-mL pots under continuous high light (600 μmol·m−2·s−1) at 25°C was ground in liquid nitrogen and extracted with 2.5 mL of acetone:water (9:1) in quadruplicate. For flower analysis, 100 mg of wild-type or idi2-1 flowers was frozen, ground, and extracted in 1.0 mL of CH2Cl2:acetone (1:1). A 300-μL aliquot of each floral extract was dried under a nitrogen stream and resuspended in tetrahydrofuran. Sterols were then derivatized with N-methyl-N-trimethylsilyltrifluoroacetamide and analyzed by GC-MS. Both leaf and flower extracts were shielded from light, and extraction was performed at 4°C using a lab quake followed by centrifugation at 16,000g (4°C). A 10-μL aliquot of each supernatant was analyzed by HPLC on an apparatus fitted with a diode array detector using a Supelcosil C18 column (7.5 cm × 4.6 mm) flowing at 1.5 mL/min. The gradient consisted of water (A) and acetone (B), running at 65% B for 4 min, then reaching 90% B at 12 min, and finally 100% B at 20 min, before returning to initial conditions. Carotenoids were monitored at 445 nm and chlorophylls at 650 nm. Both classes of compounds were identified based on their retention times and absorption spectra compared with authentic standards.
For ubiquinone analysis, the same samples were analyzed on an 1100 series HPLC apparatus (Agilent Technologies) coupled to an Esquire 6000 ESI ion-trap mass spectrometer (Bruker Daltonics) operated in positive mode in the range m/z 500 to 900. Specifics were as follows: skimmer voltage, −40 eV; capillary exit voltage, −152.4 eV; capillary voltage, −4000 V; nebulizer pressure, 35 p.s.i.; drying gas, 8 L/min; gas temperature, 330°C. Elution was accomplished using a YMC-Pack C30 column (25 cm × 4.6 mm, 5 μm; YMC) with a gradient of 99% methanol:1% water (v/v) (solvent A) and tert-butyl-methylether (solvent B) at a flow rate of 1.0 mL/min at 25°C as follows: start, 0% B; 0 to 50% (v/v) B (25 min); 50 to 0% (v/v) B (1 min); 0% B (6 min). Flow coming from the column was diverted in a ratio of 4:1 before entering the mass spectrometer electrospray chamber. Ubiquinone-9 was detected in positive ion mode and quantified by the integrated area of the extracted ion trace of the sum of m/z 795.6 + 817.6 + 833.6 ([M+H]+, [M+Na]+, [M+K]+).
5′ RACE Cloning
Total RNA isolated from flowers or leaves was used to prepare RACE-ready cDNA using the SMART RACE cDNA amplification kit (Clontech Laboratories). Following a round of PCR using UPM and gene-specific primers (IDI1RACE-R1 and IDI2RACE-R1), nested PCR was performed using NUP and nested gene-specific primers (IDI1RACE-R2 and IDI2RACE-R2). 5′ RACE PCR products were purified on a 4% agarose gel, excised, and cloned using the pCR4 TOPO-TA cloning kit (Invitrogen). Cloning reaction products were used in transformations with TOP10 cells. Resulting positive colonies were grown in 5-mL Luria-Bertani cultures containing 50 μg/mL kanamycin for plasmid mini-preps. Purified plasmids were sequenced using M13 and M13R primers.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis thaliana IDI1 (At5g16440), IDI2 (At3g02780), and RP2LS (At4g35800); Clarkia breweri IPP isomerase 1 (X82627) and IPP isomerase 2 (U48963); Escherichia coli IPP isomerase (NP_417365); and Saccharomyces cerevisiae IPP isomerase (P15496).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Scheme for the Incorporation of [2-13C]Mevalonate into Sitosterol.
Supplemental Figure 2. Sitosterol Mass Spectra from Mutant Arabidopsis Plants.
Supplemental Figure 3. Upfield Portion of the 13C NMR Spectrum of Purified Sitosterol Extracted from idi Mutant Arabidopsis Seedlings.
Supplemental Table 1. Primer Sequences Used in This Study.
Acknowledgments
We thank Jenny Wrede, Katrin Luck, Claudia Wenderoth, Jana Pastuscheck, and Martin Franke for their technical assistance and Andreas Weber and the greenhouse staff at the Max Planck Institute for Chemical Ecology (MPICE) for their assistance in caring for the Arabidopsis plants. Stefan Opitz (MPICE) and Aleš Svatoš (MPICE) are thanked for their assistance with phytosterol and ubiquinone analysis, respectively, and we thank João Pedro Marques (Institut für Pflanzenphysiologie, Halle, Germany) for providing us with the ferredoxin-NADP(H) oxidoreductase-eGFP positive control Arabidopsis line. We express our gratitude to Almut Kießlich at the Carl Zeiss Applikationszentrum (Jena, Germany) for her assistance with confocal microscopy. This work was carried out with support from the Max Planck Society and a postdoctoral fellowship from the Alexander von Humboldt Foundation (J.C.D.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Michael A. Phillips (mapgmy@ibmb.csic.es) and John C. D'Auria (dauria@ice.mpg.de).
Online version contains Web-only data.
References
- Adam, K.P., Thiel, R., and Zapp, J. (1999). Incorporation of 1-1-C-13 deoxy-D-xylulose in chamomile sesquiterpenes. Arch. Biochem. Biophys. 369 127–132. [DOI] [PubMed] [Google Scholar]
- Albrecht, M., and Sandmann, G. (1994). Light-stimulated carotenoid biosynthesis during transformation of maize etioplasts is regulated by increased activity of isopentenyl pyrophosphate isomerase. Plant Physiol. 105 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Anderson, M.S., Muehlbacher, M., Street, I.P., Proffitt, J., and Poulter, C.D. (1989). Isopentenyl diphosphate-dimethylallyl diphosphate isomerase—An improved purification of the enzyme and isolation of the gene from Saccharomyces cerevisiae. J. Biol. Chem. 264 19169–19175. [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82 259–266. [DOI] [PubMed] [Google Scholar]
- Bick, J.A., and Lange, B.M. (2003). Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: Unidirectional transport of intermediates across the chloroplast envelope membrane. Arch. Biochem. Biophys. 415 146–154. [DOI] [PubMed] [Google Scholar]
- Blanc, V.M., and Pichersky, E. (1995). Nucleotide-sequence of a Clarkia breweri cDNA clone of IPI1, a gene encoding isopentenyl pyrophosphate isomerase. Plant Physiol. 108 855–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnett, H.T., Kofer, W., Hakansson, G., and Glimelius, K. (1991). Mitochondrial involvement in petal and stamen development studied by sexual and somatic hybridization of Nicotiana species. Plant Sci. 80 119–130. [Google Scholar]
- Burke, C.C., Wildung, M.R., and Croteau, R. (1999). Geranyl diphosphate synthase: Cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc. Natl. Acad. Sci. USA 96 13062–13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, M., Hahn, F.M., Poulter, C.D., and Leustek, T. (1997). Analysis of the isopentenyl diphosphate isomerase gene family from Arabidopsis thaliana. Plant Mol. Biol. 36 323–328. [DOI] [PubMed] [Google Scholar]
- Carrigan, C.N., and Poulter, C.D. (2003). Zinc is an essential cofactor for type I isopentenyl diphosphate:dimethylallyl diphosphate isomerase. J. Am. Chem. Soc. 125 9008–9009. [DOI] [PubMed] [Google Scholar]
- Chen, F., Tholl, D., D'Auria, J.C., Farooq, A., Pichersky, E., and Gershenzon, J. (2003). Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros, M.G., and Vincens, P. (1996). Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241 779–786. [DOI] [PubMed] [Google Scholar]
- Crowell, D.N. (2000). Functional implications of protein isoprenylation in plants. Prog. Lipid Res. 39 393–408. [DOI] [PubMed] [Google Scholar]
- Cunillera, N., Boronat, A., and Ferrer, A. (1997). The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J. Biol. Chem. 272 15381–15388. [DOI] [PubMed] [Google Scholar]
- Cunningham, F.X., and Gantt, E. (2000). Identification of multi-gene families encoding isopentenyl diphosphate isomerase in plants by heterologous complementation in Escherichia coli. Plant Cell Physiol. 41 119–123. [DOI] [PubMed] [Google Scholar]
- D'Auria, J.C., and Gershenzon, J. (2005). The secondary metabolism of Arabidopsis thaliana: Growing like a weed. Curr. Opin. Plant Biol. 8 308–316. [DOI] [PubMed] [Google Scholar]
- D'Auria, J.C., Pichersky, E., Schaub, A., Hansel, A., and Gershenzon, J. (2007). Characterization of a BAHD acyltransferase responsible for producing the green leaf volatile (Z)-3-hexen-1-yl acetate in Arabidopsis thaliana. Plant J. 49 194–207. [DOI] [PubMed] [Google Scholar]
- Dudareva, N., Andersson, S., Orlova, I., Gatto, N., Reichelt, M., Rhodes, D., Boland, W., and Gershenzon, J. (2005). The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. USA 102 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich, W., Bacher, A., Arigoni, D., and Rohdich, F. (2004). Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell. Mol. Life Sci. 61 1401–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and Von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez, J.M., Cantero, A., Romero, C., Kawaide, H., Jiménez, L.F., Kuzuyama, T., Seto, H., Kamiya, Y., and León, P. (2000). Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 124 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felkai, S., Ewbank, J.J., Lemieux, J., Labbe, J.C., Brown, G.G., and Hekimi, S. (1999). CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. EMBO J. 18 1783–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge, U.I., and Gao, W. (2005). Transport of isoprenoid intermediates across chloroplast envelope membranes. Plant Biol. 7 91–97. [DOI] [PubMed] [Google Scholar]
- Gershenzon, J., and Kreis, W. (1999). Biochemistry of terpenoids: Monoterpenes, sesquiterpenes, diterpenes, sterols, cardiac glycosides and steroid saponins. In Annual Plant Reviews: Biochemistry of Plant Secondary Metabolism, Vol. 2, M. Wink, ed (Sheffield, UK: Sheffield Academic Press), pp. 222–299.
- Goad, J.L., and Akihisa, T. (1997). Analysis of Sterols. (London: Blackie Academic and Professional).
- Gutiérrez-Nava, M.D.L., Gillmor, C.S., Jiménez, L.F., Guevara-Garcia, A., and León, P. (2004). Chloroplast biogenesis genes act cell and noncell autonomously in early chloroplast development. Plant Physiol. 135 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, F., Hurlburt, A., and Poulter, D. (1999). Escherichia coli open reading frame 696 is idi, a non essential gene encoding isopentenyl diphosphate isomerase. J. Bacteriol. 181 4499–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, M.A., and Bach, T.J. (2001). Incorporation of all-trans-farnesol into sterols and ubiquinone in Nicotiana tabacum L. cv Bright Yellow-2 cell cultures. Tetrahedron Lett. 42 655–657. [Google Scholar]
- Hemmerlin, A., Hoeffler, J.F., Meyer, O., Tritsch, D., Kagan, I.A., Grosdemange-Billiard, C., Rohmer, M., and Bach, T.J. (2003). Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco Bright Yellow-2 cells. J. Biol. Chem. 278 26666–26676. [DOI] [PubMed] [Google Scholar]
- Hemmerlin, A., Tritsch, D., Hartmann, M., Pacaud, K., Hoeffler, J.F., van Dorsselaer, A., Rohmer, M., and Bach, T.J. (2006). A cytosolic Arabidopsis d-xylulose kinase catalyzes the phosphorylation of 1-deoxy-d-xylulose into a precursor of the plastidial isoprenoid pathway. Plant Physiol. 142 441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffler, J.F., Hemmerlin, A., Grosdemange-Billiard, C., Bach, T.J., and Rohmer, M. (2002). Isoprenoid biosynthesis in higher plants and in Escherichia coli: On the branching in the methylerythritol phosphate pathway and the independent biosynthesis of isopentenyl diphosphate and dimethylallyl diphosphate. Biochem. J. 366 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara, S., Fraser, P.D., Kondo, K., and Misawa, N. (1997). Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem. J. 324 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda, K., Kuzuyama, T., Takagi, M., Hayakawa, Y., and Seto, H. (2001). An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp strain CL190. Proc. Natl. Acad. Sci. USA 98 932–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY((TM)) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kreuz, K., and Kleinig, H. (1984). Synthesis of prenyl lipids in cells of spinach leaf—Compartmentation of enzymes for formation of isopentenyl diphosphate. Eur. J. Biochem. 141 531–535. [DOI] [PubMed] [Google Scholar]
- Laule, O., Furholz, A., Chang, H.S., Zhu, T., Wang, X., Heifetz, P.B., Gruissem, W., and Lange, B.M. (2003). Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100 6866–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütke-Brinkhaus, F., Liedvogel, B., and Kleinig, H. (1984). On the biosynthesis of ubiquinones in plant mitochondria. Eur. J. Biochem. 141 537–541. [DOI] [PubMed] [Google Scholar]
- Marbois, B., Gin, P., Faull, K.F., Poon, W.W., Lee, P.T., Strahan, J., Shepherd, J.N., and Clarke, C.F. (2005). Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J. Biol. Chem. 280 20231–20238. [DOI] [PubMed] [Google Scholar]
- Marques, J.P., Schattat, M.H., Hause, G., Dudeck, I., and Klösgen, R.B. (2004). In vivo transport of folded eGFP by the ΔpH/TAT-dependent pathway in chloroplasts of Arabidopsis thaliana. J. Exp. Bot. 55 1697–1706. [DOI] [PubMed] [Google Scholar]
- McGarvey, D.J., and Croteau, R. (1995). Terpenoid metabolism. Plant Cell 7 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlbacher, M., and Poulter, C.D. (1988). Isopentenyl-diphosphate isomerase: Inactivation of the enzyme with active-site-directed irreversible inhibitors and transition-state analogs. Biochemistry 27 7315–7328. [DOI] [PubMed] [Google Scholar]
- Nagata, N., Suzuki, M., Yoshida, S., and Muranaka, T. (2002). Mevalonic acid partially restores chloroplast and etioplast development in Arabidopsis lacking the non-mevalonate pathway. Planta 216 345–350. [DOI] [PubMed] [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24 34–35. [DOI] [PubMed] [Google Scholar]
- Nakamura, A., Shimada, H., Masuda, T., Ohta, H., and Takamiya, K. (2001). Two distinct isopentenyl diphosphate isomerases in cytosol and plastid are differentially induced by environmental stresses in tobacco. FEBS Lett. 506 61–64. [DOI] [PubMed] [Google Scholar]
- Ohara, K., Kokado, Y., Yamamoto, H., Sato, F., and Yazaki, K. (2004). Engineering of ubiquinone biosynthesis using the yeast coq2 gene confers oxidative stress tolerance in transgenic tobacco. Plant J. 40 734–743. [DOI] [PubMed] [Google Scholar]
- Ohara, K., Yamamoto, K., Hamamoto, M., Sasaki, K., and Yazaki, K. (2006). Functional characterization of OsPPT1, which encodes p-hydroxybenzoate polyprenyltransferase involved in ubiquinone biosynthesis in Oryza sativa. Plant Cell Physiol. 47 581–590. [DOI] [PubMed] [Google Scholar]
- Page, J.E., Hause, G., Raschke, M., Gao, W., Schmidt, J., Zenk, M.H., and Kutchan, T.M. (2004). Functional analysis of the final steps of the 1-deoxy-d-xylulose 5-phosphate (DXP) pathway to isoprenoids in plants using virus-induced gene silencing. Plant Physiol. 134 1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time PCR. Nucleic Acids Res. 29 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel, J., Donath, J., Bandemer, K., and Boland, W. (1998). Mevalonate-independent biosynthesis of terpenoid volatiles in plants: Induced and constitutive emission of volatiles. Angew. Chem. Int. Ed. 37 2478–2481. [DOI] [PubMed] [Google Scholar]
- Ramakers, C., Ruijter, J.M., Deprez, R.H.L., and Moorman, A.F.M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339 62–66. [DOI] [PubMed] [Google Scholar]
- Ramos-Valdivia, A., van der Heijden, R., and Verpoorte, R. (1997. a). Isopentenyl diphosphate isomerase: A core enzyme in isoprenoid biosynthesis. A review of its biochemistry and function. Nat. Prod. Rep. 14 591–603. [DOI] [PubMed] [Google Scholar]
- Ramos-Valdivia, A., van der Heijden, R., Verpoorte, R., and Camara, B. (1997. b). Purification and characterization of two isoforms of isopentenyl-diphosphate isomerase from elicitor-treated Cinchona robusta cells. Eur. J. Biochem. 249 161–170. [DOI] [PubMed] [Google Scholar]
- Ramos-Valdivia, A.C., van der Heijden, R., and Verpoorte, R. (1998). Isopentenyl diphosphate isomerase and prenyltransferase activities in rubiaceous and apocynaceous cultures. Phytochemistry. 48 961–969. [Google Scholar]
- Rodríguez-Concepción, M., and Boronat, A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción, M., Forés, O., Martínez-García, J.F., González, V., Phillips, M.A., Ferrer, A., and Boronat, A. (2004). Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell 16 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdich, F., Zepeck, F., Adam, P., Hecht, S., Kaiser, J., Laupitz, R., Grawert, T., Amslinger, S., Eisenreich, W., Bacher, A., and Arigoni, D. (2003). The deoxyxylulose phosphate pathway of isoprenoid biosynthesis: Studies on the mechanisms of the reactions catalyzed by IspG and IspH protein. Proc. Natl. Acad. Sci. USA 100 1586–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Satterwhite, D.M. (1985). Isopentenyldiphosphate delta-isomerase. Methods Enzymol. 110 92–99. [DOI] [PubMed] [Google Scholar]
- Schaeffer, A., Bronner, R., Benveniste, P., and Schaller, H. (2001). The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J. 25 605–615. [DOI] [PubMed] [Google Scholar]
- Shimizu, I., Nagai, J., Hatanaka, H., Saito, E., and Katsuki, H. (1971). Subcellular localization of 3-hydroxy-3-methylglutaryl-CoA reductase in Saccharomyces cerevisiae. J. Biochem. (Tokyo) 70 175–177. [DOI] [PubMed] [Google Scholar]
- Shipton, C.A., Parmryd, I., Swiezewska, E., Andersson, B., and Dallner, G. (1995). Isoprenylation of plant proteins in vivo—Isoprenylated proteins are abundant in the mitochondria and nuclei of spinach. J. Biol. Chem. 270 566–572. [DOI] [PubMed] [Google Scholar]
- Soler, E., Clastre, M., Bantignies, B., Marigo, G., and Ambid, C. (1993). Uptake of isopentenyl diphosphate by plastids isolated from Vitis vinifera L cell suspensions. Planta 191 324–329. [Google Scholar]
- Street, I.P., and Poulter, C.D. (1990). Isopentenyldiphosphate-dimethylallyldiphosphate isomerase—Construction of a high-level heterologous expression system for the gene from Saccharomyces cerevisiae and identification of an active-site nucleophile. Biochemistry 29 7531–7538. [DOI] [PubMed] [Google Scholar]
- Swiezewska, E., Dallner, G., Andersson, B., and Ernster, L. (1993). Biosynthesis of ubiquinone and plastoquinone in the endoplasmic reticulum-Golgi membranes of spinach leaves. J. Biol. Chem. 268 1494–1499. [PubMed] [Google Scholar]
- Yu, X.H., and Liu, C.J. (2006). Development of an analytical method for genome-wide functional identification of plant acyl-coenzyme A-dependent acyltransferases. Anal. Biochem. 358 146–148. [DOI] [PubMed] [Google Scholar]