Abstract
Although coculture of hematopoietic stem cells (HSCs) with stromal cells is a useful system to study hematopoiesis in the niche, little is known regarding the precise cellular and molecular mechanisms of maintaining HSCs through cell–cell interactions. The murine preadipose stromal cell line MC3T3-G2/PA6 (PA6) has been demonstrated to support HSCs in vitro. In this study, microarray analysis was performed on PA6 cells and HSC-nonsupporting PA6 subclone cells to identify genes responsible for supporting HSC activity. Comparison of gene expression profiles revealed that only 144 genes were down-regulated by more than twofold in PA6 subclone cells. Of these down-regulated genes, we selected 11 candidate genes and evaluated for the maintenance of HSC function by overexpressing these genes in PA6 subclone cells. One unknown gene, 1110007F12Rik (also named as Tmem140), which is predicted to encode an integral membrane protein, demonstrated a partial restoration of the defect in HSC-supporting activity.
Electronic supplementary material
The online version of this article (doi:10.1007/s12185-008-0048-9) contains supplementary material, which is available to authorized users.
Keywords: Hematopoietic stem cells, Stromal cells, MC3T3-G2/PA6, Coculture, Microarray
Introduction
Hematopoietic stem cells (HSCs) are capable of self-renewal and multilineage differentiation and generate all types of blood cells throughout the lifetime. A major challenge for a long time has been the establishment of culture conditions that can facilitate ex vivo expansion of HSCs. To date, a large number of attempts using combinations of multiple cytokines have resulted in limited success, with a several-fold increase in HSC numbers under the optimal culture conditions [1, 2]. A major problem is that culture conditions established thus far have induced HSC proliferation coupled with differentiation, leading to a progressive loss of in vivo long-term repopulating potential [3–8]. Therefore, a better understanding of mechanisms that regulate HSC self-renewal and differentiation is required to achieve ex vivo expansion of HSCs.
In the adult bone marrow, HSCs are thought to reside in a specific microenvironment, referred to as the niche, which is composed of stromal cells that play a vital role in determining stem cell fate. Indeed, several stromal cell lines have been established not only from bone marrow but also from fetal liver and the aorta-gonad-mesonephros region and have been shown to maintain HSCs in vitro [9–16]. Coculture of HSCs with stromal cell lines is a useful system to study hematopoiesis in the niche. Although physical contact between HSCs and stromal cell lines has been demonstrated to be essential, little is known regarding the precise cellular and molecular mechanisms of maintaining HSCs by stromal cell lines.
MC3T3-G2/PA6 (PA6) is a preadipose stromal cell line derived from newborn mouse calvaria that can support long-term hematopoiesis in vitro [17]. In addition, PA6 subclones incapable of supporting the proliferation of HSCs have been isolated [18]. It has also been shown that the hematopoiesis-supporting ability of PA6 cells is not solely conferred by their expression of c-kit ligand, stem cell factor (SCF). In this study, microarray analysis was performed to identify genes responsible for supporting HSC activity of PA6 cells. Eleven genes specifically down-regulated in the subclone cells were selected and characterized by overexpressing the corresponding cDNAs in the subclone cells.
Materials and methods
Mice
C57BL/6 (B6-Ly5.2) mice were purchased from Charles River Laboratories Japan (Yokohama, Japan). C57BL/6 mice congenic for the Ly5 locus (B6-Ly5.1) were obtained from RIKEN BRC. B6-Ly5.1/Ly5.2 F1 mice were obtained from mating pairs of B6-Ly5.1 and B6-Ly5.2 mice. All animal experiments were approved by the Animal Experiment Committee of the RIKEN Tsukuba Institute.
Stromal cell culture and lentiviral transduction
The OP9 stromal cell line was obtained from RIKEN BRC Cell Bank. PA6 and OP9 cells were maintained in Minimum Essential Medium Eagle-α (MEM-α) (Sigma-Aldrich, St Louis, MO) containing 20% fetal bovine serum (FBS) (Sigma-Aldrich) at 37°C in a 5% CO2 atmosphere. FANTOM cDNA clones (kindly provided by Dr. Y. Hayashizaki and Dr. J. Kawai, RIKEN GSC) corresponding to the genes used in this study were subcloned into the lentiviral vector plasmid pCSII-EF-MCS-IRES2-Venus. Recombinant lentiviral vectors were produced as described previously [19]. PA6 subclone cells were transduced with lentiviral vectors expressing cDNAs at a multiplicity of infection of 200 and >90% of transduction efficiency was confirmed by fluorescence-activated cell sorting (FACS) analysis for Venus expression.
Purification of CD34−KSL cells and coculture with stromal cells
CD34−/lowc-Kit+Sca-1+lineage marker− (CD34−KSL) cells were purified as described previously with minor modifications [20]. Briefly, bone marrow cells isolated from 10- to 16-week-old B6-Ly5.2 mice were stained with a lineage marker antibody cocktail consisting of biotinylated anti-Gr-1, anti-Mac-1, anti-B220, anti-IgM, anti-CD4, anti-CD8, and anti-Ter119 antibodies (eBioscience, San Diego, CA). Lineage marker+ cells were depleted using streptavidin-coupled Dynabeads M-280 (Invitrogen, Carlsbad, CA). The remaining cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-CD34, phycoerythrin (PE)-conjugated anti-Sca-1, and allophycocyanin (APC)-conjugated anti-c-Kit antibodies (all from BD Biosciences, San Jose, CA). The biotinylated antibodies were developed with streptavidin-APC-Cy7 (BD Biosciences). FACS was performed with a FACSVantage SE (BD Biosciences). CD34−KSL cells were sorted into individual wells of a 96-well plate containing stromal cells (1 × 104 cells/well) irradiated with 1.5 Gy and were cocultured in 150 μL of MEM-α containing 20% FBS.
Colony-forming cell assay
After 10 days of coculture, CD34−KSL HSCs were collected and plated in a 12-well plate containing 0.6 mL of methylcellulose medium (MethoCult GF M3434) (StemCell Technologies, Vancouver, Canada) containing 50 ng/mL rmSCF, 10 ng/mL rmIL-3, 10 ng/mL rhIL-6, and 3 units/mL rhEPO. After 12 days of incubation, colonies were recovered, cytospun onto glass slides, and then subjected to Hemacolor (Merck KGaA, Darmstadt, Germany) staining for morphological examination. Colony-forming unit-granulocyte, erythrocyte, monocyte, megakaryocyte (CFU-GEMM), CFU-granulocyte, erythrocyte, monocyte (CFU-GEM), CFU-granulocyte, monocyte (CFU-GM), CFU-granulocyte (CFU-G), CFU-monocyte (CFU-M), and burst forming unit-erythrocyte (BFU-E) were scored using standard scoring criteria. The total colony number was expressed as colony-forming cell (CFC).
Competitive repopulation assay
The competitive repopulation assay was performed by using the congenic Ly5 mouse system as described previously [8]. CD34−KSL HSCs after coculture were mixed with 2 × 105 total bone marrow competitor cells from B6-Ly5.1 mice and transplanted into lethally (9.5 Gy) irradiated B6-Ly5.1 mice. At 12 to 16 weeks after transplantation, peripheral blood cells of the recipient mice were collected by retro-orbital bleeding. After lysis of red blood cells with ammonium chloride buffer, the remaining nucleated cells were stained with FITC-conjugated anti-Ly5.2, PE-conjugated anti-Ly5.1, biotinylated anti-Mac1, and biotinylated anti-Gr1 antibodies, followed by addition of streptavidin-PerCP. The cells were stained simultaneously with APC-conjugated anti-B220 antibody or a mixture of APC-conjugated anti-CD4 and anti-CD8 antibodies. FACS analysis was performed with a FACSCalibur. Donor chimerism was determined as the percentage of Ly5.2+ cells. When the percent chimerism was >1.0% for all myeloid, B-lymphoid, and T-lymphoid lineages, recipient mice were considered to be multilineage reconstituted.
Microarray analysis
Total RNA was isolated from stromal cells using the ISOGEN reagent (Nippon Gene, Tokyo, Japan) and purified with the RNeasy MinElute cleanup kit (Qiagen, Hilden, Germany). Biotin-labeled cRNA was prepared from 1 μg of the purified total RNA with a one-cycle cDNA synthesis kit and 3′-amplification reagents for IVT labeling (Affymetrix, Santa Clara, CA) and was hybridized to an Affymetrix Gene Chip Mouse Genome 430 2.0 array (Affymetrix), which contains approximately 45,000 probe sets for analyzing the expression levels of more than 34,000 mouse genes. After washing and staining with the antibody amplification procedure, the microarrays were scanned with an Affymetrix GeneChip Scanner 3000 7G. All these procedures were carried out according to the manufacturer’s instructions. The expression value (Signal) and detection call (Present (P), Absent (A), or Marginal (M)) for each probe set were calculated using GeneChip Operating Software version 1.4 (Affymetrix). The Signal values were normalized so that their mean in each experiment was 100 in order to adjust for minor differences between the experiments. The change value (Signal Log Ratio) and change call (Increase, Marginal Increase, No Change, Marginal Decrease, or Decrease) for each probe set were calculated by Comparison Analysis of the software. All experiments were performed in duplicate using two independent cell samples. To identify differentially expressed genes, we selected probe sets that showed a change call of Decrease and a Signal Log Ratio value of ≤–1 (more than twofold down-regulation) or a change call of Increase and a Signal Log Ratio value of ≥1 (more than twofold up-regulation) in each of the two independent experiments.
Quantitative real-time polymerase chain reaction
Total RNA was isolated from stromal cells using the ISOGEN reagent, and cDNA was synthesized from 1 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen). Real-time polymerase chain reaction (PCR) amplification with two independent cell samples was carried out using the LightCycler FastStart DNA Master SYBR Green I kit (Roche, Penzberg, Germany) according to the manufacturer’s instructions. The primer sets used in this study are listed in Table 1. Data were normalized GAPDH mRNA levels. Gene-specific amplification was confirmed by determining the melting curves of the PCR products and by a single band of the expected size in agarose gel electrophoresis.
Table 1.
Primers used in quantitative real-time PCR
| Gene | Primer sequence | Product size (bp) | |
|---|---|---|---|
| 1110007F12Rik | Forward | 5′-GCCCTGTGCCTGATGTTCTAC-3′ | 111 |
| Reverse | 5′-GCCCATGTCCTCCTTCCAC-3′ | ||
| 2900064A13Rik | Forward | 5′-GTTTGACCCTGTCCGAGTCG-3′ | 205 |
| Reverse | 5′-CGGGAGAACCATCATCATAACC-3′ | ||
| Ccl2 | Forward | 5′-TTAAAAACCTGGATCGGAACCAA-3′ | 121 |
| Reverse | 5′-GCATTAGCTTCAGATTTACGGGT-3′ | ||
| Ccl9 | Forward | 5′-TCAGATTGCTGCCTGTCCTAT-3′ | 117 |
| Reverse | 5′-GAACCCCCTCTTGCTGATAAAG-3′ | ||
| Cxcl5 | Forward | 5′-TGCGTTGTGTTTGCTTAACCG-3′ | 107 |
| Reverse | 5′-AGCTATGACTTCCACCGTAGG-3′ | ||
| IL-1rn | Forward | 5′-GCTCATTGCTGGGTACTTACAA-3′ | 132 |
| Reverse | 5′-CCAGACTTGGCACAAGACAGG-3′ | ||
| IL-6 | Forward | 5′-TAGTCCTTCCTACCCCAATTTCC-3′ | 76 |
| Reverse | 5′-TTGGTCCTTAGCCACTCCTTC-3′ | ||
Results and discussion
Subclones of the PA6 cell line are defective in supporting HSCs
Although PA6 subclones 2, 12, and 14 have been isolated in terms of inability to support long-term hematopoiesis in vitro, their HSC-supporting capacity has not been precisely assessed [18]. Therefore, we evaluated the maintenance of HSC function after coculture with PA6 subclones 2 and 12 (hereinafter referred to as S-2 and S-12, respectively) by in vitro CFC and in vivo competitive repopulation assays using CD34−KSL cells as highly purified HSCs [20]. Murine bone marrow-derived OP9 stromal cell line that can support HSCs was also used as a control [13]. As shown in Table 2, the CFC frequency significantly decreased after 10 days of coculture with PA6 S-2 and S-12 cells compared with PA6 cells, and lower levels of engraftment were observed in transplanted mice. These results indicate that PA6 subclone cells have a substantial defect in supporting HSCs even after short-term coculture.
Table 2.
CFC frequency and competitive repopulation capacity
| Stromal cells | CFC frequency | No. of reconstituted mice (%) | % Chimerism |
|---|---|---|---|
| PA6 | 10.8 ± 7.51 | 6/9 (66.7) | 13.2 ± 19.9 |
| PA6 S-2 | 0.60 ± 0.97* | 4/10 (40.0) | 4.5 ± 11.2** |
| PA6 S-12 | 3.2 ± 3.05* | 3/8 (37.5) | 3.4 ± 5.9** |
| OP9 | 28.2 ± 6.61 | 5/9 (55.6) | 8.6 ± 11.5 |
A total of 80 CD34−KSL HSCs (10 cells/well) were cocultured with stromal cells for 10 days and were then subjected to the CFC assay. The CFC frequency represents the colony number per ten input CD34−KSL HSCs. For the competitive repopulation assay, 30 CD34−KSL HSCs were cocultured with stromal cells for 10 days and then transplanted into lethally irradiated mice. At 12 weeks after transplantation, peripheral blood cells of the recipient mice were analyzed. Data represent the mean ± SD of two independent experiments
*P < 0.01, **P > 0.2 versus PA6, Student’s t-test
Identification of genes that were specifically down-regulated in PA6 subclone cells
We hypothesized that genes responsible for supporting HSCs would be down-regulated in PA6 subclone cells. To identify genes specifically down-regulated in PA6 subclone cells, microarray analysis was performed on PA6 cells, PA6 S-2 and S-12 cells, and OP9 cells using an Affymetrix GeneChip array containing approximately 45,000 probe sets representing over 34,000 mouse genes. As expected, gene expression profiling showed a few differences in gene expression between PA6 cells and PA6 subclone cells. Compared to PA6 cells, only 144 genes were down-regulated by more than twofold in both PA6 subclone cells. Of these down-regulated genes, 41 genes were of unknown function, and 67 of 103 (65%) known genes encoded membrane and extracellular space proteins according to the Gene Ontology cellular component classification. On the other hand, 121 genes were up-regulated by more than twofold and 30 of 67 (45%) known genes encoded membrane and extracellular space proteins. The signal values and detection calls for this analysis are provided in supplemental data. Among the down-regulated genes, eight genes of known/predicted function including chemokine- and cytokine-related genes, which were expected to effect on the HSC-supporting activity, and three genes of unknown function, which were predicted to encode membrane proteins, were selected for further analysis (Table 3).
Table 3.
List of candidate genes down-regulated in PA6 subclone cells compared with PA6 cells
| Gene symbol | Gene title | Refseq ID | Microarray | qPCR | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| vs. PA6 | vs. OP9 | vs. PA6 | vs. OP9 | |||||||
| PA6 S-2 | PA6 S-12 | PA6 S-2 | PA6 S-12 | PA6 S-2 | PA6 S-12 | PA6 S-2 | PA6 S-12 | |||
| 1110007F12Rik | RIKEN cDNA 1110007F12 gene | NM_197986 | 0.30 | 0.46 | 0.11 | 0.19 | 0.29 | 0.23 | 0.16 | 0.13 |
| 1200009O22Rik | RIKEN cDNA 1200009O22 gene | NM_025817 | 0.30 | 0.21 | 2.79 | 1.48 | ND | ND | ND | ND |
| 2900064A13Rik | RIKEN cDNA 2900064A13 gene | NM_133749 | 0.41 | 0.37 | 1.23 | 1.16 | 0.29 | 0.33 | 0.48 | 0.55 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | NM_011333 | 0.09 | 0.13 | 0.04 | 0.06 | 0.16 | 0.07 | 0.09 | 0.04 |
| Ccl9 | Chemokine (C-C motif) ligand 9 | NM_011338 | 0.14 | 0.19 | 0.03 | 0.04 | 0.10 | 0.09 | 0.04 | 0.03 |
| Cd53 | CD53 antigen | NM_007651 | 0.23 | 0.15 | 0.28 | 0.14 | ND | ND | ND | ND |
| Cxcl5 | Chemokine (C-X-C motif) ligand 5 | NM_009141 | 0.33 | 0.23 | 0.43 | 0.38 | 0.20 | 0.13 | 0.90 | 0.60 |
| Hgf | Hepatocyte growth factor | NM_010427 | 0.41 | 0.41 | 0.85 | 0.88 | ND | ND | ND | ND |
| Il1rn(IL-1rn) | Interleukin 1 receptor antagonist | NM_031167 | 0.09 | 0.12 | 0.26 | 0.31 | <0.03 | 0.04 | <1.25 | 1.45 |
| Il6(IL-6) | Interleukin 6 | NM_031168 | 0.38 | 0.44 | 2.07 | 2.62 | 0.30 | 0.16 | 2.42 | 1.31 |
| Ppap2b | Phosphatidic acid phosphatase type 2B | NM_080555 | 0.22 | 0.22 | 0.09 | 0.09 | ND | ND | ND | ND |
Relative expression levels in PA6 subclone cells (S-2 and S-14) compared with PA6 and OP9 cells were calculated using data from microarray and quantitative real-time PCR (qPCR) analyses. Data represent the average of fold changes from two independent experiments
ND not determined
Characterization of candidate genes
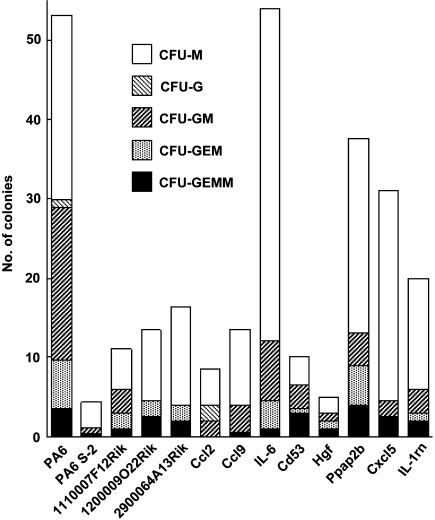
To determine the requirement of candidate genes for the supportive activity of PA6 cells, PA6 S-2 cells were transduced with lentiviral vectors containing the corresponding cDNAs. In all experiments, the transduction efficiency was more than 90% as determined by FACS analysis for Venus-positive cells. CD34−KSL HSCs were cocultured with PA6 S-2 cells expressing candidate genes for 10 days and then subjected to the CFC assay. As shown in Fig. 1, the CFC frequency increased when cocultured with PA6 S-2 cells overexpressing the candidate genes except Hgf compared with PA6 S-2 cells. Morphological evaluation of the colonies revealed that the number of CFU-GEMM, which is the most primitive colony type observed in the CFC assay, increased except Ccl2 and Ccl9. Of note, although the overexpression of IL-6 increased the number of colonies to the same level as PA6 cells, a large proportion (∼80%) of colonies were CFU-M.
Fig. 1.
Effect of over expression of candidate genes on the CFC frequency. A total of 80 CD34−KSL HSCs (10 cells/well) were cocultured with PA6 cells, PA6 S-2 cells, or PA6 S-2 cells expressing the indicated genes for 10 days and then subjected to the CFC assay. Individual colonies were scored according to their morphology. Data represent the average of two independent experiments
We next performed the competitive repopulation assay to determine the effect of overexpressing seven candidate genes including Ccl9, IL-6, Ppap2b, and Cxcl5, and three unknown genes. As shown in Table 4, successful multilineage reconstitution was observed in all mice transplanted with HSCs cocultured with PA6 S-2 cells overexpressing 1200009O22Rik or IL-6 as well as with PA6 cells. However, comparison of the chimerism showed no significant differences from that of PA6 S-2 cells, suggesting that the overexpression of neither 1200009O22Rik nor IL-6 completely restored the defective HSC-supporting activity of PA6 S-2 cells. Microarray analysis showed that the expression of IL-6 was approximately twofold up-regulated in PA6 S-2 and S-12 cells when compared with OP9 cells; this finding was confirmed by quantitative real-time PCR analysis (Table 3). It has also been demonstrated that IL-6 alone or in combination with SCF is incapable of maintaining CD34−KSL HSCs [8]. Although IL-6 is involved in various steps of hematopoiesis and has been used for the ex vivo expansion of HSCs [21], IL-6 may not be crucial for the maintenance of HSCs governed by stromal cells.
Table 4.
Effect of overexpression of candidate genes on the competitive repopulation capacity
| No. of reconstituted mice (%) | % Chimerism | ||||
|---|---|---|---|---|---|
| Total | Myeloid | B-lymphoid | T-lymphoid | ||
| PA6 | 9/9 (100) | 22.1 ± 25.1 | 30.9 ± 30.9 | 25.1 ± 26.5 | 17.4 ± 23.0 |
| PA6 S-2 | 4/10 (40) | 7.3 ± 14.0 | 4.0 ± 7.4 | 10.8 ± 18.8 | 9.2 ± 17.8 |
| 1110007F12Rik | 7/10 (70) | 16.2 ± 14.8* | 21.8 ± 26.3 | 18.7 ± 16.9 | 15.5 ± 16.7 |
| 1200009O22Rik | 8/8 (100) | 8.2 ± 10.2 | 14.5 ± 12.2 | 9.6 ± 13.4 | 8.3 ± 11.3 |
| 2900064A13Rik | 2/5 (40) | 3.3 ± 4.6 | 10.5 ± 20.6 | 3.6 ± 5.4 | 1.0 ± 2.2 |
| Ccl9 | 2/5 (40) | 11.1 ± 15.0 | 14.2 ± 19.5 | 12.7 ± 17.6 | 11.4 ± 15.7 |
| IL-6 | 9/9 (100) | 8.5 ± 13.5 | 8.1 ± 7.1 | 11.6 ± 16.9 | 9.2 ± 16.3 |
| Ppap2b | 8/10 (80) | 9.7 ± 10.4 | 20.5 ± 26.7 | 9.2 ± 9.7 | 9.6 ± 14.6 |
| Cxcl5 | 3/5 (60) | 3.0 ± 4.0 | 12.8 ± 23.6 | 2.5 ± 3.5 | 1.9 ± 1.8 |
Thirty CD34−KSL HSCs were cocultured with PA6 cells, PA6 S-2 cells, or PA6 S-2 cells expressing the indicated genes for 10 days and then transplanted into lethally irradiated mice. At 16 weeks after transplantation, peripheral blood cells of the recipient mice were analyzed. Data represent the mean ± SD
*P = 0.18 versus PA6 S-2, Student’s t-test
PA6 S-2 cells overexpressing 2900064A13Rik or CCl9 had no apparent effect on the HSC-supporting activity. In contrast, the overexpression of 1110007F12Rik, Ppap2b, or Cxcl5 resulted in a certain increase in the frequency of reconstituted mice. Although these genes as well as 1200009O22Rik and IL-6 could significantly increase the CFC frequency, the effect on the HSC-supporting activity assessed by the competitive repopulation assay was limited. This may be because 10 days of coculture was not long enough to detect a significant difference in HSC-supporting activity. It is of course well known that many growth factors and cytokines that can increase the CFC frequency have little influence on the maintenance and proliferation of HSCs. Of note, the overexpression of 1110007F12Rik showed a substantial increase in the chimerism, although the difference did not reach statistical significance (Table 4). To confirm the effect of 1110007F12Rik expression on the supportive activity of PA6 cells, we performed the following experiments. First, down-regulation of 1110007F12Rik expression in PA6 S-2 and S-12 cells compared to that in PA6 and OP9 cells was confirmed by quantitative real-time PCR analysis (Table 3). Next, PA6 S-2 cells were transduced with a myc-tagged 1110007F12Rik expression lentiviral vector and its expression was detected by Western blotting and immunofluorescence staining with anti-myc antibody. Coculture with PA6 S-2 cells overexpressing myc-tagged 1110007F12Rik resulted in an increase in the CFC frequency at a level similar to that shown in Fig. 1 (data not shown). The 1110007F12Rik cDNA encodes a predicted protein of 185 amino acids, which is rich in leucine (23%) and has no significant homology with any known protein sequences. 1110007F12Rik is also named as Tmem140 (transmembrane protein 140), which is predicted to be an integral membrane protein gene according to the Gene Ontology classification. Further studies are required to determine whether 1110007F12Rik is involved in the HSC-supporting activity of PA6 cells.
Thus far, a large number of microarray analyses of stromal cell lines derived from a variety of tissue types have been reported. Comparison of gene expression profiles between HSC-supporting and HSC-nonsupporting stromal cell lines has resulted in the identification of a number of differentially expressed genes [22]. In the present study, we expected a few differences in gene expression by comparing HSC-supporting PA6 cells with HSC-nonsupporting PA6 subclone cells and indeed, we could narrow down the candidate genes involved in maintaining HSCs. Although one of the analyzed candidate genes, 1110007F12Rik, demonstrated a partial restoration of the defect in HSC-supporting activity of PA6 S-2 cells, it is possible that other down-regulated genes in PA6 subclone cells may also be required for the maintenance of HSCs. Continuing studies to identify genes responsible for HSC-supporting activity of PA6 cells would facilitate the understanding of the mechanisms that control stem cell fate through interaction with stromal cells.
Electronic supplementary material
Below is the link to the Supplemental data.
Electronic supplementary material (XLS 388 kb)
Acknowledgments
The authors thank Dr. J. Hayashi and Dr. Y. Obata for encouragement and discussion, Dr. K. Sudo for technical advice and discussion, Dr. Y. Hayashizaki and Dr. J. Kawai for providing FANTOM cDNA clones, Dr. Miyawaki for providing the Venus cDNA, and RIKEN BRC Experimental Animal Division for animal care. N.S. is a Junior research associate at RIKEN.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12185-008-0048-9) contains supplementary material, which is available to authorized users.
References
- 1.Sauvageau G, Iscove NN, Humphries RK. In vitro and in vivo expansion of hematopoietic stem cells. Oncogene. 2004;23:7223–32. [DOI] [PubMed]
- 2.Sorrentino BP. Clinical strategies for expansion of haematopoietic stem cells. Nat Rev Immunol. 2004;4:878–88. [DOI] [PubMed]
- 3.Knobel KM, McNally MA, Berson AE, et al. Long-term reconstitution of mice after ex vivo expansion of bone marrow cells: differential activity of cultured bone marrow and enriched stem cell populations. Exp Hematol. 1994;22:1227–35. [PubMed]
- 4.Peters SO, Kittler EL, Ramshaw HS, Quesenberry PJ. Murine marrow cells expanded in culture with IL-3, IL-6, IL-11, and SCF acquire an engraftment defect in normal hosts. Exp Hematol. 1995;23:461–9. [PubMed]
- 5.Traycoff CM, Cornetta K, Yoder MC, Davidson A, Srour EF. Ex vivo expansion of murine hematopoietic progenitor cells generates classes of expanded cells possessing different levels of bone marrow repopulating potential. Exp Hematol. 1996;24:299–306. [PubMed]
- 6.Bhatia M, Bonnet D, Kapp U, Wang JC, Murdoch B, Dick JE. Quantitative analysis reveals expansion of human hematopoietic repopulating cells after short-term ex vivo culture. J Exp Med. 1997;186:619–24. [DOI] [PMC free article] [PubMed]
- 7.Glimm H, Oh IH, Eaves CJ. Human hematopoietic stem cells stimulated to proliferate in vitro lose engraftment potential during their S/G(2)/M transit and do not reenter G(0). Blood. 2000;96:4185–93. [PubMed]
- 8.Ema H, Takano H, Sudo K, Nakauchi H. In vitro self-renewal division of hematopoietic stem cells. J Exp Med. 2000;192:1281–8. [DOI] [PMC free article] [PubMed]
- 9.Collins LS, Dorshkind K. A stromal cell line from myeloid long-term bone marrow cultures can support myelopoiesis and B lymphopoiesis. J Immunol. 1987;138:1082–7. [PubMed]
- 10.Sutherland HJ, Eaves CJ, Lansdorp PM, Thacker JD, Hogge DE. Differential regulation of primitive human hematopoietic cells in long-term cultures maintained on genetically engineered murine stromal cells. Blood. 1991;78:666–72. [PubMed]
- 11.Baum CM, Weissman IL, Tsukamoto AS, Buckle AM, Peault B. Isolation of a candidate human hematopoietic stem-cell population. Proc Natl Acad Sci USA. 1992;89:2804–8. [DOI] [PMC free article] [PubMed]
- 12.Issaad C, Croisille L, Katz A, Vainchenker W, Coulombel L. A murine stromal cell line allows the proliferation of very primitive human CD34++/CD38- progenitor cells in long-term cultures and semisolid assays. Blood. 1993;81:2916–24. [PubMed]
- 13.Kodama H, Nose M, Niida S, Nishikawa S, Nishikawa S. Involvement of the c-kit receptor in the adhesion of hematopoietic stem cells to stromal cells. Exp Hematol. 1994;22:979–84. [PubMed]
- 14.Moore KA, Ema H, Lemischka IR. In vitro maintenance of highly purified, transplantable hematopoietic stem cells. Blood. 1997;89:4337–47. [PubMed]
- 15.Ohneda O, Fennie C, Zheng Z, et al. Hematopoietic stem cell maintenance and differentiation are supported by embryonic aorta-gonad-mesonephros region-derived endothelium. Blood. 1998;92:908–19. [PubMed]
- 16.Xu MJ, Tsuji K, Ueda T, et al. Stimulation of mouse and human primitive hematopoiesis by murine embryonic aorta-gonad-mesonephros-derived stromal cell lines. Blood. 1998;92:2032–40. [PubMed]
- 17.Kodama HA, Amagai Y, Koyama H, Kasai S. A new preadipose cell line derived from newborn mouse calvaria can promote the proliferation of pluripotent hemopoietic stem cells in vitro. J Cell Physiol. 1982;112:89–95. [DOI] [PubMed]
- 18.Kodama H, Nose M, Yamaguchi Y, Tsunoda J, Suda T, Nishikawa S. In vitro proliferation of primitive hemopoietic stem cells supported by stromal cells: evidence for the presence of a mechanism(s) other than that involving c-kit receptor and its ligand. J Exp Med. 1992;176:351–61. [DOI] [PMC free article] [PubMed]
- 19.Tahara-Hanaoka S, Sudo K, Ema H, Miyoshi H, Nakauchi H. Lentiviral vector-mediated transduction of murine CD34(-) hematopoietic stem cells. Exp Hematol. 2002;30:11–7. [DOI] [PubMed]
- 20.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–5. [DOI] [PubMed]
- 21.Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol. 1997;15:797–819. [DOI] [PubMed]
- 22.Charbord P, Moore K. Gene expression in stem cell-supporting stromal cell lines. Ann NY Acad Sci. 2005;1044:159–67. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the Supplemental data.
Electronic supplementary material (XLS 388 kb)