Summary
Genetic epidemiology, including twin studies, provides robust evidence that genetic variation in human populations contributes to susceptibility to infectious disease. One of the major limitations of studies that attempt to identify the genes and mechanisms that underlie this susceptibility has been lack of power caused by small sample size. With the development of novel technologies, burgeoning information on the human genome, the HapMap project, and human genetic diversity, we are at the beginning of a new era in the study of the genetics of complex diseases. This review looks afresh at the epidemiological evidence that supports a role for genetics in susceptibility to infectious disease, examines the somewhat limited achievements to date, and discusses current advances in methodology and technology that will potentially lead to translational data in the future.
Introduction
Infection is one of the leading causes of human mortality and morbidity, with much of the burden falling on children.1 Infectious diseases are a major selective pressure,2, 3, 4, 5 and the genes involved in the immune response are the most numerous and diverse in the human genome,6 indicating the evolutionary advantages of a varied immunological response to a wide range of infectious pathogens.7 This is most obvious at the HLA loci,8 the prototypical candidate genetic region for infectious disease susceptibility. For example, individuals in whom all class II HLA alleles are heterozygous are more likely to clear hepatitis B infection,9 and those with heterozygous class I alleles progress from HIV to an AIDS-defining illness more slowly and have lower mortality.10 The converse scenario, increased HLA homozygosity, may contribute to the increased susceptibility to infection in genetically isolated populations.11
There is huge variation in the individual outcomes that follow exposure to potentially life-threatening pathogens, and this differential susceptibility partly shows the functional genetic diversity of the immune response. Here we provide an overview of human genetic susceptibility in infectious diseases, review the supporting epidemiological evidence, discuss current methodological and technological issues, and highlight the somewhat limited achievements to date, especially those that have increased our understanding of outcome and treatment response for specific infectious diseases.
One gene…or many?
Susceptibility to infection and many other human diseases (including diabetes and ischaemic heart disease) arises from the complex interaction of environmental and host genetic factors. In general, many genetic loci make modest contributions to human disease susceptibility (ie, they are genetically complex), and most of the focus in the field has been on identifying these loci and their effects in infection and in other conditions. However, there are many single-gene (Mendelian) disorders affecting immune function, with over 300 primary immunodeficiencies reported. Although often rare, these profound immunodeficiencies stem from major functional aberrations at single genes and can be highly informative about immunological mechanisms and protection against specific infections.12, 13 There are also important examples of more common single-gene disorders that markedly influence infection-specific susceptibility; the best known is the protection afforded by sickle cell heterozygosity against falciparum malaria.14 A variant in the Duffy antigen gene promoter, which results in lack of erythrocyte surface expression, prevents binding by Plasmodium vivax, and is therefore protective.15, 16 More recently, a genetic variant that determines blood group secretor status has been shown to mediate susceptibility to Norwalk virus,17 and also to affect HIV progression independently of other chemokine variants.18
Hype and hyperbole
There has been substantial expectation that an increased understanding of the genetic determinants of differential susceptibility and outcome would be rapidly translated into novel therapies and preventative interventions. Much of this expectation has yet to be realised, partly because of the methodological limitations of earlier studies and a tendency to examine individual genes in isolation and without consideration of their genetic and environmental milieu. Thus many reported genetic associations have not been replicated in subsequent studies.19 Understandably, some scepticism exists as to how much a genetic epidemiological approach can actually reveal about the genetic basis of complex diseases.20 However, there have been many recent advances in technology and—more latterly—its application; such studies have identified important protective and pathogenic pathways, and have led to individually targeted anti-infective therapies. Hopefully, we are entering an era in which well designed studies will begin to deliver robust genetic data and many more opportunities for translation into better treatments.
Epidemiological evidence
The idea that human genetics may contribute to our understanding of the susceptibility to infectious diseases is not new. As early as the 18th century, differential susceptibility to infection was suggested to be a characteristic of the host, and diseases such as tuberculosis and leprosy were believed to be inherited defects.21 Infectious diseases, like other phenotypes, may exhibit familial aggregation: a greater frequency of the disease in relatives of infected individuals compared with relatives of those without disease. Studies of familial aggregation can therefore investigate the importance of shared determinants, both environmental and genetic, in disease susceptibility. They usually use a form of the recurrence risk ratio, essentially the ratio of prevalence in the relatives of the index case to the prevalence in the general population. It is important in such studies that the disease phenotype is carefully defined, particularly in the context of infectious diseases. Interpretation of recurrence risk is more straightforward if the phenotype is a binary or dichotomous trait (eg, the presence or absence of meningococcal septicaemia) than a qualitative range of clinical phenotypes, which may be less consistent (eg, measures of severity of infection determined by treatment interventions). Familial aggregation measures make no supposition about the cause of such aggregation, and recurrence risks within the same generation, such as sibling risk ratios, will identify both shared environmental and genetic effects. In infectious diseases, transmission of pathogens between family members may increase the risk of disease and contribute to the observed phenotypic aggregation. For example, in a study of sibling recurrence risk ratio for meningococcal disease, there was a 30 times increased risk of infection in siblings of cases compared with the general population risk, but the risk decreased with a longer time interval between the index case and the sibling case.22 The recurrence risk for meningococcal disease in siblings was 11·9 times the population risk when the index and sibling cases occurred more than a week apart, falling to a risk of 8·2 if a year had elapsed between cases.22 These data indicate significant genetic determinants of meningococcal susceptibility, and also the importance of common acquisition of virulent strains within families, which accounts for the more immediate increased risk in household contacts.23
Twin studies
Studies of twins can also provide an estimate of the relative contributions of shared genes and environment to disease phenotypes, by comparing the risk in genetically identical monozygotic twin pairs to that in dizygotic twins, who share on average only half their genes. Some twin studies of infectious diseases are historical,24, 25, 26 and determinations of zygosity and disease phenotype may be inaccurate.25 Notwithstanding these reservations, susceptibility to some infections show markedly increased concordance in monozygotic compared with dizygotic twins.27 Examples include tuberculosis,24, 25, 26 sinusitis,28 Helicobacter pylori-specific antibody titres,29 and leprosy.30 More recent twin and triplet data indicate that susceptibility to both acute and chronic otitis media is largely genetically determined.31, 32 The relative contributions of infection, atopy, and cranio-facial anatomy, all of which are determined in part by genetic factors, remain unclear.
Twin studies may also provide useful insights into the determinants of disease outcome. Carrier status for hepatitis B virus33 and the febrile response to (but not acquisition of) Plasmodium falciparum 34 both indicate increased concordance in monozygotic twin pairs. A study of the severity of common childhood infections, which used hospital admission as the phenotype, indicated that distinct genetic factors were likely to be involved in different infections, since the risk of admission in the second twin was only increased if the twins both had the same infectious diagnosis (Burgner D, unpublished). The concordance rates for common otolaryngological procedures such as myringotomy and adeno-tonsillectomy, which are surrogates for disease severity, also show greatly increased concordance in monozygotic twin pairs (Burgner D, unpublished). Twin studies of vaccine responses, in which the antigenic stimulus is unambiguous, indicate significant genetic influences on both humoral35 and cell-mediated36 responses. More detailed genetic studies indicate the importance of both HLA and non-HLA loci in vaccine responsiveness.37, 38 Studies in vitro of twins39 and first-degree relatives40 also show the importance of genetic factors in determining the innate immune responses to infectious stimuli.
A twin study of H pylori acquisition, which compared concordance rates for twins reared either together or apart, gave very similar measures of genetic influences in each setting, which accounted for most of the disease risk.29 However, twins and siblings are very commonly reared together, making it difficult to tease out the relative contributions of genetic factors and shared environmental exposures, particularly pathogen sharing. Intergenerational studies may circumvent this issue. A seminal study compared the risk and shared causes of premature mortality in children adopted very early in life with that of their biological and adoptive parents.41 The risk of an adopted child dying of infection was increased almost six times if their biological parent had also died prematurely of infection, whereas no increased risk was observed if the adoptive parent died of infection, suggesting that genetic rather than environmental factors are the important determinants of infectious disease mortality. Much smaller genetic effects were noted for cardiovascular disease and malignancy.41
Study design and methodological issues
These epidemiological data indicate that genetic factors influence both susceptibility to, and outcome of, infection. The various study designs and methodological issues are important in interpreting previous data and in designing contemporary and future studies in infection. As the fundamentals of genetic epidemiology have recently been reviewed in detail,42, 43, 44, 45, 46, 47, 48 only a brief overview will be provided here. Some of the terminology is explained briefly in the panel . Of note, although single nucleotide polymorphisms (SNPs) are the workhorse of current high-throughput genotyping technologies, other forms of genetic variation are also common in the human genome and may have important effects on disease risk. These include repeating sequence motifs (microsatellites and minisatellites), insertions and deletions, and differences in gene copy number.55 For example, recent data indicate that there is a strong inverse correlation between the number of copies of an HIV suppressive chemokine gene (CCL3L1) and HIV susceptibility.56
Panel. Definitions for genetic terminology.
Allelic association
Statistical analysis to determine whether disease is associated with particular allelic variants at one or more loci, usually by comparing marker allele frequencies between a disease group and a control group.
F st statistic
A statistic to describe individuals, subpopulations, and whole population structures. Used to assess evidence for selection by comparing the frequency of an allele in different populations.49
Haplotype
A combination of marker alleles at different markers along the same chromosome that tend to be inherited as a unit.
Haplotype tag
An SNP that acts as a defining marker for a haplotype by virtue of linkage disequilibrium. Analysing haplotype tags, rather than all the variants on a haplotype, reduces redundancy and genotyping costs.
Heterozygosity
A measure of genetic diversity within a population.
Linkage analysis
Statistical analysis to localise genes and markers with respect to each other in the genome, based on recombination frequency. Linkage analysis can also be used to map a disease phenotype in relation to polymorphic markers.
Linkage disequilibrium
The non-random association of alleles at two or more neighbouring loci that are inherited together more often than expected by chance, providing the basis to haplotypes and linkage disequilibrium mapping. This association can potentially derive from population admixture.
Microsatellite
A polymorphism characterised by a variable number of tandem repeats, often defined by the numbers of repeats in a row of at least two or more nucleotides. They are useful markers for family linkage studies and determining a person's unique DNA fingerprint in forensic medicine. Also known as SSR (simple sequence repeat) or STR (simple tandem repeat).
Population admixture
The recent combination of two or more previously distinct populations. Unknown admixture may result in spurious genetic associations in a case-control study design.
SNP
Single nucleotide polymorphism, single base-pair change at a specific point in the genome.
SNP chip
Array technology that allows large numbers (up to a million) of SNPs to be genotyped on a single array typically the size of a microscope slide (eg, Affymetrix50 or Illumina51)
Additional useful references: Burton et al,42 Malats and Calafell,52, 53 and Calafell and Malats.54
Genome-wide linkage and association studies
There are essentially two types of study design—genome-wide and candidate gene—that are used to localise the genes underlying human disease. Genome-wide studies have the advantage that no supposition is made about the genes involved, and potentially novel or unconsidered genes may be identified. Typically, the genome-wide approach has used linkage studies of multiply affected (or infected) pedigrees to identify regions of the entire genome that are transmitted from the parents to the offspring more often than expected under independent inheritance.43 Typically, affected sibling pairs,57, 58 or larger multicase pedigrees59 if available, are recruited and the inherited regions of the genome are defined by a few hundred microsatellite markers. The main disadvantages of linkage studies are that sufficient numbers of affected sibling pairs may be difficult to recruit for many infections, and that linkage studies are often too insensitive to pick up the relatively small contributions from individual genetic regions that are typical of complex diseases such as infection. There are examples of successful genome scans undertaken using linkage analysis for infectious diseases, including the intensity of infection with Schistosoma mansoni,59 susceptibility to H pylori,60 and to leprosy.57, 58, 61 As with association data, some linkage studies have not been replicated in different populations. In some cases this may represent a false discovery rate, an inherent problem in studies that make multiple statistical comparisons, but it is increasingly clear that many complex diseases show genetic heterogeneity, with different genetic determinants for the same disease operating in different ethnic groups. For example, mutations in the NOD2/CARD15 locus, an important susceptibility determinant of inflammatory bowel disease in white populations, are effectively absent in Asian populations with the same phenotype.62
Genome-wide association studies, in which the whole genome is interrogated using hundreds of thousands of SNPs, have recently become a reality as genotyping technology and analytical tools evolve. This approach will potentially detect more subtle genetic effects than a classic linkage study.63 At present, studies use up to 0·5 million of the approximately 11 million estimated SNPs in the human genome, but there are unresolved issues particularly relating to the optimum density and location for these SNP markers (gene-centric or evenly spaced through the genome), which have implications for how much of the genome such studies truly cover,45 as well as concerns regarding sample size and power.64 Genome-wide association studies have a high false discovery rate, and replication in independent populations is essential. There are currently no reported genome-wide association studies in infectious diseases, although such studies are underway. In particular, studies being undertaken under the banner of the Wellcome Trust Case-Control Consortium will look at up to 2000 cases and 3000 controls for eight complex diseases, including tuberculosis and malaria.65 For these two important infectious diseases, a two-stage strategy is proposed in which a 750 000 SNP chip (panel) will be used to genotype 1000 cases and 1000 controls, and regions positive at p=0·1 followed up in at least another 1000 cases and 1000 controls. This is one of the biggest projects ever undertaken to identify the genetic variations that may predispose people to, or protect them from, complex diseases. The hope is that by identifying these genetic signposts, researchers will be able to understand which people are most at risk, and also produce more effective treatments.
The genome-wide association approach has been used on a smaller scale in other non-infectious genetically complex diseases. For example, genotyping of almost 93 000 SNPs across the genome has identified a region of chromosome 6 associated with myocardial infarction. Further mapping identified a novel functional polymorphism in the lymphotoxin alpha (LTA) gene within this region and related functional variants in a ligand of LTA.66, 67 The odds ratio associated with the LTA mutation was 1·78,67 an order of magnitude expected for a complex disease. In another study, a 120 000 SNP chip was used to compare 96 cases of age-related macular degeneration with 50 controls to identify a major gene (complement factor H) that determines susceptibility.68 A common intronic SNP among the 120 000 SNPs genotyped was strongly associated with disease, with a nominal p value of less than 10−7 (after correction for multiple testing), and a genotype relative risk for homozygous individuals of 7·4 (95% CI 2·9–19·0). Re-sequencing revealed a polymorphism in linkage disequilibrium with the risk allele that caused a tyrosine to histidine change at amino acid 402 in the complement factor H protein. The numbers of cases and controls studied was small, increasing the possibility of a false positive result (although the relative risk was high compared with our usual expectation for complex diseases). Importantly, the SNP-chip study was immediately supported by two studies using four independent case-control populations.69, 70
These studies provide a major conceptual breakthrough in terms of the approaches that can be made with novel technologies, now more feasible with the recent publication of the phase I HapMap,71 and the data for phase II released in 2005 (http://www.hapmap.org). The main practical objective of the HapMap project was to identify sets of SNPs that are in linkage disequilibrium with each other, so that a subset of these can be genotyped as so-called haplotype-tagging SNPs in a genome-wide association study, thus reducing the overall amount of genotyping required. The phase I data involved the genotyping of 1 million SNPs in unrelated individuals or parent/child trios of Nigerian, European, Chinese, and Japanese descent. Phase II extends this to 3·5 million SNPs. The phase I data has already provided many predictions and tests of applicability of haplotype tagging across populations,72, 73 optimum sampling and two-stage testing strategies for whole genome association studies,74 and which commercially available SNP chips will work best. As predicted from previous studies, a greater density of SNPs on a chip is required to capture all the haplotypic diversity in the African population compared with European, Chinese, or Japanese populations, because of the greater diversity and shorter regions of linkage disequilibrium observed in the former,75 a factor that will be important in application of the technology to infectious disease association studies in Africa. Nevertheless, for 1000 cases and 1000 controls and a nominal p value of 0·01, the phase I HapMap (eg, the newly released Sentrix human Hap300 genotyping beadchip by Illimuna;51 one SNP about every 12 kb) retains 77% power to detect associations due to common (≥5%) causal alleles in the African population, compared with roughly 90% in European, Chinese, or Japanese populations.75 The 500 000 SNP chip from Affymetrix,50 which was made before the release of phase I HapMap haplotype-tagging SNP information, has about the same power as the Illumina human Hap300 chip (ie, it contains about two tags per haplotype).
Candidate gene approach
Although there will probably be many whole genome association studies in the future, to date most studies of complex diseases, including infection, have adopted a candidate gene approach. The choice of the candidate genes may come from animal data,76, 77 results of whole-genome studies,57, 58, 78 clinical data,79, 80 or simply biological plausibility. Important methodological considerations in this approach include the following: (1) an adequate density of the markers in the candidate gene to ensure that the study has sufficient sensitivity to exclude a genetic region; (2) narrow and consistent phenotypic definitions; (3) matching of cases and controls; and (4) sample size and power.
Most candidate gene studies use a case-control approach, and unrecognised ethnic differences between the groups will probably result in spurious genetic associations that are unrelated to the disease of interest. For example, there is probably a strong association between the HLA region and chopstick use in Sydney. This does not imply that the HLA region necessarily influences manual dexterity, but more likely that the HLA type identifies an Asian subpopulation. This issue of population stratification is likely to be more confounded when small genetic effects are sought in large populations.81 However, this confounding can be minimised if cases and controls are carefully matched. The presence of genetic admixture can also be quantified from the genetic data itself,82 allowing stratified analyses if the sample size permits. In paediatric populations, an alternative approach is to use the parents as the genetic controls for their affected offspring.83 This approach is intrinsically less powerful than case-control designs, since the parental controls are enriched for the genetic variants of interest, but it allows inclusion of families of mixed ethnicity and potential pooling of data across ethnic groups, if significant genetic heterogeneity is not present.
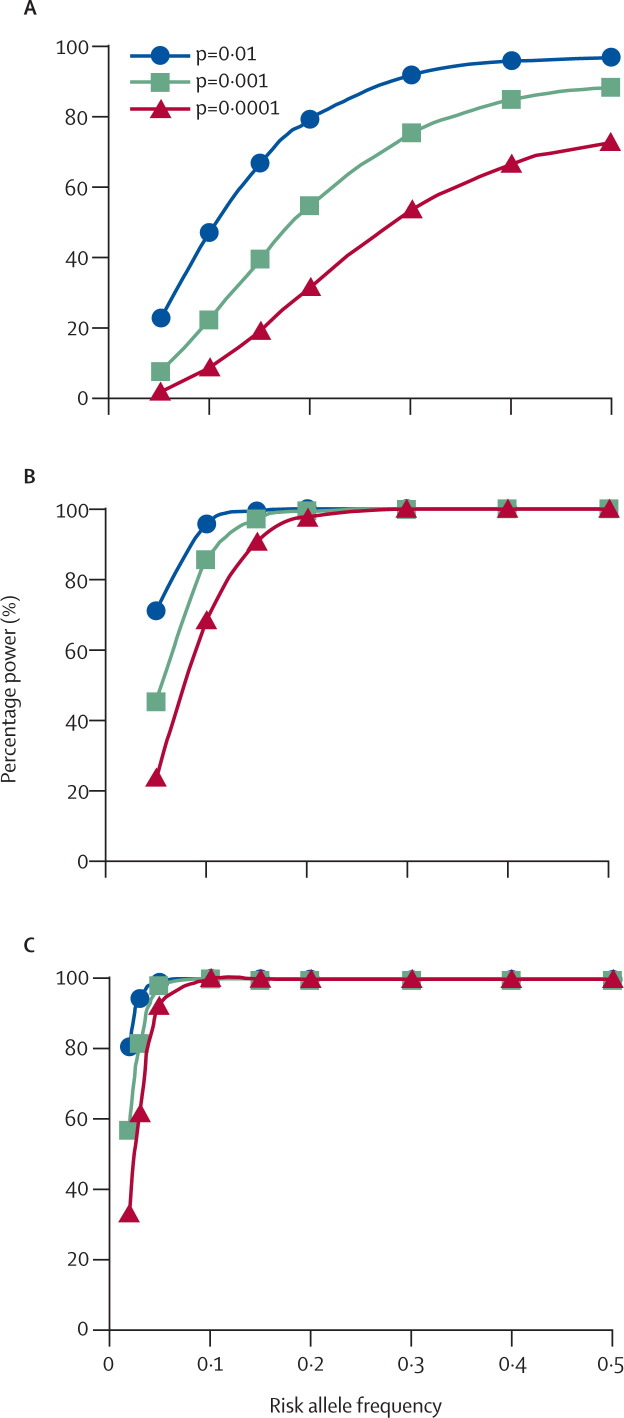
Probably the most crucial and overlooked methodological issue is sample size and power. Because the genetic effects from individual genetic variants are usually modest, the genetic model (mode of inheritance) is often unknown, and the frequency of the disease-modifying variant may vary, almost all studies of complex disease genetics to date have been underpowered. Power calculations (figure ) indicate that in excess of 1500 case-control pairs or case-parent trios are needed to provide adequate power, especially as effect sizes (odds ratios) expected for a genetic contribution to complex disease susceptibility will probably be small (about 1·5).
Figure.
Power to detect allelic association for a risk allele having an effect size (odds ratio) of 1·5
(A) 500, (B) 1500, and (C) 3000 trios or case-control pairs. Power is calculated for different frequencies of risk alleles, and for samples of 500, 1500, or 3000 trios or case-control pairs. Power approximations for trios by a standard transmission disequilibrium test84 have been made using the method of Knapp.85 Theoretical power to detect allelic association was made assuming a multiplicative model. Results are given as a first approximation of the percentage power to detect allelic association at p=0·01, p=0·001, or p=0·0001. Power calculations were essentially identical for a similar size case-control sample. The advantage of trios is that they are not influenced by population admixture. The advantage of case-control analysis is one third less genotyping to obtain equivalent power. The graphs show that 500 trios or case-control pairs have very poor power to detect allelic association even for relatively common risk alleles (frequencies >0·2). A sample of 1500 trios or case-control pairs has good power for risk alleles at frequencies >0·1. A sample of 3000 trios or case-control pairs improves power for rare risk alleles (frequencies <0·1).
The lack of power owing to small sample sizes makes it difficult to interpret current and previous data on genetic susceptibility to infection. For this reason we have not attempted a critical appraisal or value judgment of past and present studies, but have provided comprehensive tables (webtables 1, 2, 3 and 4) that list all of the studies undertaken to date for the most commonly studied infectious diseases—HIV, mycobacterial infections (leprosy and tuberculosis), malaria, and leishmaniasis. Whereas some loci (eg, HLA class I, class II, and class III alleles, SLC11A1) feature repeatedly both within and between diseases, no formal meta-analyses86 of these smaller studies have yet been undertaken to determine which may indicate statistically reliable associations. One interesting observation is the number of candidate gene studies that have focused on proteins involved in innate recognition of microbes and first-line defence innate immunity pathways (table 1 ), underscoring the likely selective pressures that have been exerted by infectious diseases. The key genes that regulate acquired immune responses also feature as candidate genes in multiple infectious diseases (table 2 ).
Table 1.
Examples of candidate genes related to innate immunity that are associated with infectious disease susceptibility or outcome
| Gene | Function | Infectious disease associations* |
|---|---|---|
| TNFA | Pro-inflammatory cytokine | HIV, hepatitis B,87 human papilloma virus,88, 89 meningococcal meningitis,90, 91 typhoid,92 leprosy, tuberculosis, malaria, leishmaniasis, Helicobacter pylori93, 94, 95 |
| IL1A, IL1B | Pro-inflammatory cytokine | HIV, hepatitis C,96 meningococcal meningitis,97H pylori,98, 99, 100 periodontitis,101, 102 tuberculosis, malaria |
| IL10 | Anti-inflammatory cytokine | HIV, Epstein-Barr virus,103, 104 herpes zoster virus,105 cytomegalovirus,106 hepatitis B and C,107H pylori,95, 99 meningococcal meningitis,91, 108 pneumonia109 and pneumococcal septic shock,110 tuberculosis, leprosy |
| NOS2A | Inducible nitric oxide synthase producing toxic nitrogen radicals | Hepatitis C,111 brucellosis,112 tuberculosis, malaria |
| FcγRIIA† | Activating Fc receptor promoting pro-inflammatory response | HIV, severe acute respiratory syndrome-associated coronavirus,113 dengue haemorrhagic fever,114 otitis media,115Streptococcus pneumoniae,116 rheumatic fever (streptococcus),117 periodontitis,118, 119, 120, 121 meningococcal meningitis and sepsis,108, 122, 123 malaria |
| FcγRIIB† | De-activating Fc receptor down regulating pro-inflammatory response | Periodontitis,124 malaria |
| SLC11A1 | Proton-coupled divalent cation transporter with multiple pleiotropic effects on macrophage function | HIV, hepatitis C,125 leprosy, tuberculosis, Buruli ulcer (Mycobacterium ulcerans),126 Kawasaki disease,127 visceral leishmaniasis |
| MBP (MBL) | Mannose binding lectin opsonises for complement activation by classic pathway | HIV, hepatitis B,128, 129 severe acute respiratory syndrome-associated coronavirus,130 Gram negative and positive bacterial infection,131 Kawasaki disease,132Chlamydia pneumoniae,133 meningococcal meningitis,134 aspergillosis,135 candidiasis,136 tuberculosis, malaria, filariasis137 |
| TLR2/TLR4 | Toll receptors 2 and 4;138 pattern recognition receptor for lipopolysaccharide and lipoproteins on bacteria and mycobacteria | Respiratory syncytial virus,139 meninogoccal meningitis,122, 140 Lyme disease (Borrelia bergdorferi),141 staphylococcal infection,142 periodontitis,143 Boutonneuse fever,144 Gram negative145, 146 and positive131 bacterial infection, brucellosis,147 Legionnaires' disease,148 tuberculosis, leprosy, malaria |
References given are only for disease associations not covered in webtables 1–4 that summarise candidate gene association studies for HIV, tuberculosis, leprosy, malaria, and leishmaniasis. References are not exhaustive, and generally provide examples of studies showing associations that have been published in the last 5 years.
These two genes are closely linked and are usually in linkage disequilibrium with each other, making it unclear which of these loci is the disease-associated gene.
Table 2.
Examples of candidate genes related to acquired immunity that are associated with infectious disease susceptibility or outcome
| Gene | Function | Infectious disease associations* |
|---|---|---|
| HLA class I | Presentation of antigen to CD8 T cells | HIV, tuberculosis, leprosy, malaria, leishmaniasis |
| HLA class II | Presentation of antigen to CD4 T cells | HIV, hepatitis C,149 tuberculosis, leprosy, malaria, leishmaniasis |
| IL4/IL13 | Cytokine product of T-helper 2 cells | HIV, hepatitis C,150 respiratory syncytial virus,151, 152H pylori,153 candidiasis,154 malaria, leishmaniasis, schistosomiasis155 |
| IFNG† | Cytokine product of T-helper 1 cells | HIV, hepatitis B,156 Epstein-Barr virus,157 respiratory syncytial virus,158, 159H pylori,95 brucellosis,160 malaria, tuberculosis, leprosy, schistosomiasis161 |
| IFNGR1† | Type 1 receptor for interferon γ on macrophages | Periodontitis,162H pylori,163 atypical mycobacterial infection,164, 165 tuberculosis, malaria, post-Kala-azar dermal leishmaniasis |
References given are only for disease associations not covered in webtables 1–4 that summarise candidate gene association studies for HIV, tuberculosis, leprosy, malaria, and leishmaniasis. References are not exhaustive, and generally provide examples of studies showing associations that have been published in the last 5 years.
Since natural killer cells make interferon γ, these genes could also be thought of as innate immunity genes.
Most of these studies also suffer from a lack of statistical power, and require replication and validation. of these immune response genes are highly polymorphic or clustered, or both, in the genome, which is an indication of evolutionary pressure and selection.166, 167 A more formal attempt to search for signals of evolutionary selection was recently undertaken in 168 genes related to immune function.3 Almost 1700 common SNPs were successfully genotyped across these genes in three population samples: 96 independent chromosomes from Utah residents with European ancestry, 120 from Han Chinese Guangxi, and 124 from the Yoruba people of Southwest Nigeria (these samples were overlapping but not identical to those used in the HapMap project). Evidence for selection was based on four tests of non-neutral evolution: (1) assessment of the percentage of SNPs within a locus with low (<10%) minor allele frequency and the percentage with high (>40%) minor allele frequency; (2) assessment of the frequency of derived (non-ancestral) alleles; (3) comparison of F ST (panel) versus heterozygosity; and (4) relative extended haplotype homozygosity. In tests of allele frequency, three of the 168 genes stood out: IL9, FUT2, and CAV2. IL9 lies in the cluster of T-helper-2 related genes encoding interleukins 4, 5, 9, and 13 on chromosome 5q31-q33, which has been implicated in many infectious disease studies (table 2; webtables 1–4). FUT2 encodes a protein responsible for determining ABO blood protein secretor status, and variation in this gene may protect against respiratory pathogens,168 H pylori,169 and HIV-1.170 CAV2 encodes one of the caveolin proteins that function as scaffolding structures in cholesterol-rich lipid rafts, which enhance immune cell function,168, 171 and are involved in pathogen entry into host cells.172 The relative extended haplotype homozygosity test provided the best evidence for selection, highlighting ABCC1 (or multidrug resistance protein MRP1, implicated in resistance to Streptococcus pneumoniae in mice173), APCS (serum amyloid P component), and VAV3 (a guanine nucleotide exchange factor expressed in haematopoietic and other cells, recently identified as a gene for type I diabetes in mice174) as genes with signatures of past selection. Although Walsh and colleagues3 discuss the limitations of their study and the need for validation, their research pilots the search for signatures of evolutionary selection that can now be undertaken in HapMap-style data, providing a real opportunity to identify the genes that have been under the strongest evolutionary pressure from infectious diseases. The advent of all these new technologies and tools offer exciting prospects for future research, allowing us to return to the drawing board in studying human genetic susceptibility to infectious disease. In so doing, it will be interesting to see how many of the candidate gene studies undertaken to date (webtables 1–4) are supported by potentially more powerful genome-wide association studies. Although a case can also be made for simply increasing sample size and power even with a more directed candidate gene approach, it is debatable whether the genes that show significant evidence of selection would have been readily chosen as obvious candidate loci. Thus, a combination of genome-wide, candidate gene, and other approaches may ultimately prove the most rewarding.
Gene–gene and gene–environment interactions
Genetic determinants obviously act in a genetic and environmental context, a fact often ignored in genetic epidemiological studies that have previously regarded the environment as something of an annoyance. A genetic variant may substantially increase the risk of avian influenza several hundred times, but if the person carrying the variant never encounters H5N1 influenza, or requires an additional polymorphism in a different gene to have a functional effect, then this first variant may be of no clinical relevance. The field of genetic epidemiology is increasingly acknowledging the importance of complex biological interactions in understanding genetic determinants. Gene–gene interactions (epistasis) have been described for HIV infection, in which an HLA class I allele is associated with more rapid progression to AIDS only when an individual also carries a specific natural killer cell (KIR) receptor.175 Gene–environment interactions have been shown in spontaneous preterm labour, which has an inflammatory basis. Although genetic variation at the tumour necrosis factor (TNFA) gene is associated with increased risk of preterm labour, this risk is increased significantly in the presence of bacterial vaginosis, itself an independent risk for the same outcome.176 In cystic fibrosis, those carrying functional variants of mannose-binding lectin (MBL) have worse pulmonary outcomes,177 and replacement MBL therapy has been reported in MBL-deficient cystic fibrosis patients.178 Not surprisingly, MBL exerts a protective effect through its innate immunological effects and in a recent small study, MBL deficiency only affected lung function when associated with Staphylococcus aureus colonisation,179 an example of a gene–enviroment interaction.
Conclusion: translation of genetic data into therapeutic interventions
Despite an increasing body of robust data identifying genetic determinants of infectious diseases and illuminating key biological pathways, the move from bench to bedside in infectious diseases and other fields remains largely an unrealised, but often made, prediction. There are a few worthy exceptions. Functional genetic variation of the P450 cytochrome system seems to mediate the efficacy of H pylori eradication by its effects on proton pump inhibitor metabolism.180 The most striking and elegant example of pharmacogenetics is in HIV medicine, in which hypersensitivity to the nucleoside reverse transcriptase inhibitor abacavir has previously limited its use. Analysis of the MHC led to the identification of a particular HLA haplotype that greatly increased the risk of abacavir hypersensitivity,181 and further mapping localised a functional variant in the peptide-binding groove of heat shock protein 70, which lies in the central MHC.182 Genotyping individuals for this variant before commencing abacavir is cost effective.183 Hopefully, further examples of clinically relevant pharmacogenetics in infectious diseases will be reported. In the face of increasing antimicrobial resistance, falling pharmaceutical enthusiasm for new antibiotic development, and the disappointing track record of adjunct biological therapies in severe sepsis, the development of novel vaccines and pharmacogenetic interventions remain among the most important goals for genetic studies of infectious disease susceptibility.
Search strategy and selection criteria
Data for this review were identified by searches of PubMed, MEDLINE, Current Contents, and references from relevant articles; numerous articles were identified through searches of the extensive files of the authors. Search terms included combinations of “susceptibility”, “genetic susceptibility”, “heritability”, “twin”, “genetic”, “genetic epidemiology”, “infection”, “infectious diseases”, as well as terms for specific infections (eg, “malaria”) and genes and gene products (eg, “mannose binding lectin”): eg, PubMed search term: “HIV AND polymorphism NOT drug”; field: text word; limits: humans. No date or language restrictions were set in these searches.
Conflicts of interest
We declare that we have have no conflicts of interest.
Acknowledgments
Acknowledgments
SEJ and JMB acknowledge financial support from The Wellcome Trust.
Conflicts of interest
We declare that we have have no conflicts of interest.
Web Extra Material
References
- 1.Black RE, Morris SS, Bryce J. Where and why are 10 million children dying every year? Lancet. 2003;361:2226–2234. doi: 10.1016/S0140-6736(03)13779-8. [DOI] [PubMed] [Google Scholar]
- 2.Frodsham AJ, Hill AV. Genetics of infectious diseases. Hum Mol Genet. 2004;13(suppl 2):R187–R194. doi: 10.1093/hmg/ddh225. [DOI] [PubMed] [Google Scholar]
- 3.Walsh EC, Sabeti P, Hutcheson HB. Searching for signals of evolutionary selection in 168 genes related to immune function. Hum Genet. 2005;119:92–102. doi: 10.1007/s00439-005-0090-0. [DOI] [PubMed] [Google Scholar]
- 4.Tishkoff SA, Dietzsch E, Speed W. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 5.Sabeti PC, Reich DE, Higgins JM. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 6.Murphy PM. Molecular mimicry and the generation of host defense protein diversity. Cell. 1993;72:823–826. doi: 10.1016/0092-8674(93)90571-7. [DOI] [PubMed] [Google Scholar]
- 7.Apanius V, Penn D, Slev PR, Ruff LR, Potts WK. The nature of selection on the major histocompatibility complex. Crit Rev Immunol. 1997;17:179–224. doi: 10.1615/critrevimmunol.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- 8.Walsh EC, Mather KA, Schaffner SF. An integrated haplotype map of the human major histocompatibility complex. Am J Hum Genet. 2003;73:580–590. doi: 10.1086/378101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thursz MR, Thomas HC, Greenwood BM, Hill AV. Heterozygote advantage for HLA class-II type in hepatitis B virus infection. Nat Genet. 1997;17:11–12. doi: 10.1038/ng0997-11. [DOI] [PubMed] [Google Scholar]
- 10.Carrington M, Nelson GW, Martin MP. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 11.Black FL, Schiffman G, Pandey JP. HLA, Gm, and Km polymorphisms and immunity to infectious diseases in South Amerinds. Exp Clin Immunogenet. 1995;12:206–216. doi: 10.1159/000424873. [DOI] [PubMed] [Google Scholar]
- 12.Casanova JL, Fieschi C, Bustamante J. From idiopathic infectious diseases to novel primary immunodeficiencies. J Allergy Clin Immunol. 2005;116:426–430. doi: 10.1016/j.jaci.2005.03.053. [DOI] [PubMed] [Google Scholar]
- 13.Casanova JL, Abel L. Inborn errors of immunity to infection: the rule rather than the exception. J Exp Med. 2005;202:197–201. doi: 10.1084/jem.20050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allison AC. Protection afforded by sickle-cell trait against subtertian malareal infection. Br Med J. 1954;4857:290–294. doi: 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michon P, Woolley I, Wood EM, Kastens W, Zimmerman PA, Adams JH. Duffy-null promoter heterozygosity reduces DARC expression and abrogates adhesion of the P vivax ligand required for blood-stage infection. FEBS Lett. 2001;495:111–114. doi: 10.1016/s0014-5793(01)02370-5. [DOI] [PubMed] [Google Scholar]
- 16.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 17.Lindesmith L, Moe C, Marionneau S. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 18.Kindberg E, Hejdeman B, Bratt G. A nonsense mutation (428G→A) in the fucosyltransferase FUT2 gene affects the progression of HIV-1 infection. AIDS. 2006;20:685–689. doi: 10.1097/01.aids.0000216368.23325.bc. [DOI] [PubMed] [Google Scholar]
- 19.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Weiss KM, Terwilliger JD. How many diseases does it take to map a gene with SNPs? Nat Genet. 2000;26:151–157. doi: 10.1038/79866. [DOI] [PubMed] [Google Scholar]
- 21.Alcais A, Fieschi C, Abel L, Casanova JL. Tuberculosis in children and adults: two distinct genetic diseases. J Exp Med. 2005;202:1617–1621. doi: 10.1084/jem.20052302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haralambous E, Weiss HA, Radalowicz A, Hibberd ML, Booy R, Levin M. Sibling familial risk ratio of meningococcal disease in UK caucasians. Epidemiol Infect. 2003;130:413–418. [PMC free article] [PubMed] [Google Scholar]
- 23.Morrow HW, Slaten DD, Reingold AL, Werner SB, Fenstersheib MD. Risk factors associated with a school-related outbreak of serogroup C meningococcal disease. Pediatr Infect Dis J. 1990;9:394–398. doi: 10.1097/00006454-199006000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Kallmann FJ, Reisner D. Twin studies on the significance of genetic factors in tuberculosis. Am Rev Tuberc. 1943;47:549–574. [Google Scholar]
- 25.Comstock GW. Tuberculosis in twins: a re-analysis of the Prophit survey. Am Rev Respir Dis. 1978;117:621–624. doi: 10.1164/arrd.1978.117.4.621. [DOI] [PubMed] [Google Scholar]
- 26.Simonds B. Tuberculosis in twins. Pitman Medical Publishing Co. Ltd; London: 1963. [Google Scholar]
- 27.Jepson A. Twin studies for the analysis of heritability of infectious diseases. Bull Inst Pasteur. 1998;96:71–81. [Google Scholar]
- 28.Marshall AG, Hutchinson EO, Honisett J. Heredity in common diseases. A retrospective survey of twins in a hospital population. Br Med J. 1962;5270:1–6. doi: 10.1136/bmj.1.5270.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malaty HM, Engstrand L, Pedersen NL, Graham DY. Helicobacter pylori infection: genetic and environmental influences. A study of twins. Ann Intern Med. 1994;120:982–986. doi: 10.7326/0003-4819-120-12-199406150-00002. [DOI] [PubMed] [Google Scholar]
- 30.Chakravartti MR, Vogel F. A twin study on leprosy. Georg Thieme; Stuttgart: 1973. [Google Scholar]
- 31.Rovers M, Haggard M, Gannon M, Koeppen-Schomerus G, Plomin R. Heritability of symptom domains in otitis media: a longitudinal study of 1,373 twin pairs. Am J Epidemiol. 2002;155:958–964. doi: 10.1093/aje/155.10.958. [DOI] [PubMed] [Google Scholar]
- 32.Casselbrant ML, Mandel EM, Fall PA. The heritability of otitis media: a twin and triplet study. JAMA. 1999;282:2125–2130. doi: 10.1001/jama.282.22.2125. [DOI] [PubMed] [Google Scholar]
- 33.Lin TM, Chen CJ, Wu MM. Hepatitis B virus markers in Chinese twins. Anticancer Res. 1989;9:737–741. [PubMed] [Google Scholar]
- 34.Jepson AP, Banya WA, Sisay-Joof F, Hassan-King M, Bennett S, Whittle HC. Genetic regulation of fever in Plasmodium falciparum malaria in Gambian twin children. J Infect Dis. 1995;172:316–319. doi: 10.1093/infdis/172.1.316. [DOI] [PubMed] [Google Scholar]
- 35.Konradsen HB, Henrichsen J, Wachmann H, Holm N. The influence of genetic factors on the immune response as judged by pneumococcal vaccination of mono- and dizygotic caucasian twins. Clin Exp Immunol. 1993;92:532–536. doi: 10.1111/j.1365-2249.1993.tb03433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jepson A, Fowler A, Banya W. Genetic regulation of acquired immune responses to antigens of Mycobacterium tuberculosis: a study of twins in West Africa. Infect Immun. 2001;69:3989–3994. doi: 10.1128/IAI.69.6.3989-3994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newport MJ, Goetghebuer T, Weiss HA, Whittle H, Siegrist CA, Marchant A. Genetic regulation of immune responses to vaccines in early life. Genes Immun. 2004;5:122–129. doi: 10.1038/sj.gene.6364051. [DOI] [PubMed] [Google Scholar]
- 38.Jepson A, Banya W, Sisay-Joof F. Quantification of the relative contribution of major histocompatibility complex (MHC) and non-MHC genes to human immune responses to foreign antigens. Infect Immun. 1997;65:872–876. doi: 10.1128/iai.65.3.872-876.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Craen AJ, Posthuma D, Remarque EJ, van den Biggelaar AH, Westendorp RG, Boomsma DI. Heritability estimates of innate immunity: an extended twin study. Genes Immun. 2005;6:167–170. doi: 10.1038/sj.gene.6364162. [DOI] [PubMed] [Google Scholar]
- 40.Westendorp RG, Langermans JA, Huizinga TW. Genetic influence on cytokine production and fatal meningococcal disease [published erratum appears in Lancet 1997; 349: 656; see comments] Lancet. 1997;349:170–173. doi: 10.1016/s0140-6736(96)06413-6. [DOI] [PubMed] [Google Scholar]
- 41.Sorensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 42.Burton PR, Tobin MD, Hopper JL. Key concepts in genetic epidemiology. Lancet. 2005;366:941–951. doi: 10.1016/S0140-6736(05)67322-9. [DOI] [PubMed] [Google Scholar]
- 43.Dawn Teare M, Barrett JH. Genetic linkage studies. Lancet. 2005;366:1036–1044. doi: 10.1016/S0140-6736(05)67382-5. [DOI] [PubMed] [Google Scholar]
- 44.Cordell HJ, Clayton DG. Genetic association studies. Lancet. 2005;366:1121–1131. doi: 10.1016/S0140-6736(05)67424-7. [DOI] [PubMed] [Google Scholar]
- 45.Palmer LJ, Cardon LR. Shaking the tree: mapping complex disease genes with linkage disequilibrium. Lancet. 2005;366:1223–1234. doi: 10.1016/S0140-6736(05)67485-5. [DOI] [PubMed] [Google Scholar]
- 46.Hattersley AT, McCarthy MI. What makes a good genetic association study? Lancet. 2005;366:1315–1323. doi: 10.1016/S0140-6736(05)67531-9. [DOI] [PubMed] [Google Scholar]
- 47.Hopper JL, Bishop DT, Easton DF. Population-based family studies in genetic epidemiology. Lancet. 2005;366:1397–1406. doi: 10.1016/S0140-6736(05)67570-8. [DOI] [PubMed] [Google Scholar]
- 48.Davey Smith G, Ebrahim S, Lewis S, Hansell AL, Palmer LJ, Burton PR. Genetic epidemiology and public health: hope, hype, and future prospects. Lancet. 2005;366:1484–1498. doi: 10.1016/S0140-6736(05)67601-5. [DOI] [PubMed] [Google Scholar]
- 49.Taylor MF, Shen Y, Kreitman ME. A population genetic test of selection at the molecular level. Science. 1995;270:1497–1499. doi: 10.1126/science.270.5241.1497. [DOI] [PubMed] [Google Scholar]
- 50.Affymetrix. GeneChip® human mapping 10K array and assay kit. http://www.affymetrix.com/products/arrays/specific/10k.affx (accessed Aug 2, 2006)
- 51.Illumina. Sentrix® humanhap300 genotyping beadchip. http://www.illumina.com/products/arraysreagents/wgghumanhap300.ilmn (accessed Aug 2, 2006)
- 52.Malats N, Calafell F. Advanced glossary on genetic epidemiology. J Epidemiol Community Health. 2003;57:562–564. doi: 10.1136/jech.57.8.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malats N, Calafell F. Basic glossary on genetic epidemiology. J Epidemiol Community Health. 2003;57:480–482. doi: 10.1136/jech.57.7.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calafell F, Malats N. Basic molecular genetics for epidemiologists. J Epidemiol Community Health. 2003;57:398–400. doi: 10.1136/jech.57.6.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crawford DC, Akey DT, Nickerson DA. The patterns of natural variation in human genes. Annu Rev Genomics Hum Genet. 2005;6:287–312. doi: 10.1146/annurev.genom.6.080604.162309. [DOI] [PubMed] [Google Scholar]
- 56.Gonzalez E, Kulkarni H, Bolivar H. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 57.Mira MT, Alcais A, Van Thuc N. Chromosome 6q25 is linked to susceptibility to leprosy in a Vietnamese population. Nat Genet. 2003;33:412–415. doi: 10.1038/ng1096. [DOI] [PubMed] [Google Scholar]
- 58.Siddiqui MR, Meisner S, Tosh K. A major susceptibility locus for leprosy in India maps to chromosome 10p13. Nat Genet. 2001;27:439–441. doi: 10.1038/86958. [DOI] [PubMed] [Google Scholar]
- 59.Marquet S, Laurent A, Hillaire D. Genetic localization of a locus controlling the intensity of infection by Schistosoma mansoni on chromosome 5q31-q33. Nat Genet. 1996;14:181–184. doi: 10.1038/ng1096-181. [DOI] [PubMed] [Google Scholar]
- 60.Thye T, Burchard GD, Nilius M, Muller-Myhsok B, Horstmann RD. Genomewide linkage analysis identifies polymorphism in the human interferon-gamma receptor affecting Helicobacter pylori infection. Am J Hum Genet. 2003;72:448–453. doi: 10.1086/367714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mira MT, Alcais A, Nguyen VT. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427:636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 62.Russell RK, Wilson DC, Satsangi J. Unravelling the complex genetics of inflammatory bowel disease. Arch Dis Child. 2004;89:598–603. doi: 10.1136/adc.2003.041046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 64.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 65.Wellcome Trust Case-Control Consortium Overview. http://www.wtccc.org.uk/info/overview.shtml (accessed Aug 2, 2006)
- 66.Ozaki K, Inoue K, Sato H. Functional variation in LGALS2 confers risk of myocardial infarction and regulates lymphotoxin-alpha secretion in vitro. Nature. 2004;429:72–75. doi: 10.1038/nature02502. [DOI] [PubMed] [Google Scholar]
- 67.Ozaki K, Ohnishi Y, Iida A. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- 68.Klein RJ, Zeiss C, Chew EY. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haines JL, Hauser MA, Schmidt S. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 70.Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 71.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stankovich J, Cox CJ, Tan RB. On the utility of data from the International HapMap Project for Australian association studies. Hum Genet. 2006;119:220–222. doi: 10.1007/s00439-005-0120-y. [DOI] [PubMed] [Google Scholar]
- 73.Ribas J, Gonzalez-Neira A, Salas A. Evaluating HapMap SNP data transferability in a large-scale genotyping project involving 175 cancer-associated genes. Hum Genet. 2006;118:669–679. doi: 10.1007/s00439-005-0094-9. [DOI] [PubMed] [Google Scholar]
- 74.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies [published erratum in Nat Genet 2006; 38: 390] Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 75.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 76.Blackwell JM, Searle S, Goswami T, Miller EN. Understanding the multiple functions of NRAMP1. Microbes Infect. 2000;2:317–321. doi: 10.1016/s1286-4579(00)00295-1. [DOI] [PubMed] [Google Scholar]
- 77.Bellamy R, Ruwende C, Corrah T, McAdam KPWJ, Whittle HC, Hill AVS. Variation in the NRAMP1 gene is associated with susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 78.Bellamy R, Beyers N, McAdam KP. Genetic susceptibility to tuberculosis in Africans: a genome-wide scan. Proc Natl Acad Sci USA. 2000;97:8005–8009. doi: 10.1073/pnas.140201897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kwiatkowski D. Tumour necrosis factor, fever and fatality in falciparum malaria. Immunol Lett. 1990;25:213–216. doi: 10.1016/0165-2478(90)90117-9. [DOI] [PubMed] [Google Scholar]
- 80.McGuire W, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D. Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature. 1994;371:508–510. doi: 10.1038/371508a0. [DOI] [PubMed] [Google Scholar]
- 81.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 82.Freedman ML, Reich D, Penney KL. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 83.Spielman RS, Ewens WJ. The TDT and other family-based tests for linkage disequilibrium and association [editorial] Am J Hum Genet. 1996;59:983–989. [PMC free article] [PubMed] [Google Scholar]
- 84.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 85.Knapp M. A note on power approximations for the transmission/disequilibrium test. Am J Hum Genet. 1999;64:1177–1185. doi: 10.1086/302334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bracken MB. Genomic epidemiology of complex disease: the need for an electronic evidence-based approach to research synthesis. Am J Epidemiol. 2005;162:297–301. doi: 10.1093/aje/kwi200. [DOI] [PubMed] [Google Scholar]
- 87.Liu Y, Guo XH, Du T. Association of TNFA polymorphisms with the outcomes of HBV infection. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2005;22:406–410. [PubMed] [Google Scholar]
- 88.Simoes RT, Goncalves MA, Donadi EA. Association of tumor necrosis factor a-2 and a-8 microsatellite alleles with human papillomavirus and squamous intraepithelial lesions among women in Brazil. J Clin Microbiol. 2005;43:3932–3937. doi: 10.1128/JCM.43.8.3932-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghaderi M, Nikitina L, Peacock CS. Tumor necrosis factor a-11 and DR15-DQ6 (B*0602) haplotype increase the risk for cervical intraepithelial neoplasia in human papillomavirus 16 seropositive women in Northern Sweden. Cancer Epidemiol Biomarkers Prev. 2000;9:1067–1070. [PubMed] [Google Scholar]
- 90.Nadel S, Newport MJ, Booy R, Levin M. Variation in the tumor necrosis factor-alpha gene promoter region may be associated with death from meningococcal disease. J Infect Dis. 1996;174:878–880. doi: 10.1093/infdis/174.4.878. [DOI] [PubMed] [Google Scholar]
- 91.Westendorp RG, Langermans JA, Huizinga TW, Verweij CL, Sturk A. Genetic influence on cytokine production in meningococcal disease. Lancet. 1997;349:1912–1913. doi: 10.1016/s0140-6736(05)63910-4. [DOI] [PubMed] [Google Scholar]
- 92.Dunstan SJ, Stephens HA, Blackwell JM. Genes of the class II and class III major histocompatibility complex are associated with typhoid fever in Vietnam. J Infect Dis. 2001;183:261–268. doi: 10.1086/317940. [DOI] [PubMed] [Google Scholar]
- 93.Goto H. Helicobacter pylori and gastric diseases. Nagoya J Med Sci. 2003;66:77–85. [PubMed] [Google Scholar]
- 94.Kunstmann E, Epplen C, Elitok E. Helicobacter pylori infection and polymorphisms in the tumor necrosis factor region. Electrophoresis. 1999;20:1756–1761. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1756::AID-ELPS1756>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 95.Zambon CF, Basso D, Navaglia F. Pro- and anti-inflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine. 2005;29:141–152. doi: 10.1016/j.cyto.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 96.Bahr MJ, el Menuawy M, Boeker KH, Musholt PB, Manns MP, Lichtinghagen R. Cytokine gene polymorphisms and the susceptibility to liver cirrhosis in patients with chronic hepatitis C. Liver Int. 2003;23:420–425. doi: 10.1111/j.1478-3231.2003.00873.x. [DOI] [PubMed] [Google Scholar]
- 97.Read RC, Camp NJ, di Giovine FS. An interleukin-1 genotype is associated with fatal outcome of meningococcal disease. J Infect Dis. 2000;182:1557–1560. doi: 10.1086/315889. [DOI] [PubMed] [Google Scholar]
- 98.Xuan J, Deguchi R, Watanabe S. Relationship between IL-1beta gene polymorphism and gastric mucosal IL-1beta levels in patients with Helicobacter pylori infection. J Gastroenterol. 2005;40:796–801. doi: 10.1007/s00535-005-1630-z. [DOI] [PubMed] [Google Scholar]
- 99.Sicinschi LA, Lopez-Carrillo L, Camargo MC. Gastric cancer risk in a Mexican population: role of Helicobacter pylori CagA positive infection and polymorphisms in interleukin-1 and -10 genes. Int J Cancer. 2006;118:649–657. doi: 10.1002/ijc.21364. [DOI] [PubMed] [Google Scholar]
- 100.Vilaichone RK, Mahachai V, Tumwasorn S, Wu JY, Graham DY, Yamaoka Y. Gastric mucosal cytokine levels in relation to host interleukin-1 polymorphisms and Helicobacter pylori cagA genotype. Scand J Gastroenterol. 2005;40:530–539. doi: 10.1080/00365520510012299. [DOI] [PubMed] [Google Scholar]
- 101.Brett PM, Zygogianni P, Griffiths GS. Functional gene polymorphisms in aggressive and chronic periodontitis. J Dent Res. 2005;84:1149–1153. doi: 10.1177/154405910508401211. [DOI] [PubMed] [Google Scholar]
- 102.Moreira PR, de Sa AR, Xavier GM. A functional interleukin-1 beta gene polymorphism is associated with chronic periodontitis in a sample of Brazilian individuals. J Periodontal Res. 2005;40:306–311. doi: 10.1111/j.1600-0765.2005.00801.x. [DOI] [PubMed] [Google Scholar]
- 103.Helminen ME, Kilpinen S, Virta M, Hurme M. Susceptibility to primary Epstein-Barr virus infection is associated with interleukin-10 gene promoter polymorphism. J Infect Dis. 2001;184:777–780. doi: 10.1086/322987. [DOI] [PubMed] [Google Scholar]
- 104.Helminen M, Lahdenpohja N, Hurme M. Polymorphism of the interleukin-10 gene is associated with susceptibility to Epstein-Barr virus infection. J Infect Dis. 1999;180:496–499. doi: 10.1086/314883. [DOI] [PubMed] [Google Scholar]
- 105.Haanpaa M, Nurmikko T, Hurme M. Polymorphism of the IL-10 gene is associated with susceptibility to herpes zoster. Scand J Infect Dis. 2002;34:112–114. doi: 10.1080/00365540110077218. [DOI] [PubMed] [Google Scholar]
- 106.Hurme M, Haanpaa M, Nurmikko T. IL-10 gene polymorphism and herpesvirus infections. J Med Virol. 2003;70(suppl 1):S48–S50. doi: 10.1002/jmv.10320. [DOI] [PubMed] [Google Scholar]
- 107.Opdal SH. IL-10 gene polymorphisms in infectious disease and SIDS. FEMS Immunol Med Microbiol. 2004;42:48–52. doi: 10.1016/j.femsim.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 108.van der Pol WL, Huizinga TW, Vidarsson G. Relevance of Fcgamma receptor and interleukin-10 polymorphisms for meningococcal disease. J Infect Dis. 2001;184:1548–1555. doi: 10.1086/324662. [DOI] [PubMed] [Google Scholar]
- 109.Gallagher PM, Lowe G, Fitzgerald T. Association of IL-10 polymorphism with severity of illness in community acquired pneumonia. Thorax. 2003;58:154–156. doi: 10.1136/thorax.58.2.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schaaf BM, Boehmke F, Esnaashari H. Pneumococcal septic shock is associated with the interleukin-10-1082 gene promoter polymorphism. Am J Respir Crit Care Med. 2003;168:476–480. doi: 10.1164/rccm.200210-1164OC. [DOI] [PubMed] [Google Scholar]
- 111.Yee LJ, Knapp S, Burgner D. Inducible nitric oxide synthase gene (NOS2A) haplotypes and the outcome of hepatitis C virus infection. Genes Immun. 2004;5:183–187. doi: 10.1038/sj.gene.6364054. [DOI] [PubMed] [Google Scholar]
- 112.Orozco G, Sanchez E, Lopez-Nevot MA. Inducible nitric oxide synthase promoter polymorphism in human brucellosis. Microbes Infect. 2003;5:1165–1169. doi: 10.1016/j.micinf.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 113.Yuan FF, Tanner J, Chan PK. Influence of FcgammaRIIA and MBL polymorphisms on severe acute respiratory syndrome. Tissue Antigens. 2005;66:291–296. doi: 10.1111/j.1399-0039.2005.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Loke H, Bethell D, Phuong CX. Susceptibility to dengue hemorrhagic fever in Vietnam: evidence of an association with variation in the vitamin D receptor and Fc gamma receptor IIa genes. Am J Trop Med Hyg. 2002;67:102–106. doi: 10.4269/ajtmh.2002.67.102. [DOI] [PubMed] [Google Scholar]
- 115.Straetemans M, Wiertsema SP, Sanders EA. Immunological status in the aetiology of recurrent otitis media with effusion: serum immunoglobulin levels, functional mannose-binding lectin and Fc receptor polymorphisms for IgG. J Clin Immunol. 2005;25:78–86. doi: 10.1007/s10875-005-0361-8. [DOI] [PubMed] [Google Scholar]
- 116.Yuan FF, Wong M, Pererva N. FcgammaRIIA polymorphisms in Streptococcus pneumoniae infection. Immunol Cell Biol. 2003;81:192–195. doi: 10.1046/j.1440-1711.2003.01158.x. [DOI] [PubMed] [Google Scholar]
- 117.Berdeli A, Celik HA, Ozyurek R, Aydin HH. Involvement of immunoglobulin FcgammaRIIA and FcgammaRIIIB gene polymorphisms in susceptibility to rheumatic fever. Clin Biochem. 2004;37:925–929. doi: 10.1016/j.clinbiochem.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 118.Tang Y, Zhang JC, Zhang WH, Pang RY. The association between Fc gamma receptor IIA gene polymorphism and susceptibility to chronic periodontitis in Chinese Han nationality. Hua Xi Kou Qiang Yi Xue Za Zhi. 2004;22:158–161. [PubMed] [Google Scholar]
- 119.Yamamoto K, Kobayashi T, Grossi S. Association of Fcgamma receptor IIa genotype with chronic periodontitis in caucasians. J Periodontol. 2004;75:517–522. doi: 10.1902/jop.2004.75.4.517. [DOI] [PubMed] [Google Scholar]
- 120.Chung HY, Lu HC, Chen WL, Lu CT, Yang YH, Tsai CC. Gm (23) allotypes and Fcgamma receptor genotypes as risk factors for various forms of periodontitis. J Clin Periodontol. 2003;30:954–960. doi: 10.1034/j.1600-051x.2003.00409.x. [DOI] [PubMed] [Google Scholar]
- 121.Loos BG, Leppers-Van de Straat FG, Van de Winkel JG, Van der Velden U. Fcgamma receptor polymorphisms in relation to periodontitis. J Clin Periodontol. 2003;30:595–602. doi: 10.1034/j.1600-051x.2003.00355.x. [DOI] [PubMed] [Google Scholar]
- 122.Emonts M, Hazelzet JA, de Groot R, Hermans PW. Host genetic determinants of Neisseria meningitidis infections. Lancet Infect Dis. 2003;3:565–577. doi: 10.1016/s1473-3099(03)00740-0. [DOI] [PubMed] [Google Scholar]
- 123.Domingo P, Muniz-Diaz E, Baraldes MA. Associations between Fc gamma receptor IIA polymorphisms and the risk and prognosis of meningococcal disease. Am J Med. 2002;112:19–25. doi: 10.1016/s0002-9343(01)01047-6. [DOI] [PubMed] [Google Scholar]
- 124.Yasuda K, Sugita N, Kobayashi T, Yamamoto K, Yoshie H. FcgammaRIIB gene polymorphisms in Japanese periodontitis patients. Genes Immun. 2003;4:541–546. doi: 10.1038/sj.gene.6364021. [DOI] [PubMed] [Google Scholar]
- 125.Romero-Gomez M, Montes-Cano MA, Otero-Fernandez MA. SLC11A1 promoter gene polymorphisms and fibrosis progression in chronic hepatitis C. Gut. 2004;53:446–450. doi: 10.1136/gut.2003.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stienstra Y, van der Werf TS, Oosterom E. Susceptibility to Buruli ulcer is associated with the SLC11A1 (NRAMP1) D543N polymorphism. Genes Immun. 2006;7:185–189. doi: 10.1038/sj.gene.6364281. [DOI] [PubMed] [Google Scholar]
- 127.Ouchi K, Suzuki Y, Shirakawa T, Kishi F. Polymorphism of SLC11A1 (formerly NRAMP1) gene confers susceptibility to Kawasaki disease. J Infect Dis. 2003;187:326–329. doi: 10.1086/345878. [DOI] [PubMed] [Google Scholar]
- 128.Chong WP, To YF, Ip WK. Mannose-binding lectin in chronic hepatitis B virus infection. Hepatology. 2005;42:1037–1045. doi: 10.1002/hep.20891. [DOI] [PubMed] [Google Scholar]
- 129.Thio CL, Mosbruger T, Astemborski J. Mannose binding lectin genotypes influence recovery from hepatitis B virus infection. J Virol. 2005;79:9192–9196. doi: 10.1128/JVI.79.14.9192-9196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang H, Zhou G, Zhi L. Association between mannose-binding lectin gene polymorphisms and susceptibility to severe acute respiratory syndrome coronavirus infection. J Infect Dis. 2005;192:1355–1361. doi: 10.1086/491479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sutherland AM, Walley KR, Russell JA. Polymorphisms in CD14, mannose-binding lectin, and Toll-like receptor-2 are associated with increased prevalence of infection in critically ill adults. Crit Care Med. 2005;33:638–644. doi: 10.1097/01.ccm.0000156242.44356.c5. [DOI] [PubMed] [Google Scholar]
- 132.Yang J, Li CR, Li YB, Huang HJ, Li RX, Wang GB. Correlation between mannose-binding lectin gene codon 54 polymorphism and susceptibility of Kawasaki disease. Zhonghua Er Ke Za Zhi. 2004;42:176–179. [PubMed] [Google Scholar]
- 133.Nagy A, Kozma GT, Keszei M, Treszl A, Falus A, Szalai C. The development of asthma in children infected with Chlamydia pneumoniae is dependent on the modifying effect of mannose-binding lectin. J Allergy Clin Immunol. 2003;112:729–734. doi: 10.1016/s0091-6749(03)02010-4. [DOI] [PubMed] [Google Scholar]
- 134.Hibberd ML, Sumiya M, Summerfield JA, Booy R, Levin M. Association of variants of the gene for mannose-binding lectin with susceptibility to meningococcal disease. Lancet. 1999;353:1049–1053. doi: 10.1016/s0140-6736(98)08350-0. [DOI] [PubMed] [Google Scholar]
- 135.Madan T, Kaur S, Saxena S. Role of collectins in innate immunity against aspergillosis. Med Mycol. 2005;43(suppl 1):S155–S163. doi: 10.1080/13693780500088408. [DOI] [PubMed] [Google Scholar]
- 136.Babula O, Lazdane G, Kroica J, Ledger WJ, Witkin SS. Relation between recurrent vulvovaginal candidiasis, vaginal concentrations of mannose-binding lectin, and a mannose-binding lectin gene polymorphism in Latvian women. Clin Infect Dis. 2003;37:733–737. doi: 10.1086/377234. [DOI] [PubMed] [Google Scholar]
- 137.Choi EH, Zimmerman PA, Foster CB. Genetic polymorphisms in molecules of innate immunity and susceptibility to infection with Wuchereria bancrofti in South India. Genes Immun. 2001;2:248–253. doi: 10.1038/sj.gene.6363767. [DOI] [PubMed] [Google Scholar]
- 138.Schroder NW, Schumann RR. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect Dis. 2005;5:156–164. doi: 10.1016/S1473-3099(05)01308-3. [DOI] [PubMed] [Google Scholar]
- 139.Tal G, Mandelberg A, Dalal I. Association between common Toll-like receptor 4 mutations and severe respiratory syncytial virus disease. J Infect Dis. 2004;189:2057–2063. doi: 10.1086/420830. [DOI] [PubMed] [Google Scholar]
- 140.Smirnova I, Mann N, Dols A. Assay of locus-specific genetic load implicates rare Toll-like receptor 4 mutations in meningococcal susceptibility. Proc Natl Acad Sci USA. 2003;100:6075–6080. doi: 10.1073/pnas.1031605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schroder NW, Diterich I, Zinke A. Heterozygous Arg753Gln polymorphism of human TLR-2 impairs immune activation by Borrelia burgdorferi and protects from late stage Lyme disease. J Immunol. 2005;175:2534–2540. doi: 10.4049/jimmunol.175.4.2534. [DOI] [PubMed] [Google Scholar]
- 142.Lorenz E, Mira JP, Cornish KL, Arbour NC, Schwartz DA. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect Immun. 2000;68:6398–6401. doi: 10.1128/iai.68.11.6398-6401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schroder NW, Meister D, Wolff V. Chronic periodontal disease is associated with single-nucleotide polymorphisms of the human TLR-4 gene. Genes Immun. 2005;6:448–451. doi: 10.1038/sj.gene.6364221. [DOI] [PubMed] [Google Scholar]
- 144.Balistreri CR, Candore G, Lio D. Role of TLR4 receptor polymorphisms in Boutonneuse fever. Int J Immunopathol Pharmacol. 2005;18:655–660. doi: 10.1177/039463200501800406. [DOI] [PubMed] [Google Scholar]
- 145.Agnese DM, Calvano JE, Hahm SJ. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. J Infect Dis. 2002;186:1522–1525. doi: 10.1086/344893. [DOI] [PubMed] [Google Scholar]
- 146.Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med. 2002;162:1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 147.Rezazadeh M, Hajilooi M, Rafiei A. TLR4 polymorphism in Iranian patients with brucellosis. J Infect. 2006;53:206–210. doi: 10.1016/j.jinf.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 148.Hawn TR, Verbon A, Janer M, Zhao LP, Beutler B, Aderem A. Toll-like receptor 4 polymorphisms are associated with resistance to Legionnaires' disease. Proc Natl Acad Sci USA. 2005;102:2487–2489. doi: 10.1073/pnas.0409831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yee LJ. Host genetic determinants in hepatitis C virus infection. Genes Immun. 2004;5:237–245. doi: 10.1038/sj.gene.6364090. [DOI] [PubMed] [Google Scholar]
- 150.Saito T, Ji G, Shinzawa H. Genetic variations in humans associated with differences in the course of hepatitis C. Biochem Biophys Res Commun. 2004;317:335–341. doi: 10.1016/j.bbrc.2004.03.056. [DOI] [PubMed] [Google Scholar]
- 151.Puthothu B, Krueger M, Forster J, Heinzmann A. Association between severe respiratory syncytial virus infection and IL13/IL4 haplotypes. J Infect Dis. 2006;193:438–441. doi: 10.1086/499316. [DOI] [PubMed] [Google Scholar]
- 152.Choi EH, Lee HJ, Yoo T, Chanock SJ. A common haplotype of interleukin-4 gene IL4 is associated with severe respiratory syncytial virus disease in Korean children. J Infect Dis. 2002;186:1207–1211. doi: 10.1086/344310. [DOI] [PubMed] [Google Scholar]
- 153.Pessi T, Virta M, Adjers K. Genetic and environmental factors in the immunopathogenesis of atopy: interaction of Helicobacter pylori infection and IL4 genetics. Int Arch Allergy Immunol. 2005;137:282–288. doi: 10.1159/000086421. [DOI] [PubMed] [Google Scholar]
- 154.Babula O, Lazdane G, Kroica J, Linhares IM, Ledger WJ, Witkin SS. Frequency of interleukin-4 (IL-4) -589 gene polymorphism and vaginal concentrations of IL-4, nitric oxide, and mannose-binding lectin in women with recurrent vulvovaginal candidiasis. Clin Infect Dis. 2005;40:1258–1262. doi: 10.1086/429246. [DOI] [PubMed] [Google Scholar]
- 155.Kouriba B, Chevillard C, Bream JH. Analysis of the 5q31-q33 locus shows an association between IL13-1055C/T IL-13-591A/G polymorphisms and Schistosoma haematobium infections. J Immunol. 2005;174:6274–6281. doi: 10.4049/jimmunol.174.10.6274. [DOI] [PubMed] [Google Scholar]
- 156.Zhu QR, Ge YL, Gu SQ. Relationship between cytokines gene polymorphism and susceptibility to hepatitis B virus intrauterine infection. Chin Med J (Engl) 2005;118:1604–1609. [PubMed] [Google Scholar]
- 157.Bogunia-Kubik K, Mlynarczewska A, Jaskula E, Lange A. The presence of IFNG 3/3 genotype in the recipient associates with increased risk for Epstein-Barr virus reactivation after allogeneic haematopoietic stem cell transplantation. Br J Haematol. 2006;132:326–332. doi: 10.1111/j.1365-2141.2005.05875.x. [DOI] [PubMed] [Google Scholar]
- 158.Gentile DA, Doyle WJ, Zeevi A, Piltcher O, Skoner DP. Cytokine gene polymorphisms moderate responses to respiratory syncytial virus in adults. Hum Immunol. 2003;64:93–98. doi: 10.1016/s0198-8859(02)00705-x. [DOI] [PubMed] [Google Scholar]
- 159.Gentile DA, Doyle WJ, Zeevi A. Cytokine gene polymorphisms moderate illness severity in infants with respiratory syncytial virus infection. Hum Immunol. 2003;64:338–344. doi: 10.1016/s0198-8859(02)00827-3. [DOI] [PubMed] [Google Scholar]
- 160.Bravo MJ, de Dios Colmenero J, Alonso A, Caballero A. Polymorphisms of the interferon gamma and interleukin 10 genes in human brucellosis. Eur J Immunogenet. 2003;30:433–435. doi: 10.1111/j.1365-2370.2003.00419.x. [DOI] [PubMed] [Google Scholar]
- 161.Chevillard C, Moukoko CE, Elwali NE. IFN-gamma polymorphisms (IFN-gamma +2109 and IFN-gamma +3810) are associated with severe hepatic fibrosis in human hepatic schistosomiasis (Schistosoma mansoni) J Immunol. 2003;171:5596–5601. doi: 10.4049/jimmunol.171.10.5596. [DOI] [PubMed] [Google Scholar]
- 162.Fraser DA, Loos BG, Boman U. Polymorphisms in an interferon-gamma receptor-1 gene marker and susceptibility to periodontitis. Acta Odontol Scand. 2003;61:297–302. doi: 10.1080/00016350310006168. [DOI] [PubMed] [Google Scholar]
- 163.Thye T, Burchard GD, Nilius M, Muller-Myhsok B, Horstmann RD. Genomewide linkage analysis identifies polymorphism in the human interferon-gamma receptor affecting Helicobacter pylori infection. Am J Hum Genet. 2003;72:448–453. doi: 10.1086/367714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jouanguy E, Altare F, Lamhamedi S. Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N Engl J Med. 1996;335:1956–1961. doi: 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 165.Newport MJ, Huxley CM, Huston S. A mutation in the interferon-g-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;26:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 166.Trowsdale J. The gentle art of gene arrangement: the meaning of gene clusters. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-comment2002. comment2002.1–2002.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Trowsdale J, Parham P. Mini-review: defense strategies and immunity-related genes. Eur J Immunol. 2004;34:7–17. doi: 10.1002/eji.200324693. [DOI] [PubMed] [Google Scholar]
- 168.Raza MW, Blackwell CC, Molyneaux P. Association between secretor status and respiratory viral illness. BMJ. 1991;303:815–818. doi: 10.1136/bmj.303.6806.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hamajima N. Persistent Helicobacter pylori infection and genetic polymorphisms of the host. Nagoya J Med Sci. 2003;66:103–117. [PubMed] [Google Scholar]
- 170.Ali S, Niang MA, N'Doye I. Secretor polymorphism and human immunodeficiency virus infection in Senegalese women. J Infect Dis. 2000;181:737–739. doi: 10.1086/315234. [DOI] [PubMed] [Google Scholar]
- 171.Meyer zum Bueschenfelde CO, Unternaehrer J, Mellman I, Bottomly K. Regulated recruitment of MHC class II and costimulatory molecules to lipid rafts in dendritic cells. J Immunol. 2004;173:6119–6124. doi: 10.4049/jimmunol.173.10.6119. [DOI] [PubMed] [Google Scholar]
- 172.Cambi A, de Lange F, van Maarseveen NM. Microdomains of the C-type lectin DC-SIGN are portals for virus entry into dendritic cells. J Cell Biol. 2004;164:145–155. doi: 10.1083/jcb.200306112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schultz MJ, Wijnholds J, Peppelenbosch MP. Mice lacking the multidrug resistance protein 1 are resistant to Streptococcus pneumoniae-induced pneumonia. J Immunol. 2001;166:4059–4064. doi: 10.4049/jimmunol.166.6.4059. [DOI] [PubMed] [Google Scholar]
- 174.Fraser H. Genetics of type 1 diabetes. Cambridge University; 2005. PhD thesis. [Google Scholar]
- 175.Martin MP, Gao X, Lee JH. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 176.Romero R, Chaiworapongsa T, Kuivaniemi H, Tromp G. Bacterial vaginosis, the inflammatory response and the risk of preterm birth: a role for genetic epidemiology in the prevention of preterm birth. Am J Obstet Gynecol. 2004;190:1509–1519. doi: 10.1016/j.ajog.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 177.Garred P, Pressler T, Madsen HO. Association of mannose-binding lectin gene heterogeneity with severity of lung disease and survival in cystic fibrosis. J Clin Invest. 1999;104:431–437. doi: 10.1172/JCI6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Garred P, Pressler T, Lanng S. Mannose-binding lectin (MBL) therapy in an MBL-deficient patient with severe cystic fibrosis lung disease. Pediatr Pulmonol. 2002;33:201–207. doi: 10.1002/ppul.10064. [DOI] [PubMed] [Google Scholar]
- 179.Carlsson M, Sjoholm AG, Eriksson L. Deficiency of the mannan-binding lectin pathway of complement and poor outcome in cystic fibrosis: bacterial colonization may be decisive for a relationship. Clin Exp Immunol. 2005;139:306–313. doi: 10.1111/j.1365-2249.2004.02690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Dickson EJ, Stuart RC. Genetics of response to proton pump inhibitor therapy: clinical implications. Am J Pharmacogenomics. 2003;3:303–315. doi: 10.2165/00129785-200303050-00002. [DOI] [PubMed] [Google Scholar]
- 181.Mallal S, Nolan D, Witt C. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- 182.Martin AM, Nolan D, Gaudieri S. Predisposition to abacavir hypersensitivity conferred by HLA-B*5701 and a haplotypic Hsp70-Hom variant. Proc Natl Acad Sci USA. 2004;101:4180–4185. doi: 10.1073/pnas.0307067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hughes DA, Vilar FJ, Ward CC, Alfirevic A, Park BK, Pirmohamed M. Cost-effectiveness analysis of HLA B*5701 genotyping in preventing abacavir hypersensitivity. Pharmacogenetics. 2004;14:335–342. doi: 10.1097/00008571-200406000-00002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.