Abstract
Background
Among Chagas disease triatomine vectors, the largest genus, Triatoma, includes species of high public health interest. Triatoma dimidiata, the main vector throughout Central America and up to Ecuador, presents extensive phenotypic, genotypic, and behavioral diversity in sylvatic, peridomestic and domestic habitats, and non-domiciliated populations acting as reinfestation sources. DNA sequence analyses, phylogenetic reconstruction methods, and genetic variation approaches are combined to investigate the haplotype profiling, genetic polymorphism, phylogeography, and evolutionary trends of T. dimidiata and its closest relatives within Triatoma. This is the largest interpopulational analysis performed on a triatomine species so far.
Methodology and Findings
Triatomines from Mexico, Guatemala, Honduras, Nicaragua, Panama, Cuba, Colombia, Ecuador, and Brazil were used. Triatoma dimidiata populations follow different evolutionary divergences in which geographical isolation appears to have had an important influence. A southern Mexican–northern Guatemalan ancestral form gave rise to two main clades. One clade remained confined to the Yucatan peninsula and northern parts of Chiapas State, Guatemala, and Honduras, with extant descendants deserving specific status. Within the second clade, extant subspecies diversity was shaped by adaptive radiation derived from Guatemalan ancestral populations. Central American populations correspond to subspecies T. d. dimidiata. A southern spread into Panama and Colombia gave the T. d. capitata forms, and a northwestern spread rising from Guatemala into Mexico gave the T. d. maculipennis forms. Triatoma hegneri appears as a subspecific insular form.
Conclusions
The comparison with very numerous Triatoma species allows us to reach highly supported conclusions not only about T. dimidiata, but also on different, important Triatoma species groupings and their evolution. The very large intraspecific genetic variability found in T. dimidiata sensu lato has never been detected in a triatomine species before. The distinction between the five different taxa furnishes a new frame for future analyses of the different vector transmission capacities and epidemiological characteristics of Chagas disease. Results indicate that T. dimidiata will offer problems for control, although dwelling insecticide spraying might be successful against introduced populations in Ecuador.
Author Summary
Chagas disease is a serious parasitic disease of Latin America. Human contamination in poor rural or periurban areas is mainly attributed to haematophagous triatomine insects. Triatoma includes important vector species, as T. dimidiata in Central and Meso-America. DNA sequences, phylogenetic methods and genetic variation analyses are combined in a large interpopulational approach to investigate T. dimidiata and its closest relatives within Triatoma. The phylogeography of Triatoma indicates two colonization lineages northward and southward of the Panama isthmus during ancient periods, with T. dimidiata presenting a large genetic variability related to evolutionary divergences from a Mexican-Guatemalan origin. One clade remained confined to Yucatan, Chiapas, Guatemala and Honduras, with extant descendants deserving species status: T. sp. aff. dimidiata. The second clade gave rise to four subspecies: T. d. dimidiata in Guatemala and Mexico (Chiapas) up to Honduras, Nicaragua, Providencia island, and introduced into Ecuador; T. d. capitata in Panama and Colombia; T. d. maculipennis in Mexico and Guatemala; and T. d. hegneri in Cozumel island. This taxa distinction may facilitate the understanding of the diversity of vectors formerly included under T. dimidiata, their different transmission capacities and the disease epidemiology. Triatoma dimidiata will offer more problems for control than T. infestans in Uruguay, Chile and Brazil, although populations in Ecuador are appropriate targets for insecticide-spraying.
Introduction
American trypanosomiasis or Chagas disease is widespread in Latin America from Mexico to Chile and southern Argentina. Although present estimates of 10 to 12 million people infected with the haemoflagellate protozoan species Trypanosoma cruzi represent 6–8 million fewer cases than those reported in the 1980s [1], it remains one of the most serious parasitic diseases of the Americas for its social and economic impact [2]. Although it can also be transmitted by blood transfusion or across the placenta from infected mothers, most human contamination is attributed to insect vectors in poor rural or periurban areas of Central and South America [1].
Chagas disease vectors are haematophagous reduviid (Hemiptera: Heteroptera) insects belonging to the subfamily Triatominae. Species of Triatominae are usually grouped into 17 genera forming five tribes, although other arrangements have been proposed. Of these, Alberproseniini, Bolboderini, Cavernicolini and Rhodniini are considered monophyletic, whereas Triatomini is considered polyphyletic [3]. Among the latter, most of the species (over 70) are included in the genus Triatoma, among which two main clades appear in ribosomal DNA (rDNA) sequence phylogenies, corresponding to species of North and Central America and species of South America separated prior to the closing of the isthmus of Panama about 3 million years ago [4]–[6]. Moreover, Triatoma species are distributed in three main groupings: the Rubrofasciata group (mainly North American and Old World species), the Phyllosoma group (mainly Mesoamerican and Caribbean), and the Infestans group (mainly South American), each including different complexes and subcomplexes in a classification which is progressively updated according to new genetic and morphometric data [7].
A priori, all of the over 130 species currently recognized within Triatominae seem capable of transmitting T. cruzi. Among the species of greatest epidemiological significance as domestic vectors, three belong to the genus Triatoma: T. infestans and T. brasiliensis from South America, and T. dimidiata, distributed in Meso- and Central America from Mexico down to Colombia, Venezuela, Ecuador and northern Peru [3].
Triatoma dimidiata can be found in sylvatic, peridomestic and domestic habitats. Non-domiciliated populations may act as reinfestation sources and become involved in the transmission of the parasite to humans [8],[9]. This species includes morphologically variable populations [10],[11]. A molecular comparison of Triatominae, including many Central American species of the Phyllosoma complex by means of rDNA second internal transcribed spacer (ITS-2) sequences demonstrated an unusual intraspecific sequence variability in a few T. dimidiata populations studied. This study even revealed differences consistent with a specific status for populations from the Yucatan peninsula, Mexico [4]–[6], thus opening a debate. A large number of recent, multidisciplinary studies using RAPD-PCR, genital structures, morphometrics of head characters, and antennal phenotypes have shown that variation within this species seems much greater than previously considered [8], [12]–[16]. Morphometric and cuticular hydrocarbon analyses suggest that a sylvatic population from Lanquin, Guatemala, is undergoing a speciation process [13],[17]. Chromosomal variation and genome size suggest that T. dimidiata may represent a complex of cryptic species (i.e. morphologically indistinguishable, yet reproductively isolated taxa) [18].
The aim of the present work is to analyze the intraspecific variability, haplotype profiling, phylogeography and genetic polymorphism of populations of the species T. dimidiata, to get a new framework able to facilitate the future understanding of the diferring peculiarities of this crucial vector species throughout its broad geographical distribution. This may also help in understanding the related differences in characteristics of Chagas disease transmission and epidemiology, as well as in responses to control initiatives in the countries concerned. After a deep analysis, it was considered that the most convenient approach would be obtained by using an appropriate marker able to furnish significant information about evolutionary trends of variation on which to construct the new baseline. This new baseline should be, whenever possible, of sufficient weight as to allow its conclusions to be reflected at systematic-taxonomic level.
For this purpose, the rDNA was preferred over mitochondrial DNA (mtDNA) because of its mendelian inheritance, evolutionary rates and overall recognized usefulness in systematics in all metazoan organism groups because of including sequences which allow to distinguish between species and between subspecies units. The better fitting of rDNA for molecular systematics has already been emphasized in large reviews on rDNA/mtDNA marker comparisons in insects [19]. Ribosomal DNA includes excellent genetic markers, because (i) the rDNA operon is tandemly repeated and present in sufficiently high quantities among the genome of an individual thus facilitating sequencing procedures; (ii) the different genes and spacers of the rDNA follow a concerted evolution which, with sufficient time, effectively homologizes the many copies of nuclear rDNA within a genome [20]; this gives rise to a uniformity of their sequences within all individuals of a population and becomes extremely useful from an applied point of view, because it is sufficient to obtain the sequence of only one individual to characterize the local population it belongs to, that is, all other individuals of that population will present the same sequence; (iii) the usefulness of rDNA genes and spacers as genetic markers at different evolutionary levels have already been verified on a large number of very different eukaryotic organism groups including insects, and consequently extensive knowledge on the different rDNA fragments is available [21]. rDNA sequence comparisons offer valuable information about the evolutionary events in triatomine lineages and, by deducing the routes of spreading of triatomine populations, they may also shed light on the ability of different species to colonize new areas [5].
Within rDNA, ITS-2 was selected as marker because of its well-known usefulness at species and subspecies levels, including the differentiation of taxa within problematic groups, as is the case of those comprising cryptic or sibling species of other insect groups [22]–[24]. Moreover, the sequences of the ITS-2 have already proved to be a useful tool in the analysis of species, subspecies, hybrids and populations, and for inferring phylogenetic relationships in Triatominae in general [4],[5],[6],[25],[26].
In order to be able to assess the ITS-2 evolutionary processes followed by T. dimidiata populations, the ITS-2 sequences of many members of the Phyllosoma, Rubrofasciata and Infestans groups were obtained and analyzed. For this purpose, a large number of rDNA ITS-2 sequences of Triatoma species from numerous geographic origins in Mexico, Guatemala, Honduras, Nicaragua, Panama, Cuba, Colombia, Ecuador, and Brazil was studied. Thus, the nucleotide divergence limits between taxa within the lineage of the genus Triatoma could be established. The present study on T. dimidiata is the largest interpopulational analysis performed on a triatomine species so far.
Materials and Methods
Triatomine materials
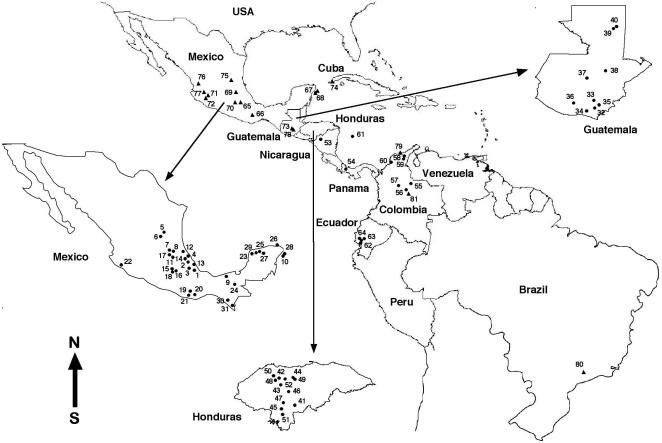
A total of 165 triatomine specimens representing 13 Triatoma species of the Phyllosoma, Rubrofasciata and Infestans groups, among which 137 specimens representing T. dimidiata from 64 different geographic origins, were used for sequencing, genetic variation and phylogenetic analyses (Table 1; Figure 1). The systematic classification recently proposed for the genus Triatoma [7] is used here throughout.
Table 1. Triatoma species and samples studied, including ITS-2 sequence length and AT composition (in percentage).
| Country | Map No. | Preliminary classification | Sampling sites | Haplotype code | Sequence length | % AT |
| PHYLLOSOMA GROUP: PHYLLOSOMA COMPLEX | ||||||
| 1) TRIATOMA DIMIDIATA : 31 different haplotype sequences/137 specimens studied: | ||||||
| MEXICO | 1 | T. dimidiata | Atoyac Tlacorrancho, Veracruz | T.dim-H18 | 496 | 75.81 |
| n = 41 | 2 | T. dimidiata | Atoyac-Manzanillo, Veracruz | T.dim-H18 | 496 | 75.81 |
| 3 | T. dimidiata | Atoyac-Cordoba, Veracruz | T.dim-H18 | 496 | 75.81 | |
| 4 | T. dimidiata | Ursulo-Galan, Veracruz | T.dim-H18 | 496 | 75.81 | |
| 5 | T. dimidiata | Tanchahuil, San Luis Potosí | T.dim-H18 | 496 | 75.81 | |
| 6 | T. dimidiata | Barrio Tzitzi, San Luis Potosí | T.dim-H18 | 496 | 75.81 | |
| 7 | T. dimidiata | Huejutla, Hidalgo (3) | T.dim-H18 | 496 | 75.81 | |
| 8 | T. dimidiata | Atlapexco, Hidalgo | T.dim-H18 | 496 | 75.81 | |
| 9 | T. dimidiata | El Rosario, Tabasco | T.dim-H18 | 496 | 75.81 | |
| 10 | T. dimidiata | Cozumel island, Quintana Roo | T.dim-H18 | 496 | 75.81 | |
| 11 | T. dimidiata | Acomul, Hidalgo | T.dim-H18 | 496 | 75.81 | |
| 12 | T. dimidiata | Mesa de Tlanchinol, Veracruz | T.dim-H19 | 494 | 75.71 | |
| 13 | T. dimidiata | La Luz, Veracruz | T.dim-H19 | 494 | 75.71 | |
| 14 | T. dimidiata | Emiliano Zapata, Veracruz | T.dim-H20 | 495 | 75.76 | |
| 15 | T. dimidiata | Morelos | T.dim-H21 | 497 | 75.85 | |
| 16 | T. dimidiata | Cajones, Morelos | T.dim-H21 | 497 | 75.85 | |
| 17 | T. dimidiata | Huehuetla, Hidalgo | T.dim-H22 | 494 | 75.71 | |
| 18 | T. dimidiata | Chalcatzingo, Morelos | T.dim-H23 | 496 | 75.60 | |
| 19 | T. dimidiata | Santiago Cuixtla, Oaxaca | T.dim-H23 | 496 | 75.60 | |
| 20 | T. dimidiata | Hierba Santa, Oaxaca | T.dim-H23 | 496 | 75.60 | |
| 21 | T. dimidiata | Nopala, Oaxaca | T.dim-H23 | 496 | 75.60 | |
| 22 | T. dimidiata | Alcaraces, Cuauhtemoc, Colima | T.dim-H24 | 496 | 75.40 | |
| 23 | T. dimidiata | Paraíso, Yucatán (3) | T.dim-H25 | 493 | 75.66 | |
| 24 | T. dimidiata | Palenque, Chiapas | T.dim-H25 | 493 | 75.66 | |
| 23 | T. dimidiata | Paraíso, Yucatán | T.dim-H26 | 489 | 75.46 | |
| 23 | T. dimidiata | Paraíso, Yucatán | T.dim-H27 | 494 | 75.51 | |
| 25 | T. dimidiata | Yaxkukul,Yucatán | T.dim-H28 | 493 | 75.66 | |
| 26 | T. dimidiata | Holbox island, Quintana Roo | T.dim-H28 | 493 | 75.66 | |
| 23 | T. dimidiata | Paraíso, Yucatán | T.dim-H28 | 493 | 75.66 | |
| 27 | T. dimidiata | Izamal, Yucatán | T.dim-H28 | 493 | 75.66 | |
| 28 | T. dimidiata | Cozumel island, Quintana Roo (3) | T.dim-H28 | 493 | 75.66 | |
| 23 | T. dimidiata | Paraíso, Yucatán | T.dim-H28 | 493 | 75.66 | |
| 29 | T. dimidiata | Chablekal, Mérida, Yucatán | T.dim-H31 | 489 | 75.25 | |
| 30 | T. dimidiata | Mapastepec, Chiapas | T.dim-H1 | 497 | 76.06 | |
| 31 | T. dimidiata | Tapachula, Chiapas | T.dim-H3 | 497 | 76.26 | |
| GUATEMALA | 32 | T. dimidiata | Jutiapa, Jutiapa (4) | T.dim-H1 | 497 | 76.06 |
| n = 37 | 33 | T. dimidiata | Agua Zarca, Jutiapa | T.dim-H1 | 497 | 76.06 |
| 34 | T. dimidiata | Pueblo Nuevo Viñas, Santa Rosa | T.dim-H1 | 497 | 76.06 | |
| 35 | T. dimidiata | Piedra Pintada, Jutiapa (3) | T.dim-H1 | 497 | 76.06 | |
| 33 | T. dimidiata | Agua Zarca, Jutiapa | T.dim-H2 | 496 | 76.01 | |
| 36 | T. dimidiata | Escuintla, Escuintla (3) | T.dim-H2 | 496 | 76.01 | |
| 37 | T. dimidiata | San Andrés Sajcabaja, Quiché | T.dim-H2 | 496 | 76.01 | |
| 34 | T. dimidiata | Pueblo Nuevo Viñas, Santa Rosa | T.dim-H2 | 496 | 76.01 | |
| 33 | T. dimidiata | Agua Zarca, Jutiapa (2) | T.dim-H3 | 497 | 76.26 | |
| 36 | T. dimidiata | Escuintla, Escuintla | T.dim-H3 | 497 | 76.26 | |
| 34 | T. dimidiata | Pueblo Nuevo Viñas, Santa Rosa | T.dim-H3 | 497 | 76.26 | |
| 37 | T. dimidiata | San Andrés Sajcabaja, Quiché | T.dim-H4 | 497 | 76.85 | |
| 34 | T. dimidiata | Pueblo Nuevo Viñas, Santa Rosa | T.dim-H8 | 497 | 76.06 | |
| 35 | T. dimidiata | Aldea Piedra Pintada, Jutiapa | T.dim-H8 | 497 | 76.06 | |
| 38 | T. dimidiata | Lanquín, Alta Verapaz (4) | T.dim-H10 | 496 | 76.01 | |
| 39 | T. dimidiata | Chultún, Yaxhá, Petén (2) | T.dim-H18 | 496 | 75.81 | |
| 37 | T. dimidiata | San Andrés Sajcabaja, Quiché (2) | T.dim-H18 | 496 | 75.81 | |
| 40 | T. dimidiata | Yaxhá, Petén | T.dim-H25 | 493 | 75.66 | |
| 40 | T. dimidiata | Yaxhá, Petén (2) | T.dim-H28 | 493 | 75.66 | |
| 40 | T. dimidiata | Yaxhá, Petén (3) | T.dim-H28 | 493 | 75.66 | |
| 40 | T. dimidiata | Yaxhá, Petén | T.dim-H30 | 491 | 75.56 | |
| HONDURAS | 41 | T. dimidiata | Güinope, El Paraiso | T.dim-H1 | 497 | 76.06 |
| n = 20 | 42 | T. dimidiata | El Tablon, Yoro (2) | T.dim-H2 | 496 | 76.01 |
| 43 | T. dimidiata | El Zapote, Yoro | T.dim-H2 | 496 | 76.01 | |
| 44 | T. dimidiata | El Salitre, Yoro | T.dim-H2 | 496 | 76.01 | |
| 45 | T. dimidiata | El Cacao, Francisco Morazán (2) | T.dim-H2 | 496 | 76.01 | |
| 46 | T. dimidiata | Orica, Francisco Morazán | T.dim-H2 | 496 | 76.01 | |
| 47 | T. dimidiata | Tegucigalpa, Francisco Morozán (2) | T.dim-H2 | 496 | 76.01 | |
| 48 | T. dimidiata | Corral Falso, Yoro (2) | T.dim-H2 | 496 | 76.01 | |
| 49 | T. dimidiata | El Salitre, Montaña, Yoro | T.dim-H2 | 496 | 76.01 | |
| 50 | T. dimidiata | Subirama, Yoro | T.dim-H2 | 496 | 76.01 | |
| 51 | T. dimidiata | San José, Choluteca | T.dim-H6 | 496 | 76.01 | |
| 48 | T. dimidiata | Corral Falso, Yoro | T.dim-H9 | 496 | 75.81 | |
| 43 | T. dimidiata | El Zapote, Yoro | T.dim-H9 | 496 | 75.81 | |
| 50 | T. dimidiata | Subirama, Yoro | T.dim-H9 | 496 | 75.81 | |
| 50 | T. dimidiata | Subirama, Yoro | T.dim-H29 | 494 | 75.71 | |
| 52 | T. dimidiata | El Paraiso, Yoro | T.dim-H29 | 494 | 75.71 | |
| NICARAGUA | 53 | T. dimidiata | Madriz | T.dim-H7 | 497 | 75.65 |
| n = 1 | ||||||
| PANAMA | 54 | T. dimidiata | Boquete, Chiriqui (3) | T.dim-H16 | 497 | 76.06 |
| n = 4 | 54 | T. dimidiata | Boquete, Chiriqui | T.dim-H17 | 495 | 75.96 |
| COLOMBIA | 55 | T. dimidiata | Pore, Casanare | T.dim-H11 | 497 | 75.85 |
| n = 31 | 56 | T. dimidiata | Boavita, Boyacá (13) | T.dim-H11 | 497 | 75.85 |
| 57 | T. dimidiata | San Joaquín, Santander (3) | T.dim-H11 | 497 | 75.85 | |
| 58 | T. dimidiata | Com. Los Kuises, SNSM Magdalena | T.dim-H12 | 495 | 75.76 | |
| 56 | T. dimidiata | Boavita, Boyacá (4) | T.dim-H12 | 495 | 75.76 | |
| 59 | T. dimidiata | Sierra Nevada, Santa Marta (4) | T.dim-H12 | 495 | 75.76 | |
| 56 | T. dimidiata | Boavita, Boyacá | T.dim-H13 | 493 | 75.66 | |
| 60 | T. dimidiata | San Onofre, Sucre (insectary) (2) | T.dim-H14 | 497 | 76.06 | |
| 56 | T. dimidiata | Boavita, Boyacá | T.dim-H15 | 497 | 75.65 | |
| 61 | T. dimidiata | Providencia island | T.dim-H1 | 497 | 76.06 | |
| ECUADOR | 62 | T. dimidiata | Guayas, Guayaquil | T.dim-H5 | 497 | 75.85 |
| n = 3 | 63 | T. dimidiata | Cerro del Carmen, Guayas, Guayaquil | T.dim-H5 | 497 | 75.85 |
| 64 | T. dimidiata | Pedro Carbo, Guayaquil | T.dim-H6 | 496 | 76.01 | |
| 2) TRIATOMA BASSOLSAE : 1 sequence/2 specimens studied: | ||||||
| MEXICO | 65 | T. bassolsae | Acatlán, Puebla (2) | T.bas-H1 | 490 | 76.94 |
| n = 2 | ||||||
| 3) TRIATOMA BOLIVARI : 1 sequence/1 specimen studied: | ||||||
| MEXICO | 66 | T. bolivari | Oaxaca, Oaxaca | T.bol-H1 | 501 | 76.85 |
| 4) TRIATOMA HEGNERI : 2 sequences/5 specimens studied: | ||||||
| MEXICO | 67 | T. hegneri | Ruinas S.Gervasio, Cozumel isl., Q. Roo | T.heg-H1 | 496 | 75.81 |
| n = 5 | 68 | T. hegneri | Cedral, Cozumel isl., Quintana Roo (3) | T.heg-H1 | 496 | 75.81 |
| 68 | T. hegneri | Cedral, Cozumel isl., Quintana Roo | T.heg-H2 | 496 | 76.01 | |
| 5) TRIATOMA MEXICANA : 1 sequence/1 specimen studied: | ||||||
| MEXICO | 69 | T.mexicana | Itatlaxco, Hidalgo | T.mex-H1 | 492 | 75.61 |
| 6) TRIATOMA PALLIDIPENNIS : 1 sequence/3 specimens studied: | ||||||
| MEXICO | 70 | T. pallidipennis | Chalcatzingo, Morelos | T.pal-H1 | 491 | 76.98 |
| n = 3 | 71 | T. pallidipennis | San Gabriel, Jalisco | T.pal-H2 | 490 | 76.94 |
| 72 | T. pallidipennis | Tecalitlan, Jalisco | T.pal-H2 | 490 | 76.94 | |
| 7) TRIATOMA RYCKMANI : 1 sequence/2 specimens studied: | ||||||
| GUATEMALA | 73 | T. ryckmani | El Progreso, El Progreso (2) | T.ryc-H1 | 500 | 76.00 |
| n = 2 | ||||||
| PHYLLOSOMA GROUP: FLAVIDA COMPLEX | ||||||
| 8) TRIATOMA FLAVIDA : 1 sequence/4 specimens studied: | ||||||
| CUBA | 74 | T. flavida | Peninsula of Guanahacabibes (4) | T.fla-H1 | 493 | 78.70 |
| n = 4 | ||||||
| RUBROFASCIATA GROUP: RUBROFASCIATA COMPLEX: LECTICULARIA SUBCOMPLEX | ||||||
| 9) TRIATOMA GERSTAECKERI : 1 sequence/1 specimen studied: | ||||||
| MEXICO | 75 | T. gerstaeckeri | Tanchahuil, S. Luis Potosí | T.ger-H1 | 483 | 76.81 |
| 10) TRIATOMA RUBIDA : 1 sequence/2 specimens studied: | ||||||
| MEXICO | 76 | T. rubida | Mocorito, Nayarit | T.rub-H1 | 516 | 77.71 |
| n = 2 | 77 | T. rubida | San Martin, Jalisco | T.rub-H1 | 516 | 77.71 |
| RUBROFASCIATA GROUP: PROTRACTA COMPLEX | ||||||
| 11) TRIATOMA NITIDA : 1 sequence/1 specimen studied: | ||||||
| GUATEMALA | 78 | T. nitida | El Progreso, El Progreso | T.nit-H1 | 490 | 76.33 |
| INFESTANS GROUP: INFESTANS COMPLEX: MACULATA SUBCOMPLEX | ||||||
| 12) TRIATOMA MACULATA : 1 sequence/4 specimens studied: | ||||||
| COLOMBIA | 79 | T. maculata | Santa Marta, Magdalena (4) | T.mac-H1 | 488 | 78.28 |
| n = 4 | ||||||
| INFESTANS GROUP: INFESTANS COMPLEX: RUBROVARIA SUBCOMPLEX | ||||||
| 13) TRIATOMA ARTHURNEIVAI : 1 sequence/2 specimens studied: | ||||||
| BRAZIL | 80 | T.arthurneivai | Espirito Santo do Pinhal | T.art-H1 | 486 | 77.98 |
| n = 2 | Sao Paulo (Fiocruz) (2) | |||||
Figure 1. Geographical distribution of the sampling sites furnishing the triatomine materials.
Numbers correspond to sampling sites listed in Table 1. • = Triatoma dimidiata; ▴ = other Triatoma species studied.
Sequencing of rDNA ITS-2
For DNA extraction, one or two legs fixed in ethanol 70% from each specimen were used and processed individually, as previously described [5],[27]. Total DNA was isolated by standard techniques [28] and stored at −20°C until use. The complete ITS-2 fragment was PCR amplified using 4–6 µl of genomic DNA for each 50 µl reaction. Amplifications were generated in a Peltier thermal cycler (MJ Research, Watertown, MA, USA), by 30 cycles of 30 sec at 94°C, 30 sec at 50°C and 1 min at 72°C, preceded by 30 sec at 94°C and followed by 7 min at 72°C. PCR products were purified with Ultra Clean™ PCR Clean-up DNA Purification System (MoBio, Solana Beach, CA, USA) according to the manufacturer's protocol and resuspended in 50 µl of 10 mM TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.6). Sequencing was performed on both strands by the dideoxy chain-termination method, and with the Taq dye-terminator chemistry kit for ABI 3730 and ABI 3700 capillary system (Perkin Elmer, Foster City, CA, USA), using the same amplification PCR primers [6].
Triatomine haplotype code nomenclature
The haplotype (H) terminology used in the present paper follows the nomenclature for composite haplotyping (CH) recently proposed [25]. Accordingly, ITS-2 haplotypes (H) are noted by numbers (Table 1).
Sequence alignment
Sequences were aligned using CLUSTAL-W version 1.83 [29] and MEGA 3.1 [30], and assembly was made with the Staden Package [31]. The alignment was carried out using the Central, Meso and South American Triatoma species studied together with other species and populations whose sequences are available in GenBank: T. phyllosoma (Accession Number AJ286881), T. pallidipennis (AJ286882), T. longipennis (AJ286883), T. picturata (AJ286884), and T. mazzotti (AJ286885) (Phyllosoma group, Phyllosoma complex); T. barberi (AJ293590) (Rubrofasciata group, Protracta complex) [5],[6]; T. rubrovaria H1 (AJ557258) [32], T. infestans CH1A (AJ576051), and T. sordida (AJ576063) [25]. The ITS-2 sequence of Rhodnius prolixus (Triatominae: Rhodniini) (AJ286882) [6] was used as outgroup.
Data deposition footnote
The GenBank (http://www.ncbi.nlm.nih.gov/Genbank) accession numbers for the new ITS-2 rDNA sequences discussed in this paper are: 31 haplotypes of T. dimidiata (AM286693–AM286723), T. bassolssae AM286724, T. bolivari (AM286725), 2 haplotypes of T. hegneri (AM286726, AM286727), T. mexicana (AM286728), 2 haplotypes of T. pallidipennis (AM286729, AM286730), T. ryckmani (AM286731), T. flavida (AM286732), T. gerstaeckeri (AM286734), T. rubida (AM286735), T. nitida (AM286733), T. maculata (AJ582027), and T. arthurneivai (AM286736).
Phylogenetic inference
Phylogenies were inferred by maximum-likelihood (ML) using PAUP*4.0b10 [33] and PHYMLv2.4.4 [34]. Maximum-likelihood parameters and the evolutionary model were determined using the hierarchical Likelihood Ratio Test (hLRTs) and the Akaike Information Criterion (AIC) [35],[36] implemented in Modeltest 3.7 [37] in conjunction with PAUP*4b10. To assess the reliability of the nodes in the ML tree, a bootstrap analysis using 1000 pseudo-replicates was made with PHYML. Since haplotype sequences for T. dimidiata individuals (populations) are quite similar and potentially subject to homoplasy and recombination, alternative procedures to phylogenetic tree reconstruction revealing their relationships were tested. Therefore, a median-joining network analysis [38] was performed using Network version 4.1.1.2 (available from Fluxus Technology Ltd., http://www.fluxus-engineering.com) with the variable positions in the multiple alignment of the different ITS-2 haplotypes from T. dimidiata populations.
Alternative methods of phylogenetic reconstruction allowing an evaluation of the support for each node were also applied. A distance-based phylogeny using the neighbor-joining (NJ) algorithm [39] with the ML pairwise distances was obtained. Statistical support for the nodes was evaluated with 1000 bootstrap replicates, with and without removal of gapped positions. Finally, a Bayesian phylogeny reconstruction procedure was applied to obtain posterior probabilities (BPP) for the nodes in the ML tree. We used the same evolutionary model as above implemented in MrBayes 3.1 [40] with four chains during 1,000,000 generations and trees were sampled every 100 generations. The last 9,000 trees were used to obtain the consensus tree and posterior probabilities.
Genetic variation studies
Genetic variation within and among populations of T. dimidiata was evaluated using DnaSP version 4 [41] and Arlequin 2000 [42]. Summary parameters include those based on the frequency of variants (haplotype number and diversity) as well as some taking genetic differences among variants into account (gene diversity, polymorphic sites). A hierarchical analysis of molecular variance (AMOVA) was performed using Arlequin. This analysis provides estimates of variance components and F-statistics [43] analogs reflecting the correlation of haplotype diversity at different levels of hierarchical subdivision. Unlike other approaches for partitioning genetic variation based on the analysis of variance of gene frequencies, AMOVA takes into account the genetic relatedness between molecular haplotypes. The hierarchical subdivision was made at three levels. At the top level, different groups were defined on the basis of the phylogenetic relationships for the different T. dimidiata haplotypes obtained. The second level corresponded to countries of sampling within each of these groups, and the third level corresponded to the different haplotypes found in each country within group. AMOVA reports components of variance at the three levels under consideration (among groups, among countries within groups, and within countries within groups) as well as F-statistics analogs. Under the present scheme, FST is viewed as the correlation of random haplotypes within countries within groups, relative to that of random pairs of haplotypes drawn from the whole species, FCT as the correlation of random haplotypes within groups, relative to that of random pairs of haplotypes drawn from the whole species, and FSC as the correlation of the molecular diversity of random haplotypes within countries within groups, relative to that of random pairs of haplotypes drawn from the corresponding group [44]. Although in the program used (only currently available for molecular variance analysis) the choice for establishing an intermediate level is fully arbitrary and has no influence on the final result of the comparison between units at the higher level, these same analyses were repeated by considering each haplotype, which may encompass several individuals, as a separate group for this intermediate level, because it could be argued that geopolitical country borders was not an appropriate choice despite its interest from the point of view of the control of Chagas disease. The statistical significance of fixation indices was tested using a non-parametric permutation approach [44]. Genetic differentiation between pairs of populations was evaluated by means of F-statistics [43]. Exact tests of population differentiation were performed [45]. Slatkin's linearized FST's [46],[47] procedure was also followed to obtain estimates of pairwise equilibrium migration rates, both among groups, among countries within groups, and within countries for those cases in which haplotypes from more than one group were present.
Results
Sequence Analyses of Triatoma dimidiata Populations
The 137 ITS-2 sequences revealed the existence of 31 different haplotypes in the T. dimidiata studied (T.dim-H1 to T.dim-H31) (see Tables 1 and 2 for localities and countries). Their length was 489–497 base pairs (bp) (mean, 495.10) with a relative AT-biased nucleotide composition of 75.25–76.85% (75.72%). Sequence similarity analysis of these 31 haplotypes revealed four distinct groupings: grouping 1 (T.dim-H1 to T.dim-H10); grouping 2 (T.dim-H11 to T.dim-H17); grouping 3 (T.dim-H18 to T. dim-H24); and grouping 4 (T. dim-H25 to T. dim-H31) (Figure 2). These four groupings appear linked to concrete wide geographical areas including neighboring countries and regions. The only exception is Providencia Island, which, although part of Colombia, is located 720 km off the northern coast of Colombia but only 240 km off the western coast of Nicaragua. No haplotype presents a very broad geographical distribution.
Table 2. Distribution of Triatoma dimidiata ITS-2 haplotypes (H) per country and locality.
| Country | Locality | Sample | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H | H |
| size | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | ||
| COLOMBIA | Dpto. Casanare | 1 | 1 | ||||||||||||||||||||||||||||||
| (n = 31) | Dpto. Boyaca | 19 | 13 | 4 | 1 | 1 | |||||||||||||||||||||||||||
| Dpto. Santander | 3 | 3 | |||||||||||||||||||||||||||||||
| Santa Marta | 5 | 5 | |||||||||||||||||||||||||||||||
| Sucre | 2 | 2 | |||||||||||||||||||||||||||||||
| Providencia Isl. | 1 | 1 | |||||||||||||||||||||||||||||||
| PANAMA (n = 4) | Chiriqui | 4 | 3 | 1 | |||||||||||||||||||||||||||||
| MEXICO | Veracruz | 7 | 4 | 2 | 1 | ||||||||||||||||||||||||||||
| (n = 41) | Oaxaca | 4 | 1 | 3 | |||||||||||||||||||||||||||||
| Morelos | 2 | 1 | 1 | ||||||||||||||||||||||||||||||
| San Luis Potosi | 1 | 2 | |||||||||||||||||||||||||||||||
| Hidalgo | 6 | 5 | 1 | ||||||||||||||||||||||||||||||
| Tabasco | 1 | 1 | |||||||||||||||||||||||||||||||
| Colima | 1 | 1 | |||||||||||||||||||||||||||||||
| Chiapas | 3 | 1 | 1 | 1 | |||||||||||||||||||||||||||||
| Yucatan | 10 | 3 | 1 | 1 | 4 | 1 | |||||||||||||||||||||||||||
| Cozumel Isl. | 4 | 1 | 3 | ||||||||||||||||||||||||||||||
| Holbox Isl. | 1 | 1 | |||||||||||||||||||||||||||||||
| HONDURAS | Yoro Yoro | 13 | 8 | 3 | 2 | ||||||||||||||||||||||||||||
| (n = 20) | El Porvenir | 2 | 2 | ||||||||||||||||||||||||||||||
| Orica | 1 | 1 | |||||||||||||||||||||||||||||||
| Tegucigalpa | 2 | 2 | |||||||||||||||||||||||||||||||
| Ginope | 1 | 1 | |||||||||||||||||||||||||||||||
| San Jose | 1 | 1 | |||||||||||||||||||||||||||||||
| ECUADOR (n = 3) | Guayaquil | 3 | 2 | 1 | |||||||||||||||||||||||||||||
| NICARAGUA (n = 1) | Madriz | 1 | 1 | ||||||||||||||||||||||||||||||
| GUATEMALA | Agua Sarca | 4 | 1 | 1 | 2 | ||||||||||||||||||||||||||||
| (n = 37) | Esquintla | 4 | 3 | 1 | |||||||||||||||||||||||||||||
| Quiche | 4 | 1 | 1 | 2 | |||||||||||||||||||||||||||||
| Pueblo Nuevo | 4 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||
| Peten | 9 | 2 | 1 | 5 | 1 | ||||||||||||||||||||||||||||
| Jutiapa | 8 | 7 | 1 | ||||||||||||||||||||||||||||||
| Lanquin | 4 | 4 | |||||||||||||||||||||||||||||||
| TOTAL | 137 | 12 | 19 | 5 | 1 | 2 | 2 | 1 | 2 | 3 | 4 | 17 | 9 | 1 | 2 | 1 | 3 | 1 | 17 | 2 | 1 | 2 | 1 | 4 | 1 | 5 | 1 | 1 | 13 | 2 | 1 | 1 |
Sequence groupings: grouping 1 (T.dim-H1 to T.dim-H10); grouping 2 (T.dim-H11 to T.dim-H17); grouping 3 (T.dim-H18 to T.dim-H24); and grouping 4 (T.dim-H25 to T.dim-H31).
Figure 2. Interhaplotype sequence differences found in the rDNA ITS-2 of the Triatoma dimidiata populations analyzed.
Numbers (to be read in vertical) refer to positions obtained in the alignments made with CLUSTAL-W 1.8 and MEGA 3.3. . = identical; * = singelton sites (7); • = parsimony informative positions (24); − = insertion/deletion. Rectangled area = microsatellite region. Horizontal lines separate the four major T. dimidiata haplotype groupings according to sequence analyses.
The alignment of the 31 T. dimidiata haplotype sequences was 501 bp-long, of which 450 characters were constant and 24 were parsimony-informative. The interrupted microsatellite (AT)4–5 TTT (AT)5–7 was detected between positions 47 and 73 in all specimens studied. Variability in this microsatellite region and their respective sequence positions are noted in Figure 2.
The 51 nucleotide variable positions detected including gaps represented a 10.18% of polymorphic sites. The seven haplotypes T.dim-H25 to T.dim-H31 are responsible for this high genetic divergence (Figure 2). This genetic divergence decreases considerably when two separate alignments are performed: (i) the first includes T.dim-H1 to T.dim-H24 from all the seven countries shows a divergence of 5.62% in a 498-bp-long alignment, including 28 nucleotide variable positions, of which 6 (1.20%) were transitions (ti), 13 (2.61%) transversions (tv) and 9 (1.81%) insertions/deletions (indels); (ii) the second includes T.dim-H25 to T.dim-H31 from only three countries (Mexico: localities of Yucatan, Chiapas, Cozumel Island and Holbox Island; Guatemala: Peten; Honduras: Yoro Yoro) shows a divergence of 2.42% in a 495-bp-long alignment, with 12 nucleotide variable positions, of which 2 ti (0.40%) and 10 are indels (2.02%).
Sequence Analyses in the Phyllosoma and Rubrofasciata Groups
ITS-2 sequences of T. bassolsae, T. bolivari, T. hegneri, T. mexicana, T. pallidipennis, T. ryckmani, T. flavida, T. nitida, T. gerstaeckeri, and T. rubida, including haplotype length and AT content are listed in Table 1. The comparison analyses which include these ITS-2 sequences and those of the Phyllosoma and Rubrofasciata groups (available in GenBank) provided 48 different haplotypes. Their alignment resulted in a total of 551 characters including gaps, of which 365 sites were constant and 99 parsimony-informative.
All the T. dimidiata haplotypes clearly differed from the Phyllosoma, Flavida, Protacta and Rubrofasciata complex species included in this analysis. Triatoma bassolsae differed in only one deletion in position 489 from T. pallidipennis of Morelos, Mexico (AJ286882). The T. pallidipennis sequence obtained represents a new haplotype (T.pal-H2) differing in only one deletion in position 31 from T. picturata and T. longipennis. The haplotype alignment of T. bassolsae, T. longipennis, T. mazzotti, T. picturata, T. pallidipennis and T. phyllosoma was 490 bp long showing a relatively small genetic diversity of 1.83%, with only 5 mutations (1.02%) and 4 indels (0.81%). The two T. hegneri haplotypes differ between each other in only 1 ti and, when compared with T. dimidiata H18 to H24 from Mexico and Guatemala, nucleotide differences found were only 1 ti and 2 tv.
Sequence Analyses in the Infestans Group
ITS-2 sequences of T. maculata and T arthurneivai, including haplotype length and AT content are listed in Table 1.
The ITS-2 of T. maculata fits very well within sequences of the Infestans complex species studied in the present work, a total of 6–19 (13.7) mutations, namely 6–11 (7.25) ti and 0–10 (6.5) tv, appearing when comparing the five Infestans complex species in question. The material of Triatoma arthurneivai here analyzed is very close to T. rubrovaria H1 (AJ557258), showing only 6 nucleotide differences (1.22%), of which only 1 ti and 5 indels.
Phylogenetic Analyses
Two different phylogenetic approaches were performed with the 31 T. dimidiata haplotypes, both yielding coincident results. A maximum likelihood tree was reconstructed using the best model of evolution as determined by the lowest AIC, which was GTR+I (−Ln = 887.089), being the proportion of invariable sites (I) of 0.166. Three groups appeared with high support values indicating that their differentiation was not due to random sampling of a low variable sequence (tree not shown). The large group 1 encompassed haplotypes from all the countries, whereas groups 2 (Mexico and Guatemala) and 3 (Mexico, Guatemala and Honduras) were more geographically restricted.
Alternatively, a median-joining network was reconstructed with the 31 different T. dimidiata sequences using the variable sites in the multiple alignment (Figure 3). This network showed the same three groups found in the ML tree. Group 1 occupies a central position in the network and is the most widespread and variable group, so that it most likely corresponds to the ancestral or source set. This is further reinforced by the direct relationship between this group and the two others, more geographically restricted and encompassing fewer variants, group 2 including samples from Mexico and Guatemala, and group 3 including samples from these two countries and Honduras. The group 1 source set would in turn be derived from group 3, which might be interpreted as a geographically restricted relict according to the phylogeographic results. Moreover, sequence variants in group 1 are clustered in two different subgroups, with genetic and geographical borders: subgroup 1A includes sequences from Colombian Providencia island, Ecuador, Guatemala, Honduras, Mexico (only South of Chiapas) and Nicaragua; subgroup 1B encompasses sequences from continental Colombia and Panama. The two closest sequences of each subgroup differ in two sites, which might correspond to haplotypes not found in this sampling.
Figure 3. Median network for Triatoma dimidiata haplotypes based on rDNA ITS-2 sequences.
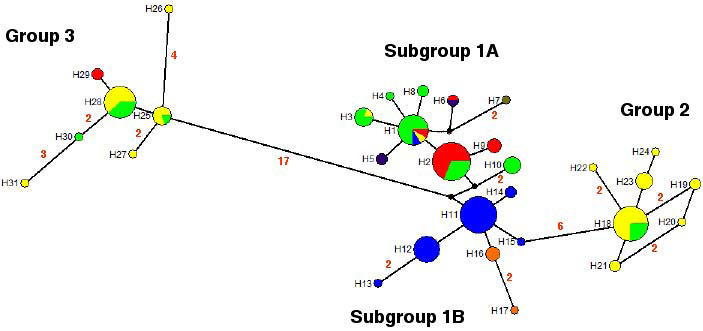
The area of each haplotype is proportional to the total sample. Small black-filled circles represent haplotypes not present in the sample. Mutational steps between haplotypes are represented by a line. More than one mutational step is represented by numbers. H = haplotype. Blue: Colombia; orange: Panama; yellow: Mexico; red: Honduras; lilac: Ecuador; ocher: Nicaragua; green: Guatemala.
The relevance of the ITS-2 differences among these T. dimidiata groups and subgroups was assessed by comparison with other Triatoma species. Therefore, a multiple, 562-nucleotide-long alignment was obtained by incorporating 22 additional ITS-2 sequences. This set includes 53 ITS-2 sequences of Triatoma species and, using R. prolixus as outgroup, a ML tree was obtained (−Ln = 2648.5129) using the HKY+G model, according to the AIC results with a gamma distribution shape parameter = 0.58. This tree (Figure 4) shows that:
Figure 4. Phylogenetic ML tree of Triatoma species and haplotypes within the Phyllosoma, Rubrofasciata and Infestans groups.
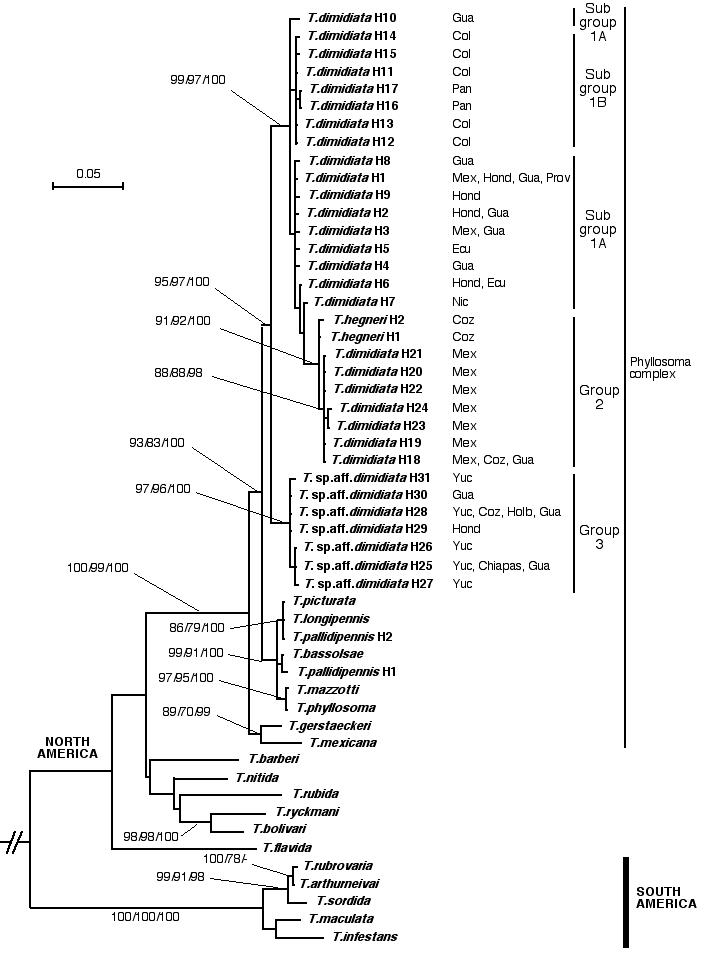
The scale bar indicates the number of substitutions per sequence position. Support for nodes a/b/c: a: bootstrap with ML reconstruction using PhyML with 1000 replicates; values larger than 70%; b: bootstrap with NJ reconstruction using PAUP with ML distance and 1000 replicates; values larger than 70%; c: Bayesian posterior probability with ML model using MrBayes; values larger than 90%.
the 31 T. dimidiata haplotypes appear within a highly supported clade (95/97/100 in ML/NJ/BPP), distributed as follows: a first large subclade, also very well supported (99/97/100), comprising subgroup 1A, subgroup 1B, group 2, and group 3 of the network analysis; subgroup 1A (sequence grouping 1 = T.dim-H1 to T.dim-H10) includes populations from Central America (Honduras, Nicaragua, Guatemala and scattered haplotypes from Mexico, Ecuador and Providence Island); interestingly, the haplotype T.dim-H10 corresponding to phenetically peculiar specimens found in cave-dwellings of Lanquin, Guatemala, appears independent although related to the rest with very high supports; subgroup 1B (sequence grouping 2 = T.dim-H11 to T.dim-H17) comprises populations from continental Colombia and Panama and appears as a monophyletic haplotype cluster; group 2 (sequence grouping 3 = T.dim-H18 to T.dim-H24) shows a well supported branch (91/92/100) and comprises populations from Mexico (Gulf coast, high plains, and Cozumel island) and Guatemala, including the two T. hegneri haplotypes; the second large clade is also highly supported (97/96/100), corresponding to group 3 (sequence grouping 4 = T.dim-H25 to T.dim-H31) and includes populations from the Yucatan peninsula, Holbox and Cozumel islands and northern Chiapas (Mexico), northern Honduras and northern Guatemala;
T. bassolsae clusters together with T. phyllosoma, T. mazzotti, T. longipennis, T. picturata and T. pallidipennis with very high support (99/91/100 in ML/NJ/BPP) in a sister clade of T. dimidiata; the separated location of the two T. pallidipennis haplotypes indicates the marked similarity of all these taxa;
T. mexicana and T. gerstaeckeri cluster together in a group basal to both T. dimidiata and T. phyllosoma clades; the extremely high values (100/99/100) supporting the monophyletic clade including T. mexicana, T. gerstaeckeri, T. phyllosoma and close species, and T. dimidiata, are worth emphasizing;
T. barberi, T. nitida, T. rubida, T. ryckmani and T. bolivari cluster in an unresolved branch, within which only T. ryckmani and T. bolivari appear related with a high support; the insular species T. flavida from Cuba appears as a basal lineage although with insufficient support values;
finally, the South American species T. rubrovaria, T. arthurneivai, T. sordida, T. maculata and T. infestans cluster together with the highest support.
Triatoma dimidiata groupings appeared well supported, with very high bootstrap proportions (BP>90%) using ML and neighbor-joining reconstruction and the highest Bayesian posterior probabilities (BPP = 100%). Similar levels were found for other well established Triatoma species, many of which showed substantially lower support values in the three statistical measurements employed. However, other species presented no ITS-2 nucleotide differences (T. picturata and T. longipennis; T. mazzotti and T. phyllosoma).
Genetic Variation Analyses
The phylogenetic analyses showed that samples from the same country may belong to different clusters. This result, on its own, is not enough to demonstrate the biological distinctiveness of the corresponding populations. Sampled individuals may represent a minor fraction of the total genetic variability in a highly heterogeneous population and the sampling procedure might have resulted, by pure chance, in the observed clustering of some variants. Given that each of these clusters holds some genetic variability of its own, the first task was to evaluate whether the observed groupings were significantly different from each other, in terms of genetic variation, by partitioning the observed genetic variability at three different levels: among groups, among populations (countries) within groups, and within populations. A hierarchical analysis of molecular variance was used to test the null hypothesis of no genetic differentiation among groups considering variation at lower levels. This procedure was first applied to T. dimidiata sequences using three levels as defined above (Table 3a). Most of the genetic variation found was allocated to the among groups level (80.24% of the total variation), with much lower portions of variation assigned to differences among populations within groups level (11.71%) and within populations level (8.05%), although both were still statistically significant after 1000 pseudo-random samples generated for testing. This indicates that, despite genetic variation within and among populations at these three levels, there is a substantial amount of genetic differentiation among them that justifies their consideration as separate groupings for further analysis. The same results were obtained, notwithstanding small numerical differences due to the different numbers of groups, when haplotypes instead of countries were considered at the intermediate level (Table S1). The geographical fitting represents in fact no surprise at all, taking into account that the distribution of T. dimidiata covers different countries which are more or less aligned following a north-south axis because of the relatively slenderness of the Central American bridge. Hence, as any of the two versions of the analyses conveys the same information and leads to the same conclusions, and which one should be reported is simply a matter of opinion, the first considering countries becomes practically more useful because Chagas disease control measures are organized at national level.
Table 3. Summary of analysis of molecular variance for Triatoma dimidiata populations.
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation | Fixation Indices |
| a) | |||||
| Among groups | 2 | 528.273 | 6.732 Va | 80.24 | FCT = 0.802*** |
| Among populations within groups | 10 | 86.820 | 0.982 Vb | 11.71 | FST = 0.920*** |
| Within populations | 123 | 83.047 | 0.675 Vc | 8.05 | FSC = 0.593*** |
| Total | 135 | 698.140 | 8.389 | ||
| b) | |||||
| Among groups | 1 | 68.257 | 1.4785 | 60.15 | FCT = 0.602* |
| Among populations within groups | 6 | 15.547 | 0.3007 | 12.23 | FST = 0.724*** |
| Within populations | 77 | 52.267 | 0.6788 | 27.62 | FSC = 0.307*** |
| Total | 84 | 136.071 | 2.4580 | ||
| c) | |||||
| Among groups | 3 | 596.530 | 5.890 | 86.84 | FCT = 0.868*** |
| Among populations within groups | 9 | 18.563 | 0.218 | 3.21 | FST = 0.900*** |
| Within populations | 123 | 83.047 | 0.675 | 9.95 | FSC = 0.244*** |
| Total | 135 | 698.140 | 6.783 | ||
(a) Three groups (1, 2, and 3), (b) two subgroups (1A vs 1B), and (c) four groups/subgroups (1A, 1B, 2 and 3) were considered as indicated in the text. Populations within groups correspond to countries of sampling. ***: P<0.001; **: P<0.01. d.f. = degrees of freedom.
The median-joining network reconstructed with the 31 different T. dimidiata ITS-2 sequences revealed the existence of three distinct groups (groups 1, 2 and 3), the first of which further subdivided into two subgroups 1A and 1B. The same AMOVA procedure was applied to ascertain whether these two subgroups could be considered as distinct populations or not. The results (Table 3b) indicate that a significant fraction (60.15%) of the total genetic variation corresponds to differences between these two subgroups which, correspondingly, could be considered as separate populations for the ensuing analyses.
Based on the four groups/subgroups previously described in the median-joining network, a summary of relevant population genetic parameters for T. dimidiata is presented in Table 4. Genetic variation in T. dimidiata populations was quite evenly distributed, with similar levels of nucleotide and haplotype diversities in the four groups/subgroups considered. Nevertheless, for all the parameters studied, subgroup 1A presented higher values than the rest, although significance of the differences was only obtained for haplotype diversity. A similar summary is shown for each country sample within groups in Table S2.
Table 4. Summary of population genetic variation parameters from ITS-2 haplotypes in the Triatoma dimidiata populations.
| Parameter | Group1 | Subgroup1A | Subgroup1B | Group2 | Group3 |
| Gene copies | 85 | 51 | 34 | 27 | 24 |
| Haplotypes | 17 | 10 | 7 | 7 | 7 |
| Polymorphic sites | 23 | 13 | 9 | 7 | 11 |
| Hap. diversity | 0.8782 | 0.797 | 0.686 | 0.6353 | 0.6775 |
| Std. error | 0.0178 | 0.040 | 0.065 | 0.0972 | 0.0902 |
| Pairwise diff. mean | 3.2398 | 1.707 | 1.524 | 1.1510 | 1.6377 |
| Std. error | 1.6872 | 1.014 | 0.938 | 0.7670 | 1.0007 |
| Nucleot diversity | 0.0065 | 0.003 | 0.003 | 0.0023 | 0.0033 |
| Std. error | 0.0037 | 0.002 | 0.002 | 0.0017 | 0.0023 |
| θ (Het) | 6.0371 | 3.105 | 1.668 | 1.3162 | 1.5990 |
| S.D. θ (Het) | 1.1075 | 0.822 | 0.523 | 0.5710 | 0.6892 |
| θ (k) | 6.1156 | 3.444 | 2.385 | 2.7281 | 2.9510 |
| 95 % C.I. for θ (k) | 3.476,10.432 | 1.668,6.785 | 1.009,5.308 | 1.134,6.223 | 1.213,6.838 |
| θ (S) | 3.1911 | 2.445 | 1.223 | 0.5189 | 0.8034 |
| S.D. θ (S) | 1.1040 | 0.976 | 0.636 | 0.3844 | 0.5094 |
| θ (π) | 3.2398 | 1.707 | 1.524 | 1.1510 | 1.6377 |
| S.D. θ (π) | 1.8694 | 1.125 | 1.043 | 0.8553 | 1.1155 |
| Tajima's D | −1.261ns | −1.572* | −1.553* | −0.536ns | −0.6435ns |
| Ewens-Watterson | 0.132ns | 0.219ns | 0.334ns | 0.388ns | 0.3507ns |
| Fu's Fs | −3.401ns | −2.601ns | −1.111ns | −2.426* | −1.4665ns |
θ = effective mutation rate estimated from equilibrium heterozygosity [θ(Het)], number of alleles [θ(k)], number of polymorphic sites [θ(S)] and nucleotide diversity [θ(π)]. The last 3 rows correspond to different statistics of neutrality at the population level. S.D. = standard deviation; C.I. = confidence interval. NS: P>0.05; * = P<0.05.
Different estimates of θ were obtained based on the expected heterozygosity, the expected number of alleles, the number of polymorphic sites and the nucleotide diversity. The four estimates were quite consistent for the four groups/subgroups and they agreed in assigning a larger value to subgroup 1A.
Differences in the genetic composition of the four groups/subgroups 1A, 1B, 2 and 3 have previously been shown to be statistically significant according to analyses of molecular variance. A further evaluation of this distinctiveness was made (Table 3c), in which the four groups/subgroups were considered for the AMOVA, in correspondence with the previous results. In this case, the amount of among-group variation rose to 86.84% of the total variation, whereas among population within groups and within population levels they were substantially lower, 3.21% and 9.95% respectively.
Genetic differences within and among the ITS-2 locus for T. dimidiata samples were further explored through pairwise comparisons, and estimates of average pairwise differences within and among the four groups/subgroups considered were obtained (Table 5). Subgroup 1A presented the largest value for within-group pairwise differences. The within-population values were much lower than among-populations comparisons. Among the latter, the smallest number of differences was found between subgroup 1A and 1B, in correspondence with their close phylogenetic relationship. Subgroup 1B was the one with the lowest overall number of pairwise differences, slightly below 1A. On the contrary, the highest value of pairwise differentiation corresponds to group 3, with almost 20 differences (corrected estimate) when compared with any other group.
Table 5. Population average pairwise differences in Triatoma dimidiata populations.
| Group 1 | Subgroup1A | Subgroup1B | Group2 | Group3 | |
| Group 1 | 3.240 | - | - | 9.953 | 20.719 |
| Subgroup1A | - | 1.707 | 4.922 | 10.325 | 21.118 |
| Subgroup1B | - | 3.307 | 1.524 | 9.397 | 20.120 |
| Group2 | 7.758 | 8.896 | 8.059 | 1.151 | 26.875 |
| Group3 | 18.280 | 19.446 | 18.539 | 25.481 | 1.638 |
Above diagonal: Average number of pairwise differences between populations (πXY). Diagonal elements: average number of pairwise differences within population (πX). Below diagonal: corrected average pairwise difference (πXY−(πX+πY)/2).
Within groups genetic differentiation was evaluated by computation of pairwise FST values for populations defined by country of origin (Table S3). Since all groups/subgroups, with the only exception of subgroup 1A, are characterized by one large (n>10) and several small (n<10) populations, significance values for test of genetic differentiation have to be interpreted cautiously. Hence, there is no apparent differentiation between two populations in subgroup 1B (Colombia2, n = 30, and Panama, n = 4) and similarly in group 2 (Mexico2, n = 23, and Guatemala2, n = 4). The only significant value found in group 3 corresponds to Honduras3 (n = 2) and Guatemala3 (n = 7), for which FST = 0.529, P<0.05. None of these two populations presented significant differentiation with respect to the largest population in this group, Mexico3 (n = 15). Subgroup 1A includes two large populations, Honduras1 (n = 18) and Guatemala1 (n = 26), which presented a highly significant FST = 0.193, P<0.001. Although this value, under the assumption of migration-drift equilibrium, corresponds to an estimate of 2.1 migrants per generation between both populations, which would be enough to prevent their complete differentiation, such estimations shall be verified by using larger samples and markers better suited for population genetics analyses. Comparisons between each of these two populations and the smaller ones in subgroup 1A revealed that Honduras1 differed from Mexico1, Guatemala1 was different from Ecuador and Nicaragua, and none of them differed from the only two individuals from Providencia island. Similar comparisons for all pairs of populations assigned to different groups/subgroups resulted in highly significant FST values (Table S4).
Discussion
Triatoma dimidiata, T. sp. aff. dimidiata and T. hegneri
The highest intraspecific ITS-2 variability (absolute nucleotide differences including indels) known in Triatomini members is 2.70% (13/482) in T. infestans specimens collected throughout the very wide geographical distribution of this species [25]. Hence, the result of 10.18% ( = 51/501) detected in T. dimidiata (Figure 2) appears to be pronouncedly outside the limits of the intraspecific variability range known for Triatoma species. Group 3 is the main responsible for such differences (Table 5) and shows a high 2.42% divergence within itself, suggesting an old origin in the light of the relatively reduced geographical area of distribution of these haplotypes in Mexico (Yucatan, Chiapas, Cozumel Island and Holbox Island), Guatemala (Peten) and Honduras (Yoro) only. The time of divergence between group 3 and other T. dimidiata populations was estimated to be of 5.9–10.5 million years ago (Mya) according to a molecular clock analysis based on rDNA evolutionary rates [4].
The divergence of 5.62% shown by the other 24 ITS-2 haplotypes (Figure 2) also appears to be too large, in spite of the wide geographical area they occupy from Mexico down to Ecuador, suggesting a speciation process. However, population average pairwise differences between subgroup 1A, subgroup 1B and group 2 are markedly lower than between these three and group 3 (Table 5), and intragroup differences do fall within the above-mentioned Triatomini range: 2.61% within subgroup 1A, 2.41% within subgroup 1B, and 2.01% within group 2.
Results indicate that several T. dimidiata populations are following different evolutionary divergences in which geographical isolation appears to have had an important influence (Figure 5). A phenotypic consequence of that process had been observed by other specialists before, who wrote about an assemblage of morphologically variable populations [10]. More recently, significant head shape differences between populations showed a separation between northern, intermediate and southern collections of T. dimidiata and also support an evolutionary divergence of populations within this species [13].
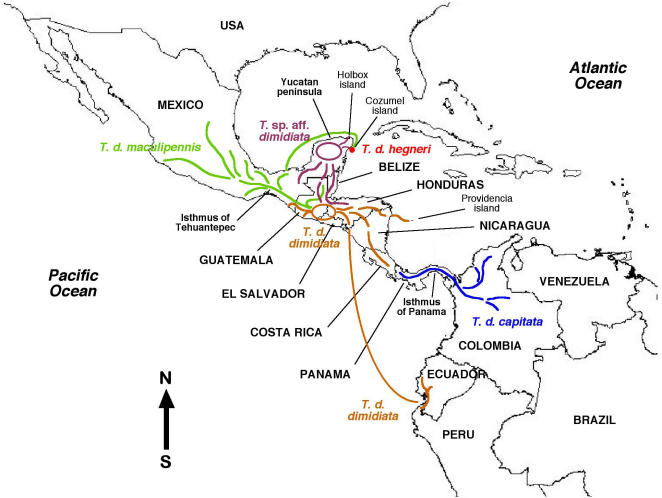
Figure 5. Phylogeography of Triatoma dimidiata sensu lato.
Distribution and spreading routes of T. d. dimidiata, T. d. capitata, T. d. maculipennis, T. d. hegneri and Triatoma sp. aff. dimidiata in Mesoamerica, Central America and the northwestern part of South America are represented according to network analyses and genetic variation studies based on rDNA ITS-2 sequences.
Three subspecies were distinguished on the basis of morphological differences [48],[49]: (i) T. d. dimidiata concerns the first description of the species in Peru (no type specimen available; no type locality assigned, but undoubtedly from northern Peru, probably around the locality of Tumbes, near Ecuador) and corresponds to most of the Central American forms; (ii) T. d. maculipennis was proposed for specimens from Mexico (type specimen in Zoologisches Museum Berlin) and corresponds to forms with relatively short heads and large eyes; and (iii) T. d. capitata was proposed for large size specimens typified by longer heads and smaller eyes originally found in Colombia (type specimen in the Academy of Sciences of California). However, these subspecies became later synonymized after results of a morphological re-examination which were interpreted as evidence of a clinal variation along a north-south axis [50],[51].
Present ITS-2 sequences and corresponding phylogenetic and genetic variation analyses support the appropriateness to (i) differentiate group 3 as a species of its own (here simply designed as T. sp. aff. dimidiata to avoid further systematic confusion with T. dimidiata, according to taxonomic rules), and (ii) re-assign subspecific status for subgroup 1A, subgroup 1B and group 2. Results of the present study do not support the rise of the above-mentioned subspecific taxa to species level for the time being, although it is evident that in the three cases relatively long divergence processes have taken place. Similar genetic studies with other molecular markers may contribute to a more complete assessment of these evolutionary isolation and speciation processes.
The taxon T. sp. aff. dimidiata concerns group 3. This species seems to represent a relatively relict species with a distribution restricted to the Mexican flat areas of the Yucatan peninsula and the northern part of Chiapas state, the northern lowland of Guatemala (and probably also Belize), and only one altitude-adapted haplotype (T.dim-H29) in its most extreme border populations in northern Honduras. The most widely spread haplotype T.dim-H28 is also present in the small island of Holbox and the large island of Cozumel, both near the Yucatan coast, suggesting that this haplotype should be considered the oldest of this species. This species is also of public health importance because of its capacity to transmit Chagas disease [52],[53] and the control problems it poses [54],[55].
The taxon T. d. dimidiata corresponds to subgroup 1A and populations mainly from Guatemala and Honduras and secondarily Mexico, Nicaragua and Ecuador. The population of the Colombian island of Providence undoubtedly derives from the most widely dispersed haplotype T.dim-H1 on the nearest Caribbean coastal area of Central America and not from continental Colombia. The present populations in Ecuador may derive from introduced specimens originally from the Guatemala-Honduras-Nicaragua region, relatively recently introduced by humans [4], very probably in the period of the early colonialization of northwestern South America by the Spanish ‘conquistadores’ in which exchange activities between Central American settlements and the Peruvian Tumbes area took place [56]. The type specimens of the original description of the species in northern Peru might also belong to populations derived from such man-made introductions from Central America. The haplotype T.dim-H10 of Lanquin, Alta Verapaz, Guatemala appears in the network analysis as directly derived from an ancestor which gave rise to the subspecies T. d. dimidiata. An isolation phenomenon in caves may explain the albinic characteristics of the specimens presenting this haplotype. These cavernicole specimens from Alta Verapaz have already shown their peculiarity in morphometric and cuticular hydrocarbon studies [13],[17].
The taxon T. d. capitata corresponds to subgroup 1B and populations from Colombia and Panama. The isthmus of Panama and the separation/joining process of South and North America towards the end of the Pliocene (3–5 Mya) [57], in a period in which several more or less closely separated islands appeared and evolved up to their fusion into the isthmus, should have played a major role in the isolation and subsequent divergence of these southernmost T. dimidiata populations. The lack of relationship between the haplotypes of Ecuador and those of Colombia is worth mentioning, as the geographical closeness of these two countries could have given rise to the erroneous hypothesis of Colombian forms having derived from Ecuadorian populations. In a recent study of three populations of sylvatic, peridomestic and domestic T. dimidiata from Colombia, the estimated low genetic distances based on RAPD analyses did not discriminate the populations studied, indicating that they maintain the genetic identity of a single recent common ancestor [9].
The taxon T. d. maculipennis corresponds to group 2 and populations mainly from Mexico, but rarely found in Guatemala. According to the network analysis, this subspecies seems to have derived from group 1 probably by isolation in the Mexican part northward from the isthmus of Tehuantepec. Similarly as for other organisms including insects [58], the mountainous Sierra Madre chain throughout southern Mexico and Guatemala areas near the Pacific coast probably played also a role in that isolation process through an area where T. sp. aff. dimidiata did not represent a competition barrier, as T. sp. aff. dimidiata appears to be preferentially a low altitude species in these two countries.
Southern Mexico (including the Yucatan peninsula and Chiapas state) and almost the whole country of Guatemala (at least ten departments) constitute a crucial evolutionary area, where a high number of taxa, including T. d. dimidiata, T. d. maculipennis, and T. sp. aff. dimidiata, overlap. In a morphometric analysis, populations from San Luis Potosi and Veracruz in Mexico were indistinguishable while clearly different from populations from Yucatan in Mexico and Peten in Guatemala [14]. The former correspond to T. d. maculipennis and the latter to T. sp. aff. dimidiata. In Guatemala, a high degree of genetic variation in T. dimidiata sensu lato was shown by RAPD-PCR [12], demonstrating a limited gene flow between different provinces, although barriers between the Atlantic and Pacific drainage slopes did not appear to be significant limiters of a gene flow, according to a hierarchical analysis.
Chromosome analyses and DNA genome size revealed the existence of three different cytotypes with different geographical distributions [18]: (i) cytotype 1 corresponds to three different taxa: T. d. maculipennis in Mexico (excluding Yucatán), T. d. dimidiata in Guatemala (excluding Petén) and probably also El Salvador; and T. d. capitata in Colombia; (ii) cytotype 2 was found in two localities (Paraiso and Chablekal) around Mérida, Yucatan, Mexico where the species T. sp. aff. dimidiata presents 5 different haplotypes (T.dim-H25, T.dim-H26, T.dim-H27, T.dim-H28 and T.dim-H31); (iii) cytotype 3 appeared in Yaxhá, Petén, Guatemala, where both T. d. maculipennis (T.dim-H18) and T. sp. aff. dimidiata (T.dim-H25, T.dim-H28 and T.dim-H30) are present. Sequencing of the same specimens studied [18] from Yaxhá showed that cytotype 3 was found in specimens of T. sp. aff. dimidiata of haplotype T.dim-H28 and T.dim-H30. Consequently, chromosomal cytotypes 2 and 3 are both found in T. sp. aff. dimidiata.
The two haplotypes of T. hegneri differ by only 3 mutations from haplotypes of T. d. maculipennis. This reduced number of nucleotide differences and the location of T. hegneri haplotypes within the clade of T. dimidiata, basal to haplotypes of group 2 (Figure 4), does not support its status as an independent species. The results obtained suggest that it is an insular form of T. d. maculipennis. Originally described from the island of Cozumel [3], a subspecific status T. d. hegneri could be maintained only if morphological characteristics allow a clear differentiation of the insular form, as the phylogenetic analysis somehow separates it in a very close but particular evolutionary line. Triatoma hegneri, although chromatically distinguishable from most forms of T. dimidiata [50], is known to produce fertile hybrids when experimentally crossed with T. dimidiata (R.E. Ryckman, unpublished). Interestingly, the most dispersed haplotypes of both T. d. maculipennis (T.dim-H18) and T. sp. aff. dimidiata (T.dim-H28) are also present on the same island, probably introduced through the intense human transport between the mainland and the island.
The distinction between T. d. dimidiata (subgroup 1A), T. d. capitata (subgroup 1B), T. d. maculipennis (group 2), T. sp. aff. dimidiata (group 3), and T. d. hegneri contributes giving systematic/taxonomic coherency to present knowledge about morphological and genetic concepts in these taxa. From an ancestral form close to T. sp. aff. dimidiata, it can be postulated that an original diversification focus of T. dimidiata forms took place most probably in Guatemala, with a southern spread into Panama and Colombia to give the capitata forms and a northwestern spread into Mexico to give the maculipennis forms (Figure 5). Thus, the results of the present paper, obtained from a large amount of samples of T. dimidiata from many different countries covering its whole latitude range, gives rise to a new frame that is different from the previous hypothesis about a clinal variation along a north-south axis, which was formerly suggested to explain both morphological data [50] and preliminary ITS-2 data from a reduced number of samples [6].
Moreover, the distinction between these five entities may facilitate the understanding of different vector transmission capacities and epidemiological characteristics of Chagas disease throughout the very large area where T. dimidiata sensu lato is distributed, from the Mexican northern latitude limit up to the Peruvian southern latitude limit [11]. Recent results obtained by means of a population dynamics model indicate that T. dimidiata in Yucatan, Mexico, is not able to sustain domestic populations, that up to 90% of the individuals found in houses are immigrants, and that consequently Chagas disease control strategies must be adapted to a transmission by non-domiciliated vectors [59]. This might be considered surprising because it does not fit the domiciliation capacity of T. dimidiata in other places, but it appears to be congruent if it is taken into account that in fact the Yucatan vector in question is not T. dimidiata but a different species T. sp. aff. dimidiata.
The results here obtained also suggest that T. d. dimidiata in Ecuador is a good candidate for the design of appropriate vector control intervention, similarly to domestic T. infestans populations in countries such as Uruguay, Chile and Brazil within the successful Southern Cone Initiative [60]. The control and even eradication of T. d. dimidiata in Ecuador by means of insecticide-spraying of its domestic habitats might be successful, if it is considered that it is merely an introduced vector species in that area, and a priori it would have difficulties in escaping from the insecticide activity because of its non-adaptativeness to the sylvatic environment in these two countries [61]. Unfortunately, such a control initiative will not be so easy to carry out in Colombia, as results prove that Colombian forms are authochthonous T. d. capitata and not T. d. dimidiata derived from the Ecuadorian introduced form. This fits with the existence of sylvatic populations in Colombia and with the high genetic similarity of sylvatic, peridomestic and domestic populations detected in that country [9]. Similarly to in Colombia, results indicate that T. dimidiata will offer, because of being authochthonous forms, more problems for insecticide-spraying control in Central American countries than introduced T. infestans in Southern Cone countries.
The other Meso- and Central American Triatoma Species
Triatoma bassolsae differs by only one deletion from T. pallidipennis and appears in the branch of the 5 species traditionally included in the Phyllosoma complex: T. longipennis, T. mazzotti, T. picturata, T. pallidipennis and T. phyllosoma. The genetic differences between these taxa are so reduced (sometimes even none at all), that there is no support to maintain them as separated species. Such a low number of nucleotide differences in the ITS is considered as pertaining to organisms able to hybridize [62]. This fully fits the capacity of these taxa to crossbreed and give fertile hybrids [63],[64] and agrees with the entomologist conclusion of applying only subspecies level to them [49]. The divergence of members of the phyllosoma complex is estimated at only 0.74–2.28 Mya by the rDNA molecular clock [4], which also seems consistent with a subspecific rank. All further ITS-2 studies have always reached the same conclusion [5],[6],[65]. By analyzing many interfertility experiments [64], it can be concluded that, in triatomines, morphological differentiation appears to be faster than the installation of reproductive or genetic barriers [66],[67]. Rapid morphological changes, associated with ecological adaptation, helps to explain discordance between phenetic and genetic differentiation. Triatomine species with consistent morphological differences would arise through divergent ecological adaptation, a vision which fits with “evolutionary units” implying a different evolutionary direction taken by some populations [67]. Until future reproductive isolation thanks to ecological isolation is reached by these morphologically different entities of the Phyllosoma complex, the subspecies concept accurately fits for all these “evolutionary units” of the Phyllosoma complex. ITS-2 results indicate that Triatoma bassolsae is one additional taxon to be included in this situation, as has already been suggested [65]. The comparison of the small genetic divergences between these taxa, their distributions exclusively restricted to regions of Mexico, and their different geographical distribution areas slightly overlapping in their bordering zones [3] suggest that genetic exchange might be impeding or delaying definitive divergence processes to reach species level.
Genetic distances between the taxa of the Phyllosoma complex found when analyzing different mtDNA genes proved to be similar to those detected in ITS-2 at the 16S [68], but higher in CytB [65],[69], and COI [69]. This agrees with the evolutionary rates of the protein-coding mtDNA genes which are pronouncedly faster than the one of ITS-2. Moreover, aminoacid sequences of the CytB and COI genes show no one difference between the Phyllosoma complex members studied (all are silent mutations or synonymous substitutions) except one aminoacid difference between two populations of the same species T. pallidipennis and one in T. picturata versus the rest [69], which also fit with an intraspecific variability. Additionally, it shall be taken into account that (i) mtDNA becomes monophyletic more rapidly than does a single nuclear gene and far more rapidly than a sample of several nuclear genes, so that mtDNA may make inferences of species-level monophyly erroneous [70], and (ii) the known great potential of mtDNA to become monophyletic by selective sweeps can decrease the time to monophyly of a clade and not be reflective of the genealogical processes in the nuclear genome, advantageous mutations occurring on mtDNA causing the entire mitochondrial genome to become monophyletic because of the little or no recombination they have [71]. The crossbreeding capacity and hybrid viability among the Phyllosoma complex taxa in question is well known and, taking into account that their geographical distributions overlap in their border areas and there are no sufficient ecological differences indicating a local spatial separation, it becomes very difficult to support them as separate species from the evolutionary, biogeographical and ecological points of view because there is apparently no barrier for a reproductive isolation. Thus, the results of both ITS-2 and mtDNA genes fit with such an evolutionary, subspecific divergence, when taking into account the peculiarities of both nuclear and mitochondrial markers.
Triatoma mexicana appears to be a good species and its location in the phylogenetic tree fully supports its ascription to the Phyllosoma complex, similarly as suggested by a phylogentic analysis by means of a mtDNA CO1 fragment [69]. Surprisingly, T. gerstaeckeri (Rubrofasciata group) clusters with T. mexicana, suggesting that it should be included in the Phyllosoma complex. All these species, i.e. T. phyllosoma (including its subspecies phyllosoma, longipennis, mazzotti, picturata, pallidipennis and bassolsae), T. dimidiata (with its three subspecies dimidiata, capitata and maculipennis, to which hegneri shall be added), T. sp. aff. dimidiata, T. mexicana and T. gerstaeckeri constitute a well defined clade for which the generic taxon Meccus, proposed long ago [72], afterwards synonymized [50] and recently tentatively revalidated [73], seem to appropriately fit. Previous molecular studies, first with complete ITS-2 sequences [74] and second with partial mtDNA 16S gene sequences [68], also indicate that Meccus might be a valid taxon.
The revalidation of Meccus, as well as that of Nesotriatoma for species of the Flavida complex, has not been accepted because of the close relationship between T. flavida and the Phyllosoma complex [7]. The results of the present study do, however, pose a serious question concerning the inclusion of species as T. bolivari and T. ryckmani in the Phyllosoma complex, as they appear to cluster with T. rubida of the Rubrofasciata group with relatively high support (83 and 96 in ML and BPP, respectively). A T. rubida - T. nitida clade previously detected with weak support under certain conditions in mitochondrial DNA marker analyses [69] does not appear to be supported in the ITS-2 phylogeny.
Although not fully resolved in the tree obtained, the location of the Cuban T. flavida as a species basal to all other North-Central American Triatoma species may be interpreted as a consequence of being a relict insular species close to the ancient first North-Central American Triatoma colonizers. Further studies with other genetic markers are needed to establish the position of T. flavida more adequately.
The South American Triatoma Species
The very scarce ITS-2 sequence differences between T. arthurneivai and T. rubrovaria, a species known in southern Brazil, Uruguay and northern Argentina [75], pose doubts on whether to keep the validity of T. arthurneivai as independent species. Recent genetic and morphometric studies have already raised several questions about T. arthurneivai, indicating that topotypes from Minas Geraes may represent a species different from populations of São Paulo State formerly also referred to T. arthurneivai and suggesting that these São Paulo populations might probably belong to T. wygodzinskyi [76]. This may explain the ITS-2 results, as the two specimens analyzed in the present paper come in fact from Espirito Santo do Pinhal, São Paulo State. Consequently, material of typical T. wygodzinskyi should be sequenced and compared to both true T. arthurneivai from Minas Geraes and T. rubrovaria to ascertain the status of these three taxa.
The South American Triatoma species cluster together with maximum support (100/100/100) and well separated from that of the North and Central American species of the same genus, thus supporting results of previous analyses which indicate an early divergence of about 23–38 Mya between species of the northern (Phyllosoma complex) and southern (T. infestans) continent [4],[6].
Supporting Information
Translation of the abstract into Spanish by S. Mas-Coma.
(0.03 MB DOC)
Summary of analysis of molecular variance for Triatoma dimidiata populations.
(0.06 MB DOC)
Summary of population genetic variation parameters from ITS-2 haplotypes in the Triatoma dimidiata populations.
(0.08 MB DOC)
Evaluation of within groups genetic differentiation by computation of pairwise FST values for populations defined by country of origin in subgroup 1A.
(0.03 MB DOC)
Summary of differentiation tests for Triatoma dimidiata populations based on ITS-2 haplotypes.
(0.06 MB DOC)
Acknowledgments
D.R. Klisiowicz was on leave from the Departamento de Patologia Basica, Universidade Federal do Paraná, Centro Politécnico Curitiba, PR, Brazil. Thanks to Drs. V.H.M. Aguilar (Quito, Ecuador), J. Moreno (Medellín, Colombia), O. Fuentes (La Habana, Cuba) and F. Brenière (Mexico DF, Mexico) for providing specimens from their respective countries. Lic. M.L. Hernandez-Viadel participated in laboratory procedures. Technical support for the automatic sequencing of triatomines was provided by the DNA Sequencing Service of the University of Valencia.
Footnotes
The authors have declared that no competing interests exist.
This work benefited from international collaboration through the ECLAT network. Financial support for DNA sequencing was obtained from the Projects “Chagas Disease Intervention Activities” (CDIA, Contract No. ICA4CT-2003-10049) and “European Commission Latin America Triatominae Network” (ECLAT, Contract No. IC18-CT98-0366) of the INCO-DEV and INCO-DC Programs of the European Commission (DG XII), Brussels, Belgium, Project No. 3042/2000 of the Dirección General de Cooperación para el Desarrollo, Presidencia de Gobierno, Generalitat Valenciana, Valencia, Spain, and the Red de Investigación de Centros de Enfermedades Tropicales - RICET (Projects No. C03/04, No. PI030545 and No. RD06/0021/0017 of the Program of Redes Temáticas de Investigación Cooperativa), FIS, Spanish Ministry of Health, Madrid, Spain. F. Panzera benefited from funding by the Conselleria de Cultura i Educació of the Valencian regional government, Spain and the University of Valencia for two working stays at the Parasitology Department of Valencia, as well as from Comisión Sectorial de Investigación Científica (CSIC), Uruguay, for sample collections. F. Guhl benefited from funding by the University of Valencia for a 6-month research stay at the Parasitology Department of Valencia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schmunis GA. Medical significance of American trypanosomiasis. In: Maudlin I, Holmes PH, Miles MA, editors. The Trypanosomiases. Wallingford, UK: CAB International; 2004. pp. 355–368. [Google Scholar]
- 2.World Bank. World Development Report 1993. Investing in Health. New York: Oxford University Press; 1993. p. 329. [Google Scholar]
- 3.Dujardin JP, Schofield JC, Panzera F. Les Vecteurs de la Maladie de Chagas. Recherches taxonomiques, biologiques et génétiques. Brussels: Académie Royale des Sciences d'Outre Mer, Classe des Sciences Naturelles et Médicales; 2000. p. 162. [Google Scholar]
- 4.Bargues MD, Marcilla A, Ramsey J, Dujardin JP, Schofield CJ, et al. Nuclear rDNA-based molecular clock of the evolution of Triatominae (Hemiptera: Reduviidae), vectors of Chagas disease. Mem Ins Oswaldo Cruz. 2000;95:567–573. doi: 10.1590/s0074-02762000000400020. [DOI] [PubMed] [Google Scholar]
- 5.Bargues MD, Marcilla A, Dujardin JP, Mas-Coma S. Triatominae vectors of Chagas disease: a molecular perspective based on nuclear ribosomal DNA markers. Trans Roy Soc Trop Med Hyg. 2002;96(S1):159–164. doi: 10.1016/s0035-9203(02)90069-6. [DOI] [PubMed] [Google Scholar]
- 6.Marcilla A, Bargues MD, Ramsey J, Magallon-Gastelum E, Salazar-Schettino PM, et al. The ITS-2 of the nuclear rDNA as a molecular marker for populations, species and phylogenetic relationships in Triatominae (Hemiptera: Reduviidae), vectors of Chagas disease. Mol Phylogen Evol. 2001;18:136–142. doi: 10.1006/mpev.2000.0864. [DOI] [PubMed] [Google Scholar]
- 7.Dujardin JP, Schofield JC. Triatominae: systematics, morphology and population biology. In: Maudlin I, Holmes PH, Miles MA, editors. The Trypanosomiases. Wallingford, UK: CAB International; 2004. pp. 181–201. [Google Scholar]
- 8.Dorn PL, Melgar S, Rouzier V, Gutierrez A, Combe C, et al. The Chagas vector, Triatoma dimidiata (Hemiptera: Reduviidae), is panmictic within and among adjacent villages in Guatemala. J Med Entomol. 2003;40:436–440. doi: 10.1603/0022-2585-40.4.436. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez CJ, Jaramillo CA, Delgado MP, Pinto NA, Aguilera G, et al. Genetic structure of sylvatic, peridomestic and domestic populations of Triatoma dimidiata (Hemiptera: Reduviidae) from an endemic zone of Boyaca, Colombia. Acta Trop. 2004;93:23–29. doi: 10.1016/j.actatropica.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Zeledon R. El Triatoma dimidiata (Latreille, 1811) y su relación con la Enfermedad de Chagas. San José, Costa Rica: Editorial Universidad Estatal a Distancia (EUNED); 1981. p. 146. [Google Scholar]
- 11.Dorn PL, Monroy C, Curtis A. Discussion – Triatoma dimidiata (Letreille, 1811): a review of its diversity across its geographic range and the relationships among populations. Inf Gen Evol. 2007;7:343–352. doi: 10.1016/j.meegid.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Calderon CI, Dorn P, Melgar S, Chavez JJ, Rodas A, et al. A preliminary assessment of genetic differentiation of Triatoma dimidiata (Hemiptera: Reduviidae) in Guatemala by Random Amplification of Polymorphic DNA-Polymerase Chain Reaction. J Med Entomol. 2004;41:882–887. doi: 10.1603/0022-2585-41.5.882. [DOI] [PubMed] [Google Scholar]
- 13.Bustamante DM, Monroy C, Menes M, Rodas A, Salazar-Schettino PM, et al. Metric variation among geographic populations of the Chagas vector Triatoma dimidiata (Hemiptera: Reduviidae: Triatominae) and related species. J Med Entomol. 2004;41:296–301. doi: 10.1603/0022-2585-41.3.296. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann P, Ordoñez R, Ojeda-Baranda R, Mendez de Liria J, Hidalgo-Sosa L, et al. Morphometric analysis of Triatoma dimidiata populations (Reduviidae: Triatominae) from Mexico and northern Guatemala. Mem Inst Oswaldo Cruz. 2005;100:477–482. doi: 10.1590/s0074-02762005000500006. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez GC, Juarez GC, Monroy C, Menes M, Bustamante DM, et al. Intraspecific variability in Triatoma dimidiata (Hemiptera: Reduviidae) populations from Guatemala based on chemical and morphometric analyses. J Med Entomol. 2005;42:29–35. doi: 10.1093/jmedent/42.1.29. [DOI] [PubMed] [Google Scholar]
- 16.Catala S, Sachetto C, Moreno M, Rosales R, Salazar-Schettino PM, et al. Antennal phenotype of Triatoma dimidiata populations and its relationship with species of phyllosoma and protracta complexes. J Med Entomol. 2005;42:719–725. doi: 10.1093/jmedent/42.5.719. [DOI] [PubMed] [Google Scholar]
- 17.Calderon Fernandez G, Juarez MP, Ramsey J, Salazar-Schettino PM, Monroy C, et al. Cuticular hydrocarbon variability among Triatoma dimidiata (Hemiptera: Reduviidae) populations from Mexico and Guatemala. J Med Entomol. 2005;42:780–788. [PubMed] [Google Scholar]
- 18.Panzera F, Ferrandis I, Ramsey J, Ordoñez R, Salazar-Schettino PM, et al. Chromosomal variation and genome size support existence of cryptic species of Triatoma dimidiata with different epidemiological importance as Chagas disease vectors. Trop Med Int Health. 2006;11:1092–1103. doi: 10.1111/j.1365-3156.2006.01656.x. [DOI] [PubMed] [Google Scholar]
- 19.Lin CP, Danforth BN. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol Phylogen Evol. 2004;30:686–702. doi: 10.1016/S1055-7903(03)00241-0. [DOI] [PubMed] [Google Scholar]
- 20.Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. Quart Rev Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 21.Caterino MS, Cho S, Sperling FAH. The current state of insect molecular systematics: a thriving tower of Babel. Ann Rev Entomol. 2000;45:1–54. doi: 10.1146/annurev.ento.45.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Collins FH, Paskewitz SM. A review of the use of ribosomal DNA to differentiate among cryptic Anopheles species. Insect Mol Biol. 1996;5:1–9. doi: 10.1111/j.1365-2583.1996.tb00034.x. [DOI] [PubMed] [Google Scholar]
- 23.Marinucci M, Romi R, Mancini P, Di Luca M, Severini C. Phylogenetic relationships of seven Palearctic members of the maculipennis complex inferred from ITS2 sequence analysis. Insect Mol Biol. 1999;8:469–480. doi: 10.1046/j.1365-2583.1999.00140.x. [DOI] [PubMed] [Google Scholar]
- 24.Proft J, Maier WA, Kampen H. Identification of six sibling species of the Anopheles maculipennis complex (Diptera: Culicidae) by a polymerase chain reaction assay. Parasitol Res. 1999;85:837–843. doi: 10.1007/s004360050642. [DOI] [PubMed] [Google Scholar]
- 25.Bargues MD, Klisiowicz DR, Panzera F, Noireau F, Marcilla A, et al. Origin and phylogeography of the Chagas disease main vector Triatoma infestans based on nuclear rDNA sequences and genome size. Inf Gen Evol. 2006;6:46–62. doi: 10.1016/j.meegid.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Bargues MD. Relojes moleculares y evolución genética de Triatomini y Rhodniini basados en el ADN ribosomal. In: Guhl F, Schofield CJ, editors. Proceedings of the Fourth International Workshop on Population Genetics and Control of Triatominae. Bogotá, Colombia: CIMPAT, Universidad de Los Andes; 2002. pp. 117–124. [Google Scholar]
- 27.Bargues MD, Mas-Coma S. Phylogenetic analysis of lymnaeid snails based on 18S rDNA sequences. Mol Biol Evol. 1997;14:569–577. doi: 10.1093/oxfordjournals.molbev.a025794. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. 2nd Ed. Vols. I, II & III. New York, USA: Cold Spring Harbor Laboratory, Cold Spring Harbor; 1989. p. 1647. [Google Scholar]
- 29.Tompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity and progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nuc Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Tamura K, Nei M. MEGA 3: Integrate software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinf. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 31.Staden R, Judge DP, Bonfield JK. Sequence assembly and finishing methods. Met Biochem Anal. 2001;43:302–322. doi: 10.1002/0471223921.ch13. [DOI] [PubMed] [Google Scholar]
- 32.Pacheco RS, Almeida CE, Costa J, Klisiowicz DR, Mas-Coma S, et al. RAPD analyses and rDNA intergenic-spacer sequences discriminate Brazilian populations of Triatoma rubrovaria (Reduviidae: Triatominae). Ann Trop Med Parasitol. 2003;97:757–768. doi: 10.1179/000349803225002372. [DOI] [PubMed] [Google Scholar]
- 33.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods). [4.0beta]. 2002. Sinauer Associates, Sunderland, MA.
- 34.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by Maximum Likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 35.Akaike H. A new look at the statistical model identification. Ieee Trans Automat Control. 1974;19:716–723. [Google Scholar]
- 36.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 37.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 38.Bandelet HJ, Foster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 39.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 41.Rozas J, Sanchez-Delbarrio JC, Meseguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 42.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinf Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 43.Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1996;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 46.Slatkin M. Inbreeding coefficients and coalescent times. Genet Res. 1991;58:167–175. doi: 10.1017/s0016672300029827. [DOI] [PubMed] [Google Scholar]
- 47.Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- 48.Usinger R. Notes and descriptions of neotropical Triatominae (Hemiptera, Reduviidae). Pan-Pacific Entomol. 1941;17:49–57. [Google Scholar]
- 49.Usinger R. The Triatominae of North and Central America and the West Indies and their public health significance. Pub Health Bull. 1944;288:1–83. [Google Scholar]
- 50.Lent H, Wygodzinsky P. Revision of the Triatominae (Hemiptera, Reduviidae) and their significance as vectors of Chagas' disease. Bull Am Mus Nat Hist. 1979;163:123–520. [Google Scholar]
- 51.Lent H, Jurberg J. Sobre a variação intra-especifica em Triatoma dimidiata (Latreille) e Triatoma infestans (Klug) (Hemiptera: Reduviidae). Mem Inst Oswaldo Cruz. 1985;80:285–299. [Google Scholar]
- 52.Dumonteil E, Gourbiere S, Barbera-Perez M, Rodriguez-Felix E, Ruiz-Piña H, et al. Geographic distribution of Triatoma dimidiata and transmission dynamics of Trypanosoma cruzi in the Yucatan peninsula of Mexico. Am J Trop Med Hyg. 2002;67:176–183. doi: 10.4269/ajtmh.2002.67.176. [DOI] [PubMed] [Google Scholar]
- 53.Dumonteil E, Gourbiere S. Predicting Triatoma dimidiata abundance and infection rate: a risk map for natural transmission of Chagas disease in the Yucatan peninsula of Mexico. Am J Trop Med Hyg. 2004;70:514–519. [PubMed] [Google Scholar]
- 54.Dumonteil E, Ruiz-Piña H, Rodriguez-Felix E, Barbera-Perez M, Ramirez-Sierra MJ, et al. Re-infestation of houses by Triatoma dimidiata after intra-domicile insecticide application in the Yucatán Peninsula, Mexico. Mem Inst Oswaldo Cruz. 2004;99:253–256. doi: 10.1590/s0074-02762004000300002. [DOI] [PubMed] [Google Scholar]
- 55.Guzman-Tapia Y, Ramirez-Sierra MJ, Escobedo-Ortegon J, Dumonteil E. Effect of hurricane Isidore on Triatoma dimidiata distribution and Chagas disease transmission risk in the Yucatan peninsula of Mexico. Am J Trop Med Hyg. 2005;73:1019–1025. [PubMed] [Google Scholar]
- 56.Lavalle B. Francisco Pizarro y la conquista del Imperio Inca. Pozuelo de Alarcon, Spain: Editorial Espasa Calpe S.A; 2005. p. 338. [Google Scholar]
- 57.Vermeij GJ. When biotas meet: understanding biotic interchange. Science. 1991;253:1099–1104. doi: 10.1126/science.253.5024.1099. [DOI] [PubMed] [Google Scholar]
- 58.Morrone JJ. Toward a synthesis of Mexican biogeography. Rev Mex Biodiv. 2006;76:207–252. [Google Scholar]
- 59.Gourbière S, Dumonteil E, Rabinovich JE, Minkoue R, Menu F. Demographic and dispersal constraints for domestic infestation by non-domiciliated Chagas disease vectors in th Yucatan Peninsula, Mexico. Am J Trop Med Hyg. 2008;78:133–139. [PubMed] [Google Scholar]
- 60.Dias JCP, Schofield CJ. Control of Triatominae. In: Maudlin I, Holmes PH, Miles MA, editors. The Trypanosomiases. Wallingford, UK: CAB International; 2004. pp. 547–563. [Google Scholar]
- 61.Abad-Franch F, Paucar CA, Carpio CC, Cuba Cuba CA, Aguilar VHM, et al. Biogeography of Triatominae (Hemiptera: Reduviidae) in Ecuador: implications for the design of control strategies. Mem Ins Oswaldo Cruz. 2001;96:611–620. doi: 10.1590/s0074-02762001000500004. [DOI] [PubMed] [Google Scholar]
- 62.Remigio EA, Blair D. Relationships among problematic North American stagnicoline snails (Pulmonata: Lymnaeidae) reinvestigated using nuclear ribosomal DNA internal transcribed spacer sequences. Can J Zool. 1997;75:1540–1545. [Google Scholar]
- 63.Mazzotti L, Osorio MT. Cruzamientos experimentales entre varias especies de triatomas. Rev Med Mex. 1940;22:215–222. [Google Scholar]
- 64.Usinger RL, Wygodzinsky P, Ryckman R. The biosystematics of Triatominae. Ann Rev Entomol. 1966;11:309–330. doi: 10.1146/annurev.en.11.010166.001521. [DOI] [PubMed] [Google Scholar]
- 65.Martinez FH, Villalobos GC, Cevallos AM, De La Torre P, Laclette JP, et al. Taxonomic study of the Phyllosoma complex and other triatomine (Insecta: Hemiptera: Reduviidae) species of epidemiological importance in the transmission of Chagas disease using ITS-2 and mtCytB sequences. Mol Phylogen Evol. 2006;41:279–287. doi: 10.1016/j.ympev.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 66.Gorla DE, Dujardin JP, Schofield CJ. Biosystematics of Old World Triatominae. Acta Trop. 1997;63:127–140. doi: 10.1016/s0001-706x(97)87188-4. [DOI] [PubMed] [Google Scholar]
- 67.Dujardin JP, Panzera F, Schofield CJ. Triatominae as a model of morphological plasticity under ecological pressure. Mem Inst Oswaldo Cruz. 1999;94(Suppl. I):223–228. doi: 10.1590/s0074-02761999000700036. [DOI] [PubMed] [Google Scholar]
- 68.Hypsa V, Tietz DF, Zrzavy J, Rego ROM, Galvão C, et al. Phylogeny and biogeography of Triatominae (Hemiptera: Reduviidae): molecular evidence of a New World origin of the Asiatic clade. Mol Phylogen Evol. 2002;23:447–457. doi: 10.1016/s1055-7903(02)00023-4. [DOI] [PubMed] [Google Scholar]
- 69.Pfeiler E, Bitler BG, Ramsey J, Palacios-Cardiel C, Markow TA. Genetic variation, population structure, and phylogenetic relationships of Triatoma rubida and T. recurva (Hemiptera: Reduviidae: Triatominae) from the Sonoran Desert, insect vectors of the Chagas' disease parasite Trypanosoma cruzi. Mol Phylogen Evol. 2006;41:209–221. doi: 10.1016/j.ympev.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 70.Hudson RR, Coyne JA. Mathematical consequences of the genealogical species concept. Evolution. 2002;56:1557–1565. doi: 10.1111/j.0014-3820.2002.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 71.Ballard JWO, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Ann Rev Ecol Evol Syst. 2005;36:621–642. [Google Scholar]
- 72.Stal C. Monographie der Gattung Conorhinus und Verwandten. Berliner Entomologische Zeitschrift. 1859;3:99–117. [Google Scholar]
- 73.Carcavallo RU, Jurberg J, Lent H, Noireau F, Galvao C. Phylogeny of the Triatominae (Hemiptera, Reduviidae). Proposals for taxonomic arrangements. Entomología y Vectores. 2000;7(S1):1–99. [Google Scholar]
- 74.Marcilla A, Bargues MD, Abad-Franch F, Panzera F, Carcavallo RU, et al. Nuclear rDNA ITS-2 sequences reveal polyphyly of Panstrongylus species (Hemiptera: Reduviidae: Triatominae), vectors of Trypanosoma cruzi. Inf Gen Evol. 2002;1:225–235. doi: 10.1016/s1567-1348(02)00029-1. [DOI] [PubMed] [Google Scholar]
- 75.Pacheco RS, Almeida CE, Klisiowicz DR, Costa J, Pires MQ, et al. Genetic variability of Triatoma rubrovaria (Reduviidae: Triatominae) from Brazil, Argentina and Uruguay as revealed by two different molecular markers. Parasite. 2007;14:231–237. doi: 10.1051/parasite/2007143231. [DOI] [PubMed] [Google Scholar]
- 76.Santos SM, Lopes CM, Dujardin JP, Panzera F, Perez R, et al. Evolutionary relationships based on genetic and phenetic characters between Triatoma maculata, Triatoma pseudomaculata and morphologically related species (Reduviidae: Triatominae). Inf Gen Evol. 2007;7:469–475. doi: 10.1016/j.meegid.2007.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Translation of the abstract into Spanish by S. Mas-Coma.
(0.03 MB DOC)
Summary of analysis of molecular variance for Triatoma dimidiata populations.
(0.06 MB DOC)
Summary of population genetic variation parameters from ITS-2 haplotypes in the Triatoma dimidiata populations.
(0.08 MB DOC)
Evaluation of within groups genetic differentiation by computation of pairwise FST values for populations defined by country of origin in subgroup 1A.
(0.03 MB DOC)
Summary of differentiation tests for Triatoma dimidiata populations based on ITS-2 haplotypes.
(0.06 MB DOC)