New insights into algal biology have recently emerged from the sequencing of the genomes of three unicellular green algae. Although green algae and plants share many of the same core metabolic pathways, analysis of the Chlamydomonas reinhardtii genome suggests retention of many genes from its heterotrophic eukaryote ancestor that have been lost from other members of the green clade of eukaryotes, such as the moss Physcomitrella patens and the angiosperm Arabidopsis thaliana. Two genomes from the prasinophycean genus Ostreococcus, O. tauri and O. lucimarinus, reveal strong synteny between species but also some surprising regions of heterogeneity within their genomes. The availability of genome sequences for these two closely related Ostreococcus species will enable identification of conserved noncoding sequences by phylogenetic footprinting, facilitate gene prediction and annotation (especially for small open reading frames), and provide insights into green algal genome evolution. Interesting features of gene loss and diversification point toward novel mechanisms for survival in the diverse environments in which representatives of the green clade thrive.
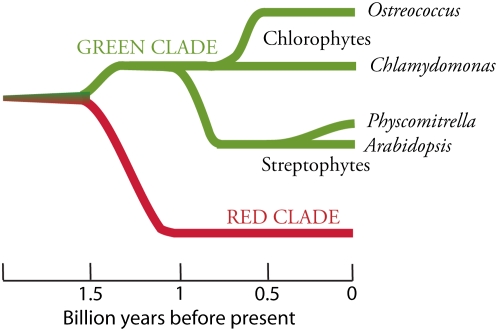
Green algae and plants are on separate evolutionary paths following ∼1 billion years of diversification (Figure 1). Chlorophytes (green algae) and Streptophytes (plants and charophycean green algae) are chlorophyll a/b–containing sister taxa that comprise the green clade of eukaryotic photosynthetic organisms. Ostreococcus spp (Figure 2) are members of the class Prasinophyceae and are relatively new to scientific inquiry, having been isolated for the first time in 1994 (Courties et al., 1994), while Chlamydomonas (Chlorophyceae) (Figure 2) has been a model organism for more than half a century (Harris, 2001). Correspondingly, the molecular tool kits and physiological information for Ostreococcus are just beginning to be developed, whereas Chlamydomonas is a nearly ideal eukaryote for forward and reverse genetic studies. Its full life cycle, from mating to dissection of meiotic products and then back to mating, takes only 2 weeks, and its haploid vegetative growth phase and rapid doubling time facilitate mutant identification and characterization (Harris, 2001). In this perspective, we highlight several prominent examples of how the sequenced genome has been used for unexpected discoveries in Chlamydomonas, along with surprising members of the gene catalog in Ostreococcus that could provide the basis for interesting biochemical studies in the near future. Before discussing specific genes in the algal catalogs, we describe briefly the impressively contrasting structures of the Chlamydomonas and Ostreococcus genomes.
Figure 1.
Evolutionary Relationships of Taxa Discussed in the Text.
The green clade of photosynthetic eukaryotes includes Chlorophytes (green algae) and Streptophytes (plants and charophycean green algae). Ostreococcus is a member of the class Prasinophyceae, whereas Chlamydomonas is classified in the Chlorophyceae. The green and red clades share a common ancestor: a heterotrophic eukaryote that engulfed a cyanobacterium to generate a plastid through primary endosymbiosis. The scale represents approximate times of divergence as taken from Yoon et al. (2004). The time of divergence of prasinophytes from other Chlorophytes is not well characterized.
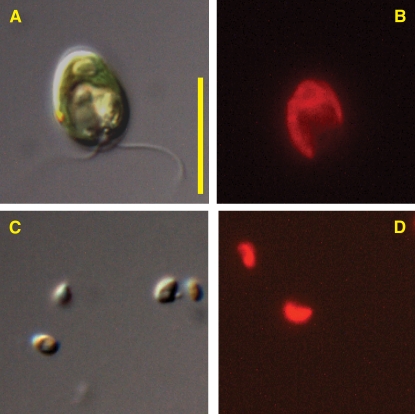
Figure 2.
C. reinhardtii and O. tauri.
(A) Differential interference contrast image of C. reinhardtii.
(B) Chlorophyll a fluorescence image of C. reinhardtii.
(C) Differential interference contrast image of O. tauri.
(D) Chlorophyll a fluorescence image of O. tauri.
The images were taken with a Zeiss Axioimager M1 fluorescence microscope. Total magnification for all images equals ×1250. Each image was captured from a different field of view, so the fluorescence images are not the same cells shown in the differential interference contrast images. Bar = 10 μm.
TALE OF THE TAPE
Measuring just under 1 μm in diameter, Ostreococcus (Figure 2) claims the title of smallest known free-living eukaryote (Courties et al., 1994). The nuclear genomes of O. tauri (Derelle et al., 2006) and O. lucimarinus (Palenik et al., 2007) are also small, weighing in at only 12.6 and 13.2 Mb, respectively, and they are predicted to have 7892 and 7651 nuclear genes packed into 20 and 21 chromosomes, respectively. These genomes contain considerably more genes than the relatively simple oxygenic prokaryote Prochlorococcus (1.7 Mb and ∼1900 protein-coding genes; Dufresne et al., 2003), but they contain only slightly more genes than are found in more complex cyanobacteria like Trichodesmium (∼7.75 Mb and ∼6300 total genes, The Institute for Genomic Research, http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org=ntte02). The structure of Ostreococcus genes illustrates the outcome of severe genome compaction. Only one-quarter of gene models in O. tauri have more than one predicted intron, and for the genome as a whole, it is predicted that there are only 1.57 exons per gene, with an average transcript size of 1257 bp. O. lucimarinus has only 20% of predicted genes containing multiple introns with similar exon number (1.27 per gene) and similar transcript size (1234 bp) as its kin.
By contrast, the 17 linkage groups of Chlamydomonas are seemingly awash in excess genetic code. The draft sequence (Merchant et al., 2007) has 15,143 predicted genes over 121 Mb, and the genes appear riddled with introns (average of 7.3 per gene) that extend the average transcript length of 1580 bp to an average gene length of 4312 bp (see Table 1 in Merchant et al. [2007] for a detailed comparison). Beyond the contrasting patterns of genome structure, Ostreococcus and Chlamydomonas exhibit unusually high G+C contents. The G+C content of the O. tauri genome is 58%, while the G+C content of Chlamydomonas is a whopping 64% (68% in coding regions!). Another notable feature of the Chlamydomonas genome is its high density of repetitive DNA sequences. Many small silencing RNAs with sequence similarity to transposable elements (TEs) have been found recently in the Chlamydomonas genome (Molnár et al., 2007), along with the first microRNAs described for a unicellular organism (Molnár et al., 2007; Zhao et al., 2007).
THE DEVIANT CHROMOSOMES OF OSTREOCOCCUS
The two Ostreococcus relatives have a high degree of synteny in their 6753 orthologous gene pairs, and 80 to 90% of the chromosomal sequences can be aligned between the two species (Palenik et al., 2007). However, this synteny is not observed in several conspicuous features found scattered on several chromosomes.
Chromosome 2
In O. tauri, Chromosome 2 (Chr2) consists of two structural blocks. One half is distinctly rich in A+T content and in predicted TEs. In fact, more than half of the TEs in the entire genome are found in the A+T-rich block (another 20% are found on Chr19, see below). The structures of the genes in this block are also considerably different. It appears as though the codon usage for the A+T-rich region deviates from the rest of the genome. The genes have conspicuously more introns and many of these are small (40 to 65 bp), with poorly conserved splice-site motifs and no clear branch-point motif (Palenik et al., 2007). These same features, with nearly identical gene content, are present in O. lucimarinus, but there have been extensive intrachromosomal rearrangements, and there is drastically different intron content between orthologous genes. The genes in this region are even more extraordinary in that many of them are arranged as convergent pairs. EST analysis shows that not only do the 3′ untranslated regions of some these genes overlap but their coding sequences can also be significantly shared.
Genes in these regions are single copy, and some encode essential housekeeping functions (such as γ-tubulin) or are required for photoautotrophic growth (PsbW). Derelle et al. (2006) originally hypothesized that this may be a sexual chromosome, and the comparative analysis showing the lack of synteny of this region (Palenik et al., 2007) strongly supports the idea that pairing of Chr2 from divergent parental strains would not be possible. The protein components of meiosis are found in O. tauri, although a sexual cycle has not been observed in culture. These factors could be important in maintaining strain-specific genetic recombination in environments where many ecotypes of Ostreococcus coexist.
Chromosome 19
Like Chr2, Chr19 in O. tauri (Chr18 in O. lucimarinus) has a lower %G+C and higher TE content compared with the rest of the genome. However, these two small chromosomes (encoding 131 and 83 predicted genes, respectively) share only nine genes between them, and only 15% of the sequences can be aligned. The majority of the predicted genes have no similarity to genes in the National Center for Biotechnology Information database, and the remainder of them appear to be faintly similar to bacteria and not to anything in the green clade. In O. tauri, these proteins are predicted to be surface membrane proteins or proteins involved in producing glucoconjugates.
Chromosome 21 of O. lucimarinus
Finally, there are several prominent examples of internal duplications in both species, but the most striking feature arising from these analyses is Chr21 of O. lucimarinus. It contains duplicated regions of Chr9 and Chr13 connected by a novel 24-bp sequence (Palenik et al., 2007). This astounding feature must be an extremely recent event, as only 5 bp deviate from the original chromosomal sequence of 321 kb.
BIOLOGICAL INSIGHTS FROM THE GREEN ALGAL GENE CATALOGS
Many of the important features described below may be attributed to the distinct ecological niches inhabited by Chlamydomonas and Ostreococcus. Chlamydomonas species can be found in a variety of aquatic environments, but the strain of C. reinhardtii used for genomic sequencing was originally isolated from soil. On a microscopic scale, the upper soil horizon has extremely heterogeneous chemical and light environments (Ranjard and Richaume, 2001). The well-documented facultative heterotrophy of Chlamydomonas is most likely an adaptation to life in this variable environment (Harris, 2001). Therefore, one could predict a priori that the genome might reveal genes derived from the heterotrophic ancestor of photosynthetic eukaryotes that have been lost in obligate photoautotrophs such as Ostreococcus and Arabidopsis.
Ostreococcus is a cosmopolitan member of ocean plankton communities, and the two species discussed here, O. tauri and O. lucimarinus, were isolated from a coastal Mediterranean lagoon and from a pier in Southern California, respectively. The two currently released genomes and an upcoming third species represent three ecologically and genetically distinct light ecotypes (Rodríguez et al., 2005). Seawater is considered to be an oligotrophic (low nutrient) environment, and the native phytoplankton are under demonstrable pressure to economize on resources, such as nitrogen and iron, that are limiting for growth (Behrenfeld et al., 1996). Differences in resource availability between these environments, ranging from light to nutrients or even oxygen levels, are likely to have left their selective imprints on the genomes of these algae.
Photosynthesis
A comparative genomics approach has been used to identify genes that encode 349 protein families that are shared by Chlamydomonas, Ostreococcus, Physcomitrella, and Arabidopsis, but absent from nonphotosynthetic organisms (Merchant et al., 2007). This study noted that a subset of these protein families in the green clade are also found in the red alga Cyanidioschyzon merolae and a diatom, resulting in a list of 90 Chlamydomonas proteins referred to as the PlastidCut. Some of these proteins have an inferred function based on motif content or reassuringly are known to be specific to the plastid, but most have completely unknown functions. Using these protein subsets as roadmaps for reverse genetic studies promises the discovery of many novel proteins that are necessary for photosynthesis or other critical plastid functions.
Interestingly, few components of the photosynthetic light-harvesting antennae were identified by comparative genomics. This is because the peripheral system of pigments and antenna proteins that serve the reaction centers of photosynthesis show significant differences between algal classes and plants (Green and Durnford, 1996). For instance, in plants, a large family of LHCB genes encodes the chlorophyll a/b binding proteins that are associated primarily with photosystem II, but these genes generally do not have clear orthologs in green algae. Chlamydomonas has nine LHCBM genes encoding polypeptides that comprise the major trimeric antenna of photosystem II (Elrad and Grossman, 2004; Minagawa and Takahashi, 2004), but these genes have diversified independently of the LHCB genes in plants (Koziol et al., 2007). Only the minor antenna proteins LHCB4, LHCB5, and LHCB7 are conserved in plants and Chlamydomonas. By contrast, there is a striking absence of either LHCB or LHCBM genes in Ostreococcus, which instead has LHCP (P for prasinophyte) genes like other prasinophytes (Six et al., 2005). Investigation of how these different antenna proteins participate in the photophysiology of green algae is just beginning.
Carbon-Concentrating Mechanisms
Fixing CO2 in an aquatic environment can be problematic because of variable local concentrations of CO2, pH-dependent species behavior in solution, and its slow diffusion rate in water relative to air. To maintain high photosynthetic rates when inorganic carbon is scarce, many algae have evolved a series of enzymatic steps to transport CO2 from the external milieu and release it within close proximity of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). The Chlorophyte genomes have revealed some unexpected pathways for these carbon-concentrating mechanisms (CCMs).
Chlamydomonas has been an important organism for establishing the role of carbonic anhydrase (CA) in eukaryotic CCMs. CA catalyzes the interconversion of CO2 and HCO3−, and plants use it to facilitate the transport and accumulation of carbonate species within particular organelles. While five CAs were previously known in Chlamydomonas, the draft genome reveals an additional four new CAs with unknown function (Grossman et al., 2007). One isoform, CAH6, appears to be targeted to the chloroplast stroma where it could assist in trapping freely diffusing CO2 as bicarbonate, but this is only speculation (Grossman et al., 2007). In stark contrast with Chlamydomonas, each Ostreococcus species has one clearly predicted CA, which appears to be most closely related to Chlamydomonas CAH6. Both Ostreococcus species also have genes with relatively weak similarity to δ-CAs from diatoms. Additionally, the Ostreococcus genomes appear to encode some homologs of proteins that influence the CCM in Chlamydomonas, but their exact physiological roles have yet to be determined in either system. These include (using Chlamydomonas gene names): (1) low-CO2-inducible genes (LCIB, LCIC, LCID, and LCIE), a family of soluble, plastid-localized proteins that somehow influence inorganic carbon transport, and (2) putative plastid envelope transporter genes (LCI1 and YCF10).
In a surprising turn, Ostreococcus has all the machinery necessary to perform C4 photosynthesis. This includes a plastid-targeted NADP(+)-dependent malic enzyme and a phosphoenolpyruvate carboxylase (see Table 10 in Derelle et al. [2006] for a full listing of carbon fixation and assimilation enzymes found in O. tauri). In C4 plants, Kranz anatomy provides a physical separation between phosphoenolpyruvate carboxylation and the Calvin-Benson cycle. This cellular division of labor is clearly not possible in a unicellular eukaryote, but the concept of unicellular C4 photosynthesis is beginning to gain hold in the literature (Edwards et al., 2004). The convergent evolution of these biochemical mechanisms suggests a significant pressure to balance the oxygenation versus carboxylation reactions of Rubisco across taxa. While all of the features listed above may influence each alga's success at concentrating CO2 from the water, other inorganic nutrients may also be limiting factors in the growth of these algae.
Nitrogen
Eukaryotic algae can use a wide variety of compounds for their nitrogen nutrition. These include inorganic molecules such as nitrate and ammonia, but they also use urea and small, organic nitrogenous molecules (Antia et al., 1991). Both Ostreococcus and Chlamydomonas can grow on oxidized and reduced forms of inorganic nitrogen, and their genomes encode complete suites of transporters and assimilation proteins. Several nitrate assimilation genes are clustered in the Chlamydomonas genome (Quesada et al., 1998) and several urea uptake and utilization genes (DUR1, -2, and -3A) are clustered on Linkage Group VIII. O. tauri also has four clustered genes encoding its urea assimilation components. The arrangements of functionally related genes may be examples of eukaryotic operons with regulation by a single promoter or cis-acting regulatory element (see Ben-Shahar et al., 2007).
Metals
Numerous micronutrient metals are required for photosynthesis and general metabolism in the green clade. Chlamydomonas has been used extensively to study the import, utilization, and interactions of trace metals, particularly in the plastid (Merchant et al., 2006). Analysis of the Chlamydomonas genome has revealed numerous metal transporter families, including the ZIP, Cu-transporting P-type ATPase, ferrireductase, and Nramp families (Merchant et al., 2006). While some of these families have homologs in Ostreococcus, the genus appears to lack Fe transporters like those seen in other unicellular eukaryotes (Palenik et al., 2007). This finding is surprising because the concentration of Fe in the seawater in which Ostreococcus thrives is extremely low. To survive in this environment, phytoplankton need extremely high-affinity Fe acquisition strategies (Morel and Price, 2003). In an unexpected twist, Ostreococcus may have access to the majority of soluble Fe in the upper ocean, which is strongly bound to small organic ligands that are believed to be siderophores of prokaryotic origin. Phytosiderophores (secreted by grasses) are structurally similar to nicotinamide, while prokaryotic siderophores are based on hydroxymate or catecholate moieties (Morel and Price, 2003). Not only does Ostreococcus have the potential to synthesize catecholates, but there are also putative prokaryote-type siderophore uptake proteins encoded in the genome (Palenik et al., 2007). Given the high abundance of prasinophytes in surface waters (Worden et al., 2004), their influence on metal cycling in the ocean may be significant.
Selenoproteins
Selenocysteine (Sec)-containing enzymes appear to be absent from vascular plants, but the genomes of green algae reveal numerous putative examples of enzymes containing Sec residues. Sec proteins are identified by the presence of a SECIS element located in the 3′ untranslated region of the gene. This element allows recognition of a TGA codon by a Sec tRNA instead of as a stop codon. Green algal selenoproteins include enzymes involved in redox signaling (thioredoxin reductase), oxidative stress response (glutathione peroxidase), and a membrane protein of unknown function that is unique to green algae (Grossman et al., 2007). Kim et al. (2006) identified a Sec-containing methionine-S-sulfoxide reductase (MsrA) in Chlamydomonas and Ostreococcus, among many other eukaryotes. MsrA is key enzyme for the repair of oxidatively damaged peptides. They found that a mutation changing the Sec to a Cys reduced catalytic efficiency of this repair protein in vitro. Conversely, the introduction of Sec into a mouse Cys-containing MsrA homolog correspondingly increased the activity of the mammalian enzyme. By employing catalytically efficient Sec enzymes, Palenik et al. (2007) propose a trade-off between increased Se requirements but decreasing N requirements for peptide synthesis. Lobanov et al. (2007) noted that the genomes of aquatic organisms had more predicted Se-containing enzymes than terrestrial organisms, but why these enzymes appear to have been completely lost from terrestrial plants remains an open question.
Anoxia: Fermentation and Hydrogen in Chlamydomonas
Under natural conditions some algae can experience low concentrations of oxygen. While rare in the sunlit areas of open water or streams, pockets of anoxia are predicted to be common, microscale habitats formed in soils or in biofilms when high rates of respiration consume molecular oxygen (Ranjard and Richaume, 2001). During anoxia, Chlamydomonas is able to activate fermentative pathways to generate compounds such as ethanol and formate and to consume reducing equivalents through the production of molecular hydrogen (Hemschemeier and Happe, 2005). A proteomic investigation of isolated mitochondria revealed the presence of pyruvate formate lyase, a protein widely used for ATP production in anaerobic prokaryotes but found in only a few eukaryotes (Atteia et al., 2006). The same proteomic analysis identified numerous other surprising anaerobic metabolism enzymes, including a pyruvate ferredoxin oxidoreductase. Additional genomic analyses have revealed a putative lactate dehydrogenase homolog that could regenerate NAD+ (Grossman et al., 2007).
Cilia (Flagella)
Chlamydomonas retains many of the properties of the ancestral heterotrophic cell that engulfed the future plastid. This includes the most prominent ultrastructural difference between cells of angiosperms and Chlamydomonas: the presence of cilia (flagella) in the latter. It is important to realize that aside from fungi and flowering plants, nearly every eukaryotic division still retains this organelle. Aside from playing an important role in motility, cilia are essential components of the sensory systems of animals and for developing cellular polarity. Using an early release of the Chlamydomonas genome, Li et al. (2004) identified a suite of novel proteins involved in ciliary structure and function by subtracting Arabidopsis proteins from the shared proteome of Chlamydomonas and humans. RNA interference studies of several candidates from this pool revealed impaired motility phenotypes, showing the utility of this approach to identify proteins of interest. Furthermore, two proteins identified by this approach were found to have homologs encoded within a region of the human genome that contained an unknown locus implicated in Bardet-Biedl syndrome (a disease affecting the development of numerous anatomical features in humans), leading to the identification of the human BBS5 gene that encodes a basal body–localized protein. Merchant et al. (2007) expanded this comparative approach by identifying 195 protein families (CiliaCut) that are found in Chlamydomonas, human, and a Phytophthora species but absent in organisms that lack cilia. While not quite doubling the list compiled by Li et al. (2004), 35% of these proteins were found in the proteome of isolated flagella (see Pazour et al., 2005 for the proteomic analyses of these organelles). These additional predictions provide a guide for future studies using reverse genetics.
The comparative approach has also led to insights regarding the structure of cilia in relatively uncharacterized members of the green clade. For instance, Merchant et al. (2007) predict that Physcomitrella has a 9+2 axoneme structure that lacks outer dynein arms, as observed in electron micrographs of sperm flagella from a related moss. Flagellated cells have not been observed in either Ostreococcus species, consistent with the absence of most of the CiliaCut genes. A related prasinophycean alga, Micromonas pusilla, does have an active flagellum (Omoto and Witman, 1981), suggesting that Ostreococcus might have recently lost its flagella.
A GREEN FUTURE
The three genomes discussed in this perspective are just the beginning of a deluge of sequence information for Division Chlorophyta. Genome sequences of Volvox carteri (http://genome.jgi-psf.org/Volca1/Volca1.home.html), Dunaliella salina, two Chlorella species, two Micromonas species, and a third species of Ostreococcus are on the way. The third Ostreococcus species is a low-light-adapted strain, which will enable a genomic comparison of different light-adapted species of eukaryotic phytoplankton. The two Micromonas genomes will add to the sequence information available for the Prasinophyceae, while the sequencing of two Chlorella genomes will be the first representatives of the Trebouxiophyceae. Chlorella is also an important organism for algal biotechnology. V. carteri is a close relative of Chlamydomonas and is an important organism for the study of multicellularity and cell differentiation. Unlike unicellular Chlamydomonas, Volvox forms colonies of up to several thousand flagellate cells that surround 16 reproductive gonidia. D. salina, an algal extremophile that thrives in high salt environments will also be sequenced. It survives salt concentrations of 10% and accumulates very high internal concentrations of carotenoids, making it an important crop for production of high-value biomolecules such as β-carotene. In addition to these upcoming genomes, numerous EST libraries from a wider range of green algae are being produced and will no doubt hasten further genome-based progress in understanding the biology of the green clade.
Acknowledgments
We thank Brian Palenik and Sabeeha Merchant for providing preprints of their manuscripts and Steve Ruzin of the Biological Imaging Facility for assistance with microscopy. Research on green algae in the authors' laboratory is supported by grants from the National Science Foundation, the National Institutes of Health, and the Department of Energy.
References
- Antia, N.J., Harrison, P.J., and Oliveira, L. (1991). The role of dissolved organic nitrogen in phytoplankton nutrition, cell biology and ecology. Phycologia 30 1–89. [Google Scholar]
- Atteia, A., van Lis, R., Gelius-Dietrich, G., Adrait, A., Garin, J., Joyard, J., Rolland, N., and Martin, W. (2006). Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J. Biol. Chem. 281 9909–9918. [DOI] [PubMed] [Google Scholar]
- Behrenfeld, M.J., Bale, A.J., Kolber, Z.S., Aiken, J., and Falkowski, P.G. (1996). Confirmation of iron limitation of phytoplankton photosynthesis in the equatorial Pacific Ocean. Nature 383 508–511. [Google Scholar]
- Ben-Shahar, Y., Nannapaneni, K., Casavant, T.L., Scheetz, T.E., and Welsh, M.J. (2007). Eukaryotic operon-like transcription of functionally related genes in Drosophila. Proc. Natl. Acad. Sci. USA 104 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courties, C., Vaquer, A., Troussellier, M., Lautier, J., Chrétiennot-Dinet, M.J., Neveux, J., Machado, C., and Claustre, H. (1994). Smallest eukaryotic organism. Nature 370 255. [Google Scholar]
- Derelle, E., et al. (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA 103 11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne, A., et al. (2003). Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. USA 100 10020–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, G.E., Franceschi, V.R., and Voznesenskaya, E.V. (2004). Single-cell C4 photosynthesis versus the dual-cell (Kranz) paradigm. Annu. Rev. Plant Biol. 55 173–196. [DOI] [PubMed] [Google Scholar]
- Elrad, D., and Grossman, A.R. (2004). A genome's-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr. Genet. 45 61–75. [DOI] [PubMed] [Google Scholar]
- Green, B.R., and Durnford, D.G. (1996). The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 685–714. [DOI] [PubMed] [Google Scholar]
- Grossman, A.R., Croft, M., Gladyshev, V.N., Merchant, S.S., Posewitz, M.C., Prochnik, S., and Spalding, M.H. (2007). Novel metabolism in Chlamydomonas through the lens of genomics. Curr. Opin. Plant Biol. 10 190–198. [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (2001). Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 363–406. [DOI] [PubMed] [Google Scholar]
- Hemschemeier, A., and Happe, T. (2005). The exceptional photofermentative hydrogen metabolism of the green alga Chlamydomonas reinhardtii. Biochem. Soc. Trans. 33 39–41. [DOI] [PubMed] [Google Scholar]
- Kim, H.-Y., Fomenko, D.E., Yoon, Y.-E., and Gladyshev, V.N. (2006). Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry 45 13697–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol, A.G., Borza, T., Ishida, K.-I., Keeling, P., Lee, R.W., and Durnford, D.G. (2007). Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol. 143 1802–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.B., et al. (2004). Comparative and basal genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117 541–552. [DOI] [PubMed] [Google Scholar]
- Lobanov, A.V., Fomenko, D.E., Zhang, Y., Sengupta, A., Hatfield, D.L., and Gladyshev, V.N. (2007). Evolutionary dynamics of eukaryotic selenoproteomes: Large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol. 8 R198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, S.S., Allen, M.D., Kropat, J., Moseley, J.L., Long, J.C., Tottey, S., and Terauchi, A.M. (2006). Between a rock and a hard place: Trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763 578–594. [DOI] [PubMed] [Google Scholar]
- Merchant, S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa, J., and Takahashi, Y. (2004). Structure, function and assembly of Photosystem II and its light-harvesting proteins. Photosynth. Res. 82 241–263. [DOI] [PubMed] [Google Scholar]
- Molnár, A., Schwach, F., Studholme, D.J., Thuenemann, E.C., and Baulcombe, D.C. (2007). miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447 1126–1129. [DOI] [PubMed] [Google Scholar]
- Morel, F.M.M., and Price, N.M. (2003). The biogeochemical cycles of trace metals in the oceans. Science 300 944–947. [DOI] [PubMed] [Google Scholar]
- Omoto, C.K., and Witman, G.B. (1981). Functionally significant central-pair rotation in a primitive eukaryotic flagellum. Nature 290 708–710. [DOI] [PubMed] [Google Scholar]
- Palenik, B., et al. (2007). The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA 104 7705–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Agrin, N., Leszyk, J., and Witman, G.B. (2005). Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada, A., Gómez, I., and Fernández, E. (1998). Clustering of the nitrite reductase gene and a light-regulated gene with nitrate assimilation loci in Chlamydomonas reinhardtii. Planta 206 259–265. [DOI] [PubMed] [Google Scholar]
- Ranjard, L., and Richaume, A.S. (2001). Quantitative and qualitative microscale distribution of bacteria in soil. Res. Microbiol. 152 707–716. [DOI] [PubMed] [Google Scholar]
- Rodríguez, F., Derelle, E., Guillou, L., Le Gall, F., Vaulot, D., and Moreau, H. (2005). Ecotype diversity in the marine picoeukaryote Ostreococcus (Chlorophyta, Prasinophyceae). Environ. Microbiol. 7 853–859. [DOI] [PubMed] [Google Scholar]
- Six, C., Worden, A.Z., Rodríguez, F., Moreau, H., and Partensky, F. (2005). New insights into the nature and phylogeny of prasinophyte antenna proteins: Ostreococcus tauri, a case study. Mol. Biol. Evol. 22 2217–2230. [DOI] [PubMed] [Google Scholar]
- Worden, A.Z., Nolan, J.K., and Palenik, B. (2004). Assessing the dynamics and ecology of marine picophytoplankton: The importance of the eukaryotic component. Limnol. Oceanogr. 49 168–179. [Google Scholar]
- Yoon, H.S., Hackett, J.D., Cinglia, C., Pinto, G., and Bhattacharya, D. (2004). A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21 809–818. [DOI] [PubMed] [Google Scholar]
- Zhao, T., Li, G., Mi, S., Li, S., Hannon, G.J., Wang, X.-J., and Qi, Y. (2007). A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 21 1190–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]