Abstract
Successful reproduction in vertebrates depends upon the actions of gonadotropin-releasing hormone (GnRH). Despite the wide presence of GnRH in Phylum Chordata, GnRH has not been isolated in protostomes other than the common octopus. To provide information on the evolution of this critical hormone, we isolated the full-length cDNA of a GnRH-like molecule from the central nervous system of a gastropod mollusk, the sea hare Aplysia californica. The open reading frame of this cDNA encodes a protein of 147 amino acids. The molecular architecture of the deduced protein is highly homologous to that reported for the prepro-octopus GnRH (oct-GnRH) and consists of a putative signal peptide, a GnRH dodecapeptide, a downstream processing site, and a GnRH-associated peptide (GAP). The deduced amino acid sequence of the Aplysia GnRH (ap-GnRH) is QNYHFSNGWYAG and differs from oct-GnRH by only two amino acids. The transcript for ap-GnRH is widely expressed in the central nervous system (CNS), the ovotestis, and the atrial gland, an exocrine gland. Immunocytochemistry (ICC) using an antiserum against oct-GnRH detected immunoreactive neurons in all CNS ganglia examined, and the staining was abolished by the preadsorption of the antiserum with synthetic ap-GnRH. In sum, ap-GnRH sequence is the first gastropod GnRH-like molecule to be elucidated. Further, it represents one of the only two GnRH-like molecules found outside Phylum Chordata. These data refute the possibility that oct-GnRH arose singly in cephalopods by convergent evolution and provide valuable support for an ancient origin of GnRH during metazoan evolution.
Keywords: Aplysia californica, sea hare, GnRH, cDNA cloning, mRNA expression, gastropod, nervous system
Introduction
In vertebrates, gonadotropin-releasing hormone (GnRH) is a decapeptide hormone critical for reproduction. Its presence was once thought to be restricted to members of Phylum Chordata, but increasing evidence suggests otherwise. A large number of studies reported the presence of GnRH-immunoreactivity (ir) and bioactivity in the gastropods (Goldberg et al., 1993; Young et al., 1999; Zhang et al., 2000; Tsai et al., 2003), bivalves (Pazos and Mathieu, 1999; Nakamura et al., 2007), polyplacophorans (Gorbman et al., 2003), cephalopods (Di Cosmo and Di Cristo, 1998; Di Cristo et al., 2002), cnidarians (Anctil, 2000; Twan et al., 2006), and platyhelminthes (Anctil and Tekaya., 2005). Importantly, a dodecapeptide containing the structural core of chordate GnRH was isolated from the central nervous system (CNS) of the common octopus (Iwokoshi et al., 2002). Although the octopus GnRH (oct-GnRH) deviates from the common chordate GnRH motif of having ten amino acids, it contains several features common to all forms of chordate GnRH. These include an N-terminal pyroglutamyl residue and a C-terminal amidated glycine residue, the general conservation of the N-terminal amino acids (when Asn2 and Tyr3 are removed), and the conservation of the C-terminal Pro11 and Gly12 residues (see Fig. 1). Further, the oct-GnRH prohormone contains the highly conserved dibasic cleavage site (Lys14-Arg15) downstream of Gly12 (Iwakoshi et al., 2002), another universal feature of chordate GnRH prohormones.
Figure 1.

Amino acid sequences of oct-GnRH aligned with mammalian GnRH (mGnRH) for comparison. Amino acids highly conserved within Phylum Chordata are shaded.
Until now, oct-GnRH was the only non-chordate GnRH sequence elucidated. To ensure oct-GnRH did not arise in a single group as the result of convergent evolution, GnRH sequences in additional groups of protostomes must be examined. Characterization of GnRH in other protostomes will also allow us to decipher functional and structural features that are critical enough to warrant strong conservation. As a first step towards these goals, we elucidated the sequence of a GnRH-like molecule in a marine gastropod mollusk, Aplysia californica. Several features make A. californica an ideal model for this study. First, GnRH-ir is present in the CNS and hemolymph of A. californica (Zhang et al., 2000; Tsai et al., 2003). Second, A. californica possess a well-identified CNS and reproductive axis (Kandel, 1979) that should greatly facilitate our understanding of neural and reproductive roles of GnRH. Lastly, the recent publication of A. californica CNS transcriptome (Moroz et al., 2006) is a tremendous resource that aids in the search for vertebrate neuropeptide homologues in mollusks. With the help of the CNS transcriptome database, we cloned the first gastropod GnRH-like molecule (ap-GnRH) in A. californica. Further, we examined the pattern of ap-GnRH transcript expression and peptide localization to provide an initial characterization of the distribution of this neuropeptide.
Materials and Methods
Animals and tissue collection
Sexually mature wild-caught A. californica (100–200 g in mass) were purchased from Alacrity Marine Biological Services (Redondo Beach, CA). Animals were kept in a 400-gal tank with circulating artificial seawater (Instant Ocean) that was continuously filtered through biological and chemical filters. Animals were maintained between 15–18°C and fed Romaine lettuce daily. All animals were anesthetized by an injection of 1/3 body volume of isotonic magnesium chloride before sacrifice. For RNA preparation, CNS ganglia and peripheral tissues were collected, snap-frozen on dry ice, and stored at −70°C until RNA isolation. For immunocytochemistry (ICC), the CNS ganglia were dissected, pinned out on a Sylgard-lined dish, and immersion fixed overnight in Bouin's fixative. Ganglia were stored in 70% ethanol until ICC.
RNA isolation
RNA from pooled CNS, individual ganglia, and peripheral tissues were isolated using the lithium chloride method previously described (Querat et al., 1991). RNA samples were quantified based on absorbance at 260 nm. All samples were pretreated with RNase-free DNase (Promega, Madison, WI) to eliminate genomic DNA contamination prior to reverse transcription (RT).
Oligonucleotide primers
In silico searches of Aplysia CNS transcriptome was conducted with TBLASTN program using the PAM30 matrix. Searches using oct-GnRH dodecapeptide as the query sequence revealed two A. californica cDNA fragments (Accession# EB190103 and EB187791) containing deduced amino acid sequences highly similar to oct-GnRH. We denoted these sequences as ap-GnRH1 (562 bp) and ap-GnRH2 (653 bp). Both sequences contained an open reading frame but lacked a portion of the 5'-untranlated region (UTR) and the majority of 3'-UTR. The two sequences differ from one another by only 5 nucleotides. The gene-specific oligonucleotide primers used for 5' and 3' rapid amplification of cDNA ends (RACE) and RT-polymerase chain reaction (PCR) were designed based on the common regions of these two sequences and listed in Table 1. Searches using vertebrate GnRH decapeptide motif failed to produce any positive hits. Primers for A. californica actin (ap-actin) were designed based on the corresponding cDNA sequence (Accession# U01352).
Table 1.
Primers used to amplify ap-GnRH cDNA
| Primer | Sequence |
|---|---|
| BRL-A1 | 5’- GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTTTTT-3’ |
| Smart III | 5’-AAGCAGTGGTATCAACGCAGAGTGGCCATTATGGCCGGG-3’ |
| BRL-A2 | 5’-GGCCACGCGTCGACTAGTAC-3’ |
| Smart P1 | 5’-AAGCAGTGGTATCAACGCAGAGT-3 |
| Smart P2 | 5’- ATCAACGCAGAGTGGCCATTATG-3’ |
| ApGS1 | 5’-GTAGTAGTTAGACGCCAGGAA-3’ |
| ApGS2 | 5’-AGCAGTAACAGTGGCTTGGACG-3’ |
| ApGS3 | 5’-GAGGCAGCGAGAATACAGAGG-3’ |
| ApGS4 | 5’-CCGCCACCACCACTCTTTTC-3’ |
| ApGA1 | 5’-CGAAACCACGCCCACTCAAGC-3’ |
| ApGA2 | 5’-GCTGCTGTCGGCGTCTGTGA-3’ |
| ApGA3 | 5’-CCAAGTTGTCTGCGAGGCTGT-3’ |
| ApGA4 | 5’-CTCACCAACGCCGAAACCAC-3’ |
| ActinS1 | 5’-GGTATTGTGTTGGACTCTGG-3’ |
| ActinA1 | 5’-TGATGGAGTTGAAGGTGGTC-3’ |
Cloning of full-length cDNA for prepro-ap-GnRH
The full-length cDNA sequence for the prepro-ap-GnRH was obtained by 5’ and 3’ RACE. Total RNA isolated from the CNS (5 µg) was reverse-transcribed using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) in a 20 µl reaction that contained 0.5 µM BRL-A1, 0.5 mM Smart III adapter primer, 0.5 mM dNTP, 1X first-strand reaction buffer, 10 mM dithiothreitol, and 200 U reverse transcriptase. The reaction was incubated at 42°C for 60 min, and the reverse transcriptase was heat-inactivated at 70°C for 15 min.
An initial PCR was performed to amplify the open reading frame of EB190103 and EB187791 using primers ApGS1 and ApGA3. PCR was performed in a 25-µl reaction mixture containing 1 µl of the first-strand cDNA, 1X Ex Taq Buffer, 2.0 mM MgCl2, 0.2 mM dNTP, 0.4 µM of each primer, and 0.625 U Ex Taq DNA polymerase (TaKaRa, DaLian, China). The conditions for PCR were 0.5 min at 94°C, 0.5 min at 55°C, and 1.0 min at 72°C for 35 cycles, followed by a final extension for 7 min at 72°C. The resulting PCR product (~600 bp) was subcloned into pGEM-T Easy Vector (Promega) and sequenced.
For 3’ RACE, two rounds of PCR were performed. For the first round, 1 µl of the first-strand cDNA was amplified with ApGS2 and BRL-A2 primers using PCR conditions identical to those described above. For the nested PCR, 1 µl of the first PCR product (diluted 1:30) was amplified with ApGS3 and BRL-A2 primers. The PCR conditions were identical to the first-round PCR except the annealing temperature was elevated to 56°C. The major amplicons (250bp, 1000bp and 1200bp) were subcloned and sequenced.
For 5' RACE, two rounds of PCR were also performed. For the first round, 1 µl of the firststrand cDNA were amplified with ApGA1 and Smart P1 primers. For the second-round PCR, 1 µl of the first PCR product (diluted 1:30) was amplified with ApGA2 and Smart P2 primers. The PCR conditions were identical to those in 3’ RACE. The resulting PCR product (~400 bp) was subcloned and sequenced.
Sequence analysis
The deduced amino acid sequence of the prepro-ap-GnRH was compared with those of other species using the MegAlign feature of the DNAstar software package. Alignment of deduced amino acid sequences of prepro-ap-GnRH and prepro-oct-GnRH was performed with ClustalX1.8.
Phylogenetic analysis
The phylogenetic analysis was performed using the UPGMA method (Sneath and Sokal, 1973) of MEGA4 (Tamura et al., 2007). The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). The protein sequences of other species were downloaded from Entrez (NCBI).
RT-PCR analysis of ap-GnRH expression in different tissues
Total RNA (1 µg) isolated from the whole CNS, ovotestis, cerebral ganglia, pleural/pedal ganglia, atrial gland, and abdominal ganglia was reverse-transcribed with Superscript III reverse transcriptase (Invitrogen) using the oligo(dT) primer. Amplification of ap-GnRH and ap-actin was carried out using 2 µl of the first-strand cDNA and the following primer sets: ApGS4 and ApGA4 for ap-GnRH, and ActinS1 and ActinA1 for ap-actin (see Table 1). Using these primer sets, fragments of 248 bp and 400 bp were amplified for ap-GnRH and ap-actin, respectively. All reaction mixtures consisted of 1.5 mM MgCl2, 0.2 mM dNTP, 0.2 µM of each primer, and 2.5 U Taq DNA Polymerase (New England Biolabs, Ipswich, MA). Cycling conditions were 0.5 min at 94°C, 0.5 min at 58°C, and 0.5 min at 72°C, followed by a final extension for 7 min at 72°C. Cycle numbers were 35 and 30 for ap-GnRH and ap-actin, respectively. PCR products were resolved on a 1% agarose gel and visualized by ethidium bromide staining.
Immunocytochemistry (ICC)
ICC was performed on sections of paraffin-embedded CNS ganglia using a rabbit anti-oct-GnRH antiserum (AS9779). AS9779 was generated against a synthetic dodecapeptide CNYHFSNGWHPGamide (BioSource, Hopkinton, MA) in which the N-terminal pyroglutamyl residue of oct-GnRH has been replaced with a cysteine to facilitate its conjugation to ovalbumin. This antiserum was used at 1:500 in an ICC protocol described previously (Tsai et al., 2003). For preadsorption studies, AS9779 was pre-incubated with 10 µM of synthetic ap-GnRH (pQNYHFSNGWYAGamide; Genscript, Piscataway, NJ) overnight at 4°C and applied to the sections. All reactions were visualized with diaminobenzidine as the chromagen. Some sections were counterstained with hematoxylin to facilitate visualization of nuclei. ICC was conducted on four animals, and the results were consistent in all cases.
Results
Cloning and sequence analysis of prepro-ap-GnRH
Although TBLASTN searches of A. californica CNS transcriptome revealed two potential candidates for ap-GnRH, we detected only one ap-GnRH cDNA species (corresponding to ap-GnRH1) out of 50 cDNA fragments we have cloned and sequenced. This suggests ap-GnRH2 (EB187791) in the published transcriptome may have arisen due to a sequencing error. The cloned prepro-ap-GnRH cDNA spans 1712 bp and includes a 171-bp 5’-untranslated region (UTR), a 441-bp open reading frame that encodes a putative preprohormone of 147 amino acids, and a 1100-bp 3’-UTR (Fig. 2). The consensus ATTAAA polyadenylation signal is located 15 bases upstream of the poly (A) tail. The sequence of prepro-ap-GnRH has been deposited into Genbank (Accession# EU204144). Similar to other GnRH precursors, the molecular architecture of prepro-ap-GnRH includes a signal peptide, the GnRH peptide, a putative processing site, and a GnRH-associated peptide (GAP). The deduced ap-GnRH peptide, similar to oct-GnRH, also consists of twelve amino acids. Of note, the tribasic cleavage (KKR) site in A. californica replaces the dibasic motif in vertebrates and octopus (Fig. 3). A high level of sequence identity exists between oct-GnRH and ap-GnRH in the dodecapeptide region (83.3%). Outside this region, identity is reduced to 11.1% in the signal peptide region and 37.2% in the GAP region, but considerable homology still exists between prepro-oct-GnRH and prepro-ap-GnRH (Fig. 3). Within the dodecapeptide region, sequence variation exists between oct-GnRH and ap-GnRH in amino acids 10 and 11, but Gly12 remains invariant (Fig. 3). A comparative sequence analysis reveals the ap-GnRH dodecapeptide shares 60%, 50%, and 40% homologies with the decapeptide region of chicken GnRH II, mammalian GnRH, and salmon GnRH, respectively.
Figure 2.
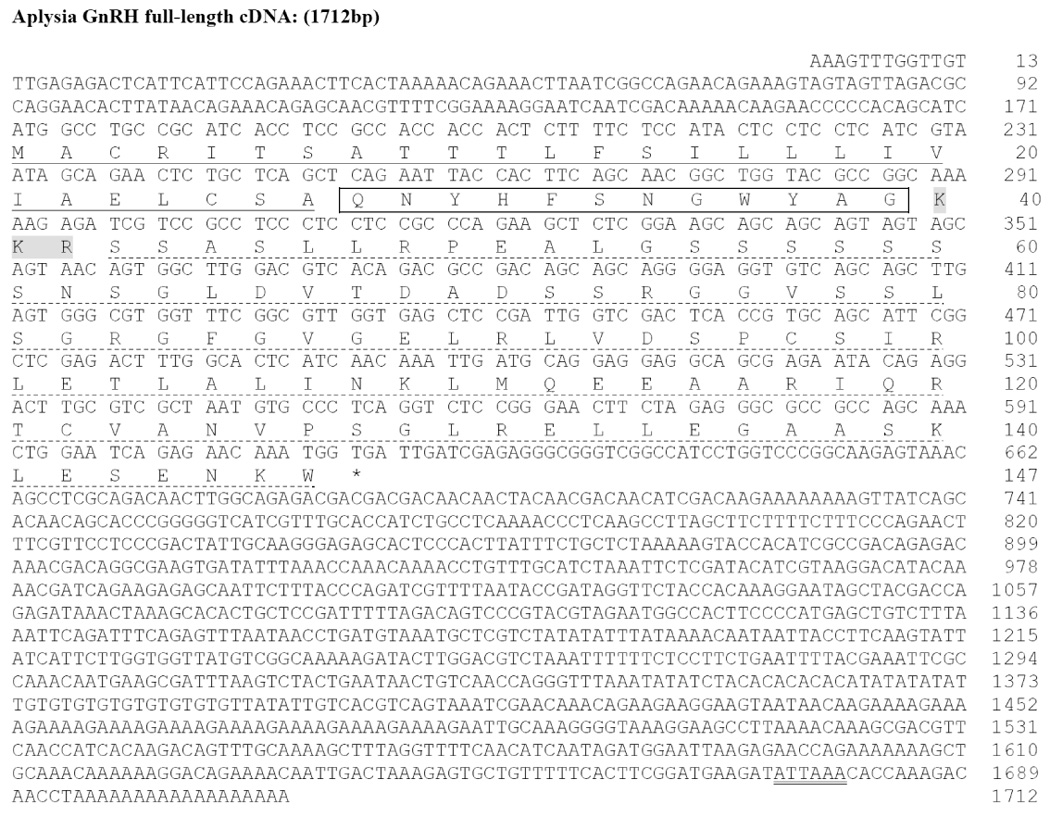
Nucleotide and deduced amino acid sequences of prepro-ap-GnRH (Genbank Accession# EU204144). The nucleotides (upper row) and amino acids (lower row) are numbered accordingly. The putative signal peptide region is underlined (solid). The putative ap-GnRH dodecapeptide is boxed, and the KKR tribasic cleavage site is shaded. The putative GAP is underlined (dashed). The asterisk (*) denotes the stop codon. The nucleotides corresponding to the polyadenylation signal (ATTAAA) are double-underlined.
Figure 3.

Sequence alignment of deduced amino acids of prepro-ap-GnRH and prepro-oct-GnRH. Both prepro-GnRH sequences are numbered in accordance with the prepro-ap-GnRH from the putative N-terminal. The signal peptide, GnRH peptide, and GAP are indicated. The GnRH dodecapeptide is shaded. The cleavage site is in bold. The identical, highly conserved, and less conserved amino acid residues are indicated by *, :, and ., respectively.
Phylogenetic analysis of ap-GnRH
Phylogenetic analysis segregates prepro-GnRHs of vertebrates into four groups: GnRH-I (consists of several hypothalamic/preoptic forms of GnRH), GnRH-II (consists of the mesencephalic form of GnRH), a fish-specific GnRH-III (consists of the terminal nerve form of GnRH), and a lamprey-specific GnRH-IV (consists of two lamprey forms of GnRH; Fig. 4). The two tunicate sequences were clustered into a separate group that is not part of this nomenclature. The analysis clusters prepro-ap-GnRH and prepro-oct-GnRH into a monophyletic group that is segregated from all chordate prepro-GnRHs (Fig. 4). We propose the term "GnRH-V" for this lineage (Fig. 4).
Figure 4.
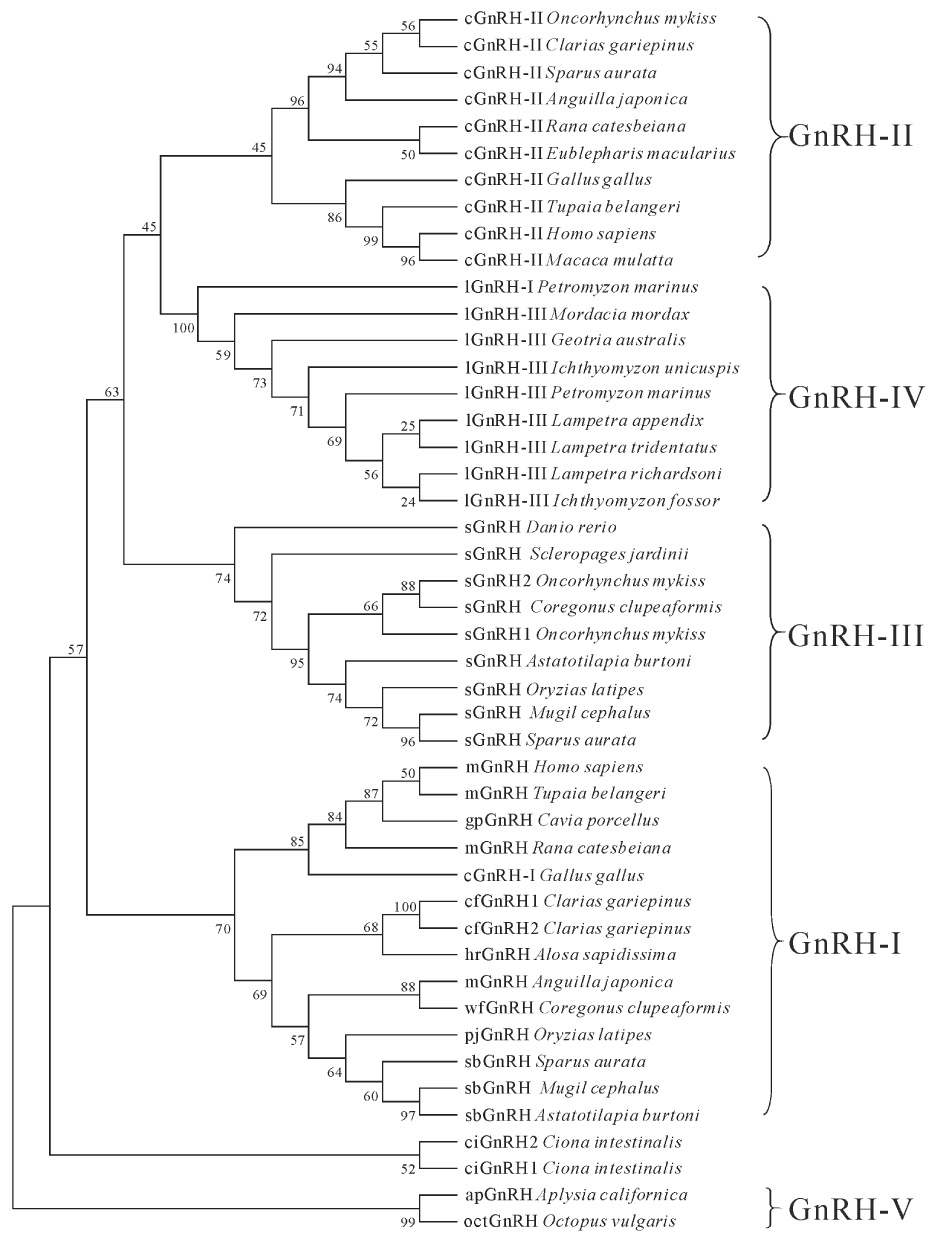
Phylogenetic analysis of prepro-GnRHs in Phylum Chordata and Phylum Mollusca. The bootstrap values (in %) from 1000 replicas are given at each branch point. Prepro-GnRHs are clustered into five groups (GnRH-I to GnRH V). Each sequence is denoted by the name of its GnRH peptide form followed by the species from where it was isolated (in italics). Arabic numbers at the end of vertebrate sequences are used to indicate different precursors that encode the same GnRH peptide. Arabic numbers at the end of ciGnRH (tunicate) indicate different molecular forms of GnRH. The following abbreviations are used to denote the GnRH peptide forms: cGnRH-I (chicken GnRH-I), cGnRH-II (chicken GnRH-II), cfGnRH (catfish GnRH), ciGnRH (Ciona GnRH), gpGnRH (guinea pig GnRH), hrGnRH (herring GnRH), lGnRH-I (lamprey GnRH-I), lGnRH-III (lamprey GnRH-III), mGnRH (mammalian GnRH), pjGnRH (pejerry GnRH), sGnRH (salmon GnRH), sbGnRH (sea bream GnRH), and wfGnRH (whitefish GnRH).
RT-PCR analysis of ap-GnRH expression in several tissues
The ap-GnRH transcript is wide spread in the central and peripheral tissues (Fig. 5). In addition to the CNS ganglia examined (pleural/pedal, cerebral, and abdominal), ap-GnRH is also expressed in the ovotestis and an exocrine gland (atrial gland; Fig. 5). Negative controls in which RNA samples had not been reverse transcribed (No RT controls) did not yield positive signals.
Figure 5.
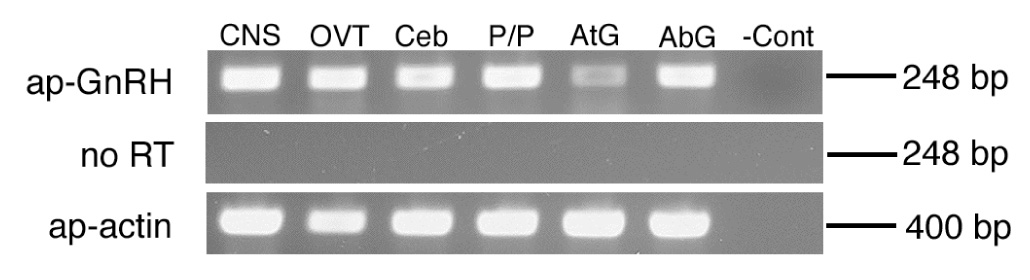
Expression of ap-GnRH in different A. californica tissues examined by RT-PCR (top panel). ap-actin was used as a control to ensure the quality of RNA samples (bottom panel). No RT negative controls used RNA samples that have not been reverse transcribed (middle panel). PCR products were stained with ethidium bromide. The sizes of the PCR products are shown on the right. CNS, central nervous system; OVT, ovotestis; Ceb, cerebral ganglia; P/P, pedal/pleural ganglia; AtG, atrial gland; AbG, abdominal ganglia; -Cont, negative water control.
Distribution of ap-GnRH-ir in A. californica CNS
ICC reveals the presence of ap-GnRH-ir in all CNS ganglia. The pedal ganglia contain the largest number of ap-GnRH immunopositive neurons (Fig. 6A), and this ir was completely abolished by the preadsorption with synthetic ap-GnRH (Fig. 6B). The pedal ganglia also contain abundant ap-GnRH positive fibers, and the majority of fiber staining was abolished by preadsorption with ap-GnRH (Figs. 6A, B). Neurons positive for ap-GnRH are also observed in the cerebral ganglia (Figs. 6C, D), abdominal ganglia (Figs. 6E, F), buccal ganglia, and pleural ganglia (data not shown). The presence of ap-GnRH immunopositive neurons in the latter two ganglia is relatively sparse; only 1–3 positive neurons could be found within a given ganglion. Lastly, thick bundles of ap-GnRH immunoreactive fibers are found in the pleurovisceral connective nerves (Fig. 6G), a pair of nerves responsible for transmitting signals (including those for egg-laying) from the head ganglia to the neuroendocrine bag cell neurons. However, bag cell neurons are devoid of ap-GnRH-ir (Fig. 6H).
Figure 6.
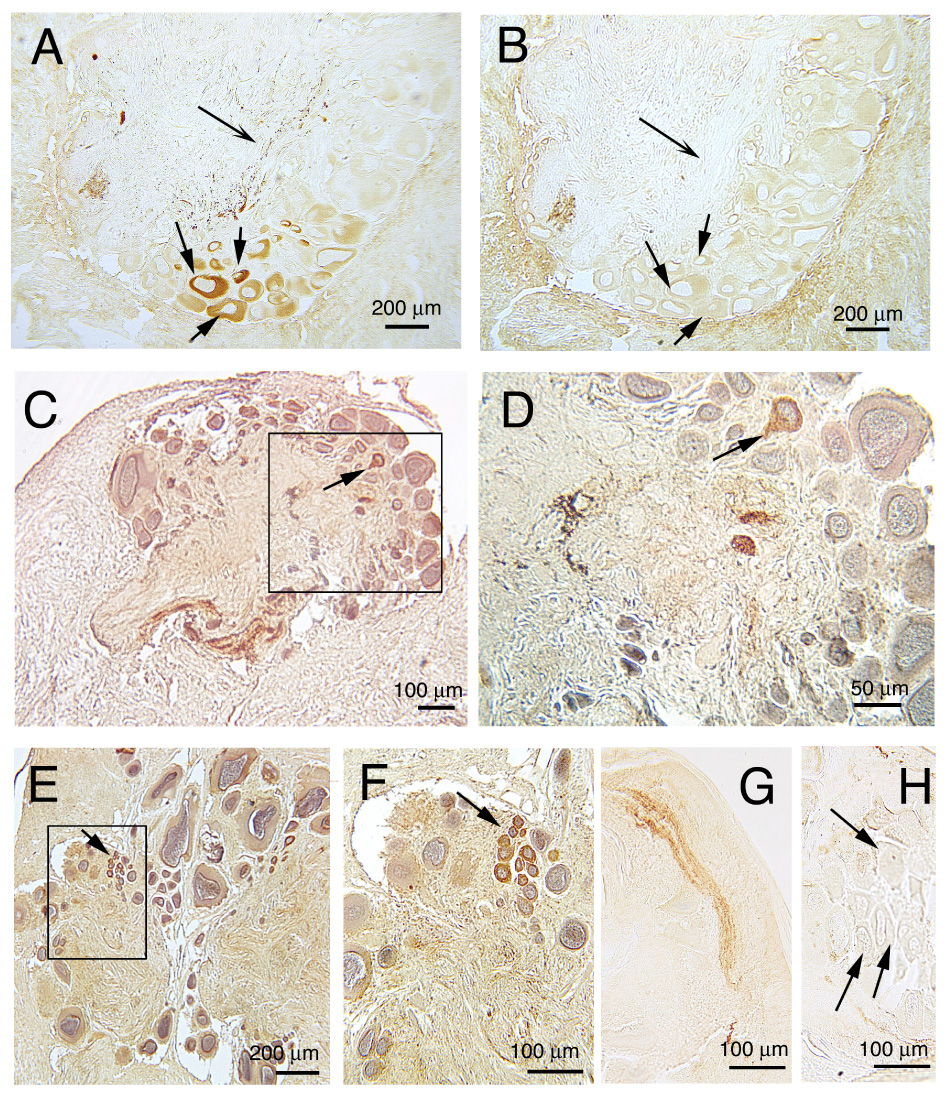
Immunocytochemical localization of ap-GnRH-ir in the central ganglia of A. californica using AS9779, an anti-oct-GnRH antiserum. A, B) Numerous immunoreactive neurons were observed in the medial (bottom) and superior (right) quadrant of the pedal ganglia (A), and this staining was abolished by the preadsorption of the antiserum with synthetic ap-GnRH in an adjacent section (B). Solid arrows in A and B point to the same neurons with preadsorbed staining. Concave arrow in A and B points to immunoreactive fibers in the neuropil region that have been preadsorbed. C, D) Low (C) and high (D) magnification photomicrographs of the cerbral ganglia. Box in C is magnified in D to demonstrate the presence of an immunoreactive neuron (arrow). E, F) Low (E) and high (F) magnification photomicrographs of the abdominal ganglia. Box in E is magnified in F to demonstrate a cluster of immunoreactive neurons (arrow). G) Immunoreactive fibers within the pleurovisceral connective nerve. H) Bag cell neurons are negative for ap-GnRH-ir. Arrows point to representative bag cell neurons with no signal. Brown staining indicates ap-GnRH-ir. Blue staining indicates hematoxylin-counterstained nuclei.
Discussion
This study reports the first full-length cDNA sequence of a gastropod prepro-GnRH. The wide spread presence of ap-GnRH-ir and transcript also suggests this neuropeptide may assume a wide range of central and peripheral functions. Importantly, these data demonstrate the existence of a gastropod peptide highly homologous to oct-GnRH. As cephalopods and gastropods diverged ca. 520 million years ago (Lieb et al., 2000), the extraordinary conservation of ap-GnRH dodecapeptide suggests a role so important that little structural change could occur to it during gastropod evolution. This scenario parallels the structural conservation of GnRH in Phylum Chordata.
Comparison between the deduced amino acid sequences of prepro-ap-GnRH and prepro-oct-GnRH reveals several shared and divergent features within the translated region. First, they both share the common molecular architecture of a signal peptide, a GnRH dodecapeptide, followed by a GAP. This general architecture is also observed in chordate prepro-GnRHs (Seeburg and Adelman, 1984; Suzuki et al., 1992; Dunn et al., 1993; Hayes et al., 1994; Adam et al., 2003). Second, a moderate level of similarity (37.2%) exists in the GAP regions, but the greatest sequence similarity exists in the GnRH dodecapeptide region (83.3%; Fig. 3). The selective conservation of the dodecapeptide region suggests this region is evolutionarily constrained by its function, which is likely tied to its ability to bind to the cognate receptor. Third, the highly conserved Pro11 residue immediately upstream of the C-terminal Gly12, present universally in all chordate and oct-GnRH forms (see Fig. 1), does not exist in ap-GnRH. Instead, it is replaced by an Ala11 residue. This is the first documented case of a GnRH-like molecule with a substitution in one of the two invariant C-terminal residues. As the conserved C-terminal of vertebrate GnRH is critical for its gonadotropin-releasing activity (Millar et al., 1989), the Pro11 to Ala11 substitution could explain, in part, why partially purified A. californica CNS extracts failed to stimulate gonadotropin release in a mouse pituitary bioassay (Tsai et al., 2003).
Another notable difference between ap-GnRH and oct-GnRH preprohormones is that the familiar dibasic GKR cleavage motif downstream of the dodecapeptide is replaced by the tribasic KKR motif. A number of prohormone convertases target regions associated with the KR site (Rockwell et al., 2002, von Eggelkraut-Gottanka and Beck-Sickinger, 2004), and KKR is a common processing site for many neuropeptides (see Mori and Fujino, 2004). At present, the functional implication of this substitution is unclear. Lastly, ap-GnRH cDNA possesses a 3'-UTR that spans 1100 bp, which is approximately three times longer than that of oct-GnRH. The significance of this feature in relation to mRNA stability and translational efficiency remains to be investigated.
The results of our phylogenetic analysis agree well with many other studies (see Guilgur et al., 2006). We used an extension of the GnRH nomenclature proposed by Fernald and White (1999) to delineate the four prepro-GnRH lineages in vertebrates (Fig. 4). That GnRH-II variants failed to form a monophyletic group is likely due to the great divergence of mammalian and avian sequences from the relatively conserved fish and basal tetrapod sequences (Silver et al, 2004; Vickers et al, 2004; Guilgur et al, 2007). Although the definition of several vertebrate GnRH clades is still under debate (Guilgur et al., 2006), the two non-chordate prepro-GnRHs form a tightly clustered monophyletic group. The evolutionary history of this molluscan lineage, termed "GnRH-V", will become clearer with the elucidation of additional non-chordate GnRH sequences.
RT-PCR and ICC analyses reveal that ap-GnRH is widely distributed in the CNS and peripheral tissues. This type of wide spread distribution has been reported for oct-GnRH (Iwakoshi-Ukena et al., 2004). Of interest is the absence of GnRH-ir in bag cell neurons and the presence of prominent GnRH fibers within the pleurovisceral connective nerves. When A. californica are ready to ovulate, stimulatory signals originating in the head ganglia are relayed to the bag cell neurons via the pleurovisceral connective nerves (Brown et al., 1989; Ferguson et al., 1989). Upon stimulation, bag cell neurons undergo a prolonged and highly characteristic pattern of electrical discharge called afterdischarge (AD; Kupfermann and Kandel, 1970). AD triggers the bag cell neurons to secrete a neurohormone, egg-laying hormone (ELH), that is required for the release of eggs from the ovotestis and the display of stereotypical egg laying behaviors (Kupfermann and Kandel, 1970). Our current ICC data are consistent with our previous observations that bag cell neurons respond to vertebrate GnRH (Zhang et al., 2000) but do not produce detectable levels of immunoreactive GnRH (Tsai et al., 2003). Overall, these observations indicate that bag cell neurons are a target, not a source, of ap-GnRH and suggest a functional link between ap-GnRH and A. californica reproduction.
Although we acknowledge a possible reproductive role of ap-GnRH, the function of ap-GnRH is unlikely limited to reproduction based on its wide distribution in the CNS and peripheral tissues. Deuterostomes and protostomes diverged 630 million years ago. Thus, under different selection pressures, ap-GnRH may have been recruited as a general neuropeptide to regulate diverse functions. In other words, the considerable specialization of GnRH as a reproductive activator may be a phenomenon specific to chordates or deuterostomes. This notion is supported by the powerful cardioexcitatory effect of oct-GnRH in the octopus (Iwakoshi-Ukena et al., 2004), an effect originally used in a bioassay to isolate oct-GnRH (Iwakoshi et al., 2002). Although it is important to investigate the reproductive role of ap-GnRH, one must exercise caution when inferring the function of a protostome GnRH based on the function of chordate GnRHs. Elucidating the non-reproductive functions of protostome GnRH is an important next step towards understanding the functional evolution of this hormone and represents a conceptual shift that could open the door to many avenues of investigation.
In this study, we report the full-length cDNA sequence of ap-GnRH as well as its mRNA and peptide distribution. The ap-GnRH sequence represents one of the only two non-chordate GnRH sequences known to date. For an in-depth understanding of GnRH evolution, additional studies must be conducted to 1) establish that non-chordate and chordate GnRHs share a common ancestor, 2) examine GnRH function in protostomes to understand functional conservation and diversification, and 3) elucidate the structure of protostome GnRH receptor to understand how GnRH receptor co-evolved with its ligands. Based on the observation that ap-GnRH immunoreactive fibers innervate bag cell neurons (present study) and bag cell neurons respond to vertebrate GnRH (Zhang et al., 2000), it would be tantalizing to speculate that ap-GnRH has been recruited to provide a neural link to the reproductive system in a fashion analogous to its regulation of the vertebrate pituitary (Gorbman and Sower, 2003; Kah et al., 2007). However, the wide distribution of ap-GnRH in both central and peripheral tissues also suggests it may assume a wide range of roles far beyond just reproductive regulation.
Acknowledgements
We thank Yumei Yang, Katie Wahr, and Sarah Moyle for technical assistance. We also thank Dr. Sue Moenter for the helpful discussions and editorial comments. This work was supported by NIH HD042634 and NSF IBN-9996398 to P.-S.T., and by the Natural Science Foundation of China (30570228) to L.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams BA, Tello JA, Erchegyi J, Warby C, Hong DJ, Akinsanya KO, Mackie GO, Vale W, Rivier JE, Sherwood NM. Six novel gonadotropin-releasing hormones are encoded as triplets on each of two genes in the protochordate, Ciona intestinalis. Endocrinology. 2003;144:1907–1919. doi: 10.1210/en.2002-0216. [DOI] [PubMed] [Google Scholar]
- Anctil M. Evidence for gonadotropin-releasing hormone-like peptides in a cnidarian nervous system. Gen Comp Endocrinol. 2000;119:317–328. doi: 10.1006/gcen.2000.7524. [DOI] [PubMed] [Google Scholar]
- Anctil M, Tekaya S. Gonadotropin-releasing hormone-like immunoreactivity in the planarian Bdelloura candida (Platyhelminthes, Tricladida) Invert Biol. 2005;124:11–17. [Google Scholar]
- Brown RO, Pulst SM, Mayeri E. Neuroendocrine bag cells of Aplysia are activated by bag cell peptide-containing neurons in the pleural ganglion. J Neurophysiol. 1989;61:1142–1152. doi: 10.1152/jn.1989.61.6.1142. [DOI] [PubMed] [Google Scholar]
- Di Cosmo A, Di Cristo C. Neuropeptidergic control of the optic gland of Octopus vulgaris: FMRF-amide and GnRH immunoreactivity. J Comp Neurol. 1998;398:1–12. doi: 10.1002/(sici)1096-9861(19980817)398:1<1::aid-cne1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Di Cristo C, Paolucci M, Iglesias J, Sanchez J, Di Cosmo A. Presence of two neuropeptides in the fusiform ganglion and reproductive ducts of Octopus vulgaris FMRFamide and gonadotropin-releasing hormone (GnRH) J Exp Zool. 2002;292:267–276. doi: 10.1002/jez.90000. [DOI] [PubMed] [Google Scholar]
- Dunn IC, Chen Y, Hook C, Sharp PJ, Sang HM. Characterization of the chicken preprogonadotrophin-releasing hormone-I gene. J Mol Endocrinol. 1993;11:19–29. doi: 10.1677/jme.0.0110019. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Ferguson GP, Ter Maat A, Pinsker HM. Egg Laying in Aplysia. II. Organization of central and peripheral pathways for initiating neurosecretory activity and behavioral patterns. J Comp Physiol. 1989;164A:849–857. doi: 10.1007/BF00616756. [DOI] [PubMed] [Google Scholar]
- Fernald RD, White RB. Gonadotropin-releasing hormone genes: phylogeny, structure and function. Front Neuroendocrinol. 1999;20:224–240. doi: 10.1006/frne.1999.0181. [DOI] [PubMed] [Google Scholar]
- Goldberg JI, Garofalo R, Price CJ, Chang JP. Presence and biological activity of a GnRH-like factor in the nervous system of Helisoma trivolvis. J Comp Neurol. 1993;336:571–582. doi: 10.1002/cne.903360409. [DOI] [PubMed] [Google Scholar]
- Gorbman A, Sower SA. Evolution of the role of GnRH in animal (Metazoan) biology. Gen Comp Endocrinol. 2003;134:207–213. doi: 10.1016/j.ygcen.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Gorbman A, Whiteley A, Kavanaugh S. Pheromonal stimulation of spawning release of gametes by gonadotropin releasing hormone in the chiton. Mopalia sp. Gen Comp Endocrinol. 2003;131:62–65. doi: 10.1016/s0016-6480(02)00647-0. [DOI] [PubMed] [Google Scholar]
- Guilgur LG, Moncaut NP, Canario AVM, Somoza GM. Evolution of GnRH ligands and receptors in gnathostomata. Comp Biochem Physiol A. 2006;144:272–283. doi: 10.1016/j.cbpa.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Guilgur LG, Orti G, Strobl-Mazzulla PH, Fernandino JI, Miranda LA, Somoza GM. Characterization of the cDNAs encoding three GnRH forms in the pejerrey fish Odontesthes bonariensis (Atheriniformes) and the evolution of GnRH precursors. J Mol Evol. 2007;64:614–627. doi: 10.1007/s00239-006-0125-8. [DOI] [PubMed] [Google Scholar]
- Hayes WP, Wray S, Battey JF. The frog gonadotropin-releasing hormone-I (GnRH-I) gene has a mammalian-like expression pattern and conserved domains in GnRH-associated peptide, but brain onset is delayed until metamorphosis. Endocrinology. 1994;134:1835–1845. doi: 10.1210/endo.134.4.8137750. [DOI] [PubMed] [Google Scholar]
- Iwakoshi E, Takuwa-Kuroda K, Fujisawa Y, Hisada M, Ukena K, Tsutsui K, Minakata H. Isolation and characterization of a GnRH-like peptide from Octopus vulgaris. Biochem Biophys Res Commun. 2002;291:1187–1193. doi: 10.1006/bbrc.2002.6594. [DOI] [PubMed] [Google Scholar]
- Iwakoshi-Ukena E, Ukena K, Takuwa-Kuroda K, Kanda A, Tsutsui K, Minakata H. Expression and distribution of octopus gonadotropin-releasing hormone in the central nervous system and peripheral organs of the octopus (Octopus vulgaris) by in situ hybridization and immunohistochemistry. J Comp Neurol. 2004;477:310–323. doi: 10.1002/cne.20260. [DOI] [PubMed] [Google Scholar]
- Kah O, Lethimonier C, Somoza G, Guilgur LG, Vaillant C, Lareyre JJ. GnRH and GnRH receptors in metazoa: a historical, comparative, and evolutive perspective. Gen Comp Endocrinol. 2007;153:346–364. doi: 10.1016/j.ygcen.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Kandel ER. Behavioral Biology of Aplysia. San Francisco: W.H. Freeman and Company; 1979. [Google Scholar]
- Kupfermann I, Kandel ER. Electrophysiological properties and functional interconnections of two symmetrical neurosecretory clusters (bag cells) in abdominal ganglion of Aplysia. J Neurophysiol. 1970;33:865–876. doi: 10.1152/jn.1970.33.6.865. [DOI] [PubMed] [Google Scholar]
- Lieb B, Altenhein B, Markl J. The sequence of a gastropod hemocyanin (HtH1 from Haliotis tuberculata) J Biol Chem. 2000;275:5675–5681. doi: 10.1074/jbc.275.8.5675. [DOI] [PubMed] [Google Scholar]
- Millar RP, Flanagan CA, Milton RC, King JA. Chimeric analogues of vertebrate gonadotropin-releasing hormones comprising substitutions of the variant amino acids in positions 5, 7, and 8. Characterization of requirements for receptor binding and gonadotropin release in mammalian and avian pituitary gonadotropes. J Biol Chem. 1989;264:21007–21013. [PubMed] [Google Scholar]
- Mori M, Fujino M. Urotensin II-related peptide, the endogenous ligand for the urotensin II receptor in the rat brain. Peptides. 2004;25:1815–1818. doi: 10.1016/j.peptides.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Ha T, Heyland A, Knudsen B, Sahni A, Yu F, Liu L, Jezzini S, Lovell P, Iannucculli W, Chen M, Nguyen T, Sheng H, Shaw R, Kalachikov S, Panchin YV, Farmerie W, Russo JJ, Ju J, Kandel ER. Neuronal transcriptome of aplysia: neuronal compartments and circuitry. Cell. 2006;127:1453–1467. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Osada M, Kijima A. Involvement of GnRH neuron in the spermatogonial proliferation of the scallop, Patinopecten yessoensiss. Mol Reprod Dev. 2007;74:108–115. doi: 10.1002/mrd.20544. [DOI] [PubMed] [Google Scholar]
- Pazos AJ, Mathieu M. Effects of five natural gonadotropin-releasing hormones on cell suspensions of marine bivalve gonad: stimulation of gonial DNA synthesis. Gen Comp Endocrinol. 1999;113:112–120. doi: 10.1006/gcen.1998.7186. [DOI] [PubMed] [Google Scholar]
- Querat B, Hardy A, Fontaine YA. Regulation of the type-II gonadotrophin alpha and beta subunit mRNAs by oestradiol and testosterone in the European eel. J Mol Endocrinol. 1991;7:81–86. doi: 10.1677/jme.0.0070081. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Krysan DJ, Komiyama T, Fuller RS. Precursor processing by kex2/furin proteases. Chem Rev. 2002;102:4525–4548. doi: 10.1021/cr010168i. [DOI] [PubMed] [Google Scholar]
- Seeburg PH, Adelman JP. Characterization of cDNA for precursor of human luteinizing hormone releasing hormone. Nature. 1984;311:666–668. doi: 10.1038/311666a0. [DOI] [PubMed] [Google Scholar]
- Silver MR, Kawauchi H, Nozaki M, Sower SA. Cloning and analysis of the lamprey GnRH-III cDNA from eight species of lamprey representing the three families of Petromyzoniformes. Gen Comp Endocrinol. 2004;139:85–94. doi: 10.1016/j.ygcen.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Sneath PHA, Sokal RR. Numerical Taxonomy. San Francisco: Freeman; 1973. [Google Scholar]
- Suzuki M, Hyodo S, Kobayashi M, Aida K, Urano A. Characterization and localization of mRNA encoding the salmon-type gonadotrophin-releasing hormone precursor of the masu salmon. J Mol Endocrinol. 1992;9:73–82. doi: 10.1677/jme.0.0090073. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Maldonado TA, Lunden JB. Localization of gonadotropin-releasing hormone in the central nervous system and a peripheral chemosensory organ of Aplysia californica. Gen Comp Endocrinol. 2003;130:20–28. doi: 10.1016/s0016-6480(02)00519-1. [DOI] [PubMed] [Google Scholar]
- Twan WH, Hwang JS, Lee YH, Jeng SR, Yueh WS, Tung YH, Wu HF, Dufour S, Chang CF. The presence and ancestral role of gonadotropin-releasing hormone in the reproduction of scleractinian coral, Euphyllia ancora. Endocrinology. 2006;147:397–406. doi: 10.1210/en.2005-0584. [DOI] [PubMed] [Google Scholar]
- Vickers ED, Laberge F, Adams BA, Hara TJ, Sherwood NM. Cloning and localization of three forms of gonadotropin-releasing hormone, including the novel whitefish form, in a salmonid, Coregonus clupeaformis. Biol Reprod. 2004;70:1136–1146. doi: 10.1095/biolreprod.103.023846. [DOI] [PubMed] [Google Scholar]
- von Eggelkraut-Gottanka R, Beck-Sickinger AG. Biosynthesis of peptide hormones derived from precursor sequences. Curr Med Chem. 2004;11:2651–2665. doi: 10.2174/0929867043364405. [DOI] [PubMed] [Google Scholar]
- Young KG, Chang JP, Goldberg JI. Gonadotropin-releasing hormone neuronal system of the freshwater snails Helisoma trivolvis and Lymnaea stagnalis: possible involvement in reproduction. J Comp Neurol. 1999;404:427–437. doi: 10.1002/(sici)1096-9861(19990222)404:4<427::aid-cne1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wayne NL, Sherwood NM, Postigo HR, Tsai PS. Biological and immunological characterization of multiple GnRH in an opisthobranch mollusk, Aplysia californica. Gen Comp Endocrinol. 2000;118:77–89. doi: 10.1006/gcen.2000.7457. [DOI] [PubMed] [Google Scholar]


