The transcriptional regulator CmeR from C. jejuni has been purified and crystallized and X-ray diffraction data have been collected to a resolution of 2.2 Å.
Keywords: CmeR, transcriptional regulators, gene regulation, multi-ligand recognition
Abstract
In Campylobacter jejuni, a Gram-negative bacterial pathogen causing gastroenteritis in humans, the CmeR regulatory protein controls transcription of the multidrug transporter gene operon cmeABC. CmeR belongs to the TetR family of transcriptional regulators. The 210-residue CmeR consists of two functional motifs: an N-terminal DNA-binding domain and a C-terminal ligand-binding domain. It is predicted that the DNA-binding domain interacts directly with target promoters, while the C-terminal motif interacts with inducing ligands (such as bile salts). As an initial step towards confirming this structural model, recombinant CmeR protein containing a 6×His tag at the N-terminus was crystallized. Crystals of ligand-free CmeR belonged to space group P21212, with unit-cell parameters a = 37.4, b = 57.6, c = 93.3 Å. Diffraction was observed to at least 2.2 Å at 100 K. Analysis of the detailed CmeR structure is currently in progress.
1. Introduction
Campylobacter jejuni is the leading bacterial cause of foodborne diarrhea in the United States and other developed countries (Friedman et al., 2000 ▶). It is also a significant enteric pathogen for young children in developing countries. This Gram-negative enteric organism colonizes the intestinal tracts of animals and has become increasingly resistant to antimicrobials owing to its possession of multidrug efflux transporters and its acquisition of various resistance mechanisms, compromising the effectiveness of antibiotic treatment. According to the genomic sequence of NCTC 11168, C. jejuni harbors 13 putative antibiotic efflux transporters of the ATP-binding cassette (ABC), resistance–nodulation–division (RND), multidrug and toxic compound extrusion (MATE), major facilitator (MF) and small multidrug-resistance (SMR) families (Parkhill et al., 2000 ▶; Lin, Akiba & Zhang, 2005 ▶). At present, CmeABC and CmeDEF, which belong to the RND family, are the only two antibiotic efflux transporters that have been functionally characterized in Campylobacter (Akiba et al., 2006 ▶; Lin et al., 2002 ▶; Pumbwe & Piddock, 2002 ▶).
The CmeABC efflux system consists of three members, including an outer membrane channel (CmeC), an inner membrane drug transporter (CmeB) and a periplasmic membrane-fusion protein (CmeA). These three proteins are encoded by a three-gene operon (cmeABC) and form an efflux system that extrudes a variety of toxic compounds directly out of C. jejuni (Lin et al., 2002 ▶). The substrates extruded by CmeABC include commonly used antibiotics (e.g. fluoroquinolones, macrolides, ampicillin, tetracycline, chloramphenicol, cefotaxime and rifampin), ions (e.g. Co2+ and Cu2+) and lipophilic compounds (e.g. SDS and various bile salts). Thus, CmeABC contributes significantly to the intrinsic and acquired resistance of Campylobacter to structurally diverse antimicrobials (Cagliero et al., 2005 ▶; Lin et al., 2002 ▶; Luo et al., 2003 ▶; Pumbwe & Piddock, 2002 ▶). In addition, this efflux system is also essential for Campylobacter colonization of the animal intestinal tract by conferring resistance to bile acids (Lin et al., 2003 ▶), which are normally present in the animal intestinal tract and have bactericidal effect.
The expression of cmeABC is controlled by the transcriptional regulator CmeR (Lin, Akiba, Sahin et al., 2005 ▶). The cmeR gene is located immediately upstream of the cmeABC operon and encodes a 210-residue protein that shares N-terminal sequence and structural similarities with members of the TetR family of transcriptional repressors (Grkovic et al., 2002 ▶; Ramos et al., 2005 ▶). Like other members of the TetR family, the N-terminal domain of CmeR contains a predicted DNA-binding helix–turn–helix (HTH) motif, while its C-terminal region contains unique sequences and is expected to be involved in the binding of inducing ligands (Grkovic et al., 2002 ▶; Lin, Akiba, Sahin et al., 2005 ▶). cmeR is transcribed in the same direction as cmeABC and the intergenic region between cmeR and cmeA contains the inverted-repeat (IR) operator site for cmeABC. As a transcriptional regulator, CmeR binds directly to the IR operator and represses the transcription of cmeABC (Lin, Akiba, Sahin et al., 2005 ▶). Deletion of cmeR or mutations in the IR operator release the repression, resulting in the overexpression of CmeABC, which in turn leads to the enhanced resistance to multiple antibiotics. In addition, bile compounds, including both conjugated (e.g. taurodeoxycholate) and nonconjugated (e.g. cholate), induce the expression of cmeABC by inhibiting the binding of CmeR to the promoter of cmeABC (Lin, Cagliero et al., 2005 ▶), suggesting that bile compounds are inducing ligands of CmeR.
The hypothesis is that binding of inducing ligands to the C-terminal domain of CmeR triggers conformational change in the N-terminal DNA-binding region. This change in conformation results in the release of CmeR from its operator DNA and thus allows transcription from its cognate promoter. As an initial step to elucidate the mechanisms that CmeR uses to regulate gene expression, we here report the crystallization and preliminary X-ray diffraction analysis of the CmeR repressor.
2. Materials and methods
2.1. Protein purification and crystallization
Recombinant CmeR was produced in Escherichia coli using the pQE30 vector (Qiagen). The cloning, expression and purification procedures have been described previously (Lin et al., 2002 ▶, 2003 ▶; Lin, Akiba, Sahin et al., 2005 ▶). This recombinant CmeR, containing a 6×His tag at the N-terminus, is fully functional in DNA binding in vitro and is inducible by bile salts. After purification, the purity of the protein was judged using 10% SDS–PAGE stained with Coomassie Brilliant Blue. The purified protein was extensively dialyzed against buffer containing 10 mM sodium phosphate pH 7.2 and 100 mM NaCl and then concentrated to 10 mg ml−1.
Selenomethionyl-CmeR (SeMet-CmeR) containing a 6×His tag at the N-terminus was overproduced in E. coli JM109 cells. Briefly, a 10 ml LB overnight pre-culture was transferred into 100 ml LB medium containing 100 µg ml−1 ampicillin. The cell culture was grown at 310 K and 210 rev min−1. When the OD600 value was around 1.2, cells were harvested by centrifugation at 6000 rev min−1 for 10 min and then washed two times with 5 ml M9 solution. The cells were resuspended in 10 ml M9 solution and then transferred into 1 l pre-warmed M9 solution containing 100 µg ml−1 ampicillin. The cell culture was incubated at 310 K and 210 rev min−1. When the OD600 reached 0.4, 100 mg lysine, phenylalanine and threonine, 50 mg isoleucine, leucine and valine and 60 mg l-selenomethionine were added. The culture was induced with 1 mM IPTG after 15 min. Cells were harvested within 4 h and were frozen and stored at 193 K. The purification procedure for the SeMet-CmeR protein was the same as that for native CmeR. The 210-amino-acid CmeR contains three methionines; replacement of these methionine sulfurs with seleniums in the SeMet-CmeR protein was confirmed by MALDI time-of-flight mass spectrometry.
The 6×His CmeR protein was crystallized in 24-well plates using the hanging-drop vapor-diffusion method at 293 K. Initial crystallization screening was performed using commercially available sparse-matrix screens (Jancarik & Kim, 1991 ▶) from Hampton Research. Briefly, a 2 µl mixture consisting of 1 µl protein solution (10 mg ml−1 CmeR in 10 mM sodium phosphate pH 7.2 and 100 mM NaCl) and 1 µl reservoir solution was equilibrated against 500 µl reservoir solution. Crystals of CmeR appeared within two weeks. After optimization, the best native 6×His CmeR crystals were obtained from reservoir solution containing 22% PEG 4000, 0.1 M Tris pH 8.5, 0.2 M MgCl2 and 4% glycerol. Cryoprotection was achieved by raising the PEG 4000 concentration stepwise to 35% with a 5% increment in each step. Crystals of the SeMet-CmeR protein were grown under the same conditions.
2.2. Data collection and processing
For data collection, a single native crystal was flash-cooled in a cryoprotectant solution containing 35% PEG 4000, 7% glycerol, 0.2 M MgCl2 and 0.1 M Tris buffer pH 8.5 at 100 K. The best diffraction of the native CmeR crystal was obtained to at least 2.2 Å resolution at a cryogenic temperature of 100 K using a synchrotron light source (Table 1 ▶). Multiple-wavelength anomalous diffraction (MAD) data were collected from a single SeMet-CmeR crystal to a maximum resolution of 2.1 Å (Table 1 ▶). Diffraction data sets from both the native and SeMet-CmeR crystals were taken at the Advanced Light Source (beamline 8.2.2) at cryogenic temperature (100 K) using an ADSC Quantum 315 CCD-based detector. The beam size was 140 × 150 µm. Diffraction data sets were processed with DENZO and scaled with SCALEPACK (Otwinowski & Minor, 1997 ▶).
Table 1. Data collection and crystallographic analysis of CmeR.
Values in parentheses are for the highest resolution shell.
| SeMet | ||||
|---|---|---|---|---|
| Native | Inflection point | Peak | Remote | |
| Data collection | ||||
| Wavelength (Å) | 0.9795 | 0.9798 | 0.9795 | 0.9662 |
| Space group | P21212 | P21212 | ||
| Unit-cell parameters (Å) | a = 37.4, b = 57.6, c = 93.3 | a = 37.3, b = 57.5, c = 93.0 | ||
| Resolution (Å) | 2.24 (2.33–2.24) | 2.10 (2.18–2.10) | 2.07 (2.28–2.07) | 2.07 (2.14–2.07) |
| Completeness (%) | 99.6 (97.7) | 97.6 (85.1) | 93.4 (84.1) | 97.2 (80.3) |
| Total No. of reflections | 297224 | 168174 | 179712 | 179712 |
| No. of unique reflections | 10172 | 12493 | 14087 | 12897 |
| Rsym (%) | 5.5 (26.2) | 5.8 (25.0) | 5.2 (26.4) | 5.1 (27.3) |
| Average I/σ(I) | 16.6 (4.7) | 23.3 (3.8) | 22.3 (5.3) | 18.2 (3.2) |
| Phasing | ||||
| Se-atom sites | 3 | |||
| Resolution range of data used (Å) | 50–2.80 | |||
| Overall figure of merit | 0.59 | |||
3. Results and discussion
Using hanging-drop vapor diffusion, we have successfully grown a large quantity of CmeR crystals. The dimensions of the crystals were typically 0.4 × 0.4 × 0.1 mm (Fig. 1 ▶). The crystals of 6×His CmeR belonged to space group P21212, with unit-cell parameters a = 37.4, b = 57.6, c = 93.3 Å, α = β = γ = 90°.
Figure 1.
Image of the CmeR crystal. The native crystal was grown using 22% PEG 4000, 0.1 M Tris pH 8.5, 0.2 M MgCl2 and 4% glycerol.
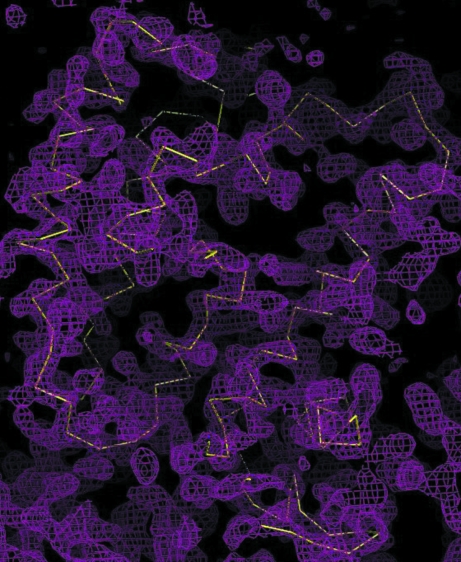
The SeMet crystal belonged to space group P21212, with unit-cell parameters very similar to those of the native crystal (Table 1 ▶). The three-wavelength anomalous dispersion (MAD) data were scaled with SCALEPACK (Otwinowski & Minor, 1997 ▶). Phase calculation was carried out at 2.8 Å resolution using the program BnP (Weeks et al., 2005 ▶) after finding and refinement of all three selenium sites. The electron-density map obtained was improved by density modification (DM) using RESOLVE (Terwilliger, 2001 ▶). Fig. 2 ▶ shows the MAD electron-density map improved by DM and the initial Cα protein trace.
Figure 2.
MAD electron-density map of CmeR contoured at 1σ. The Cα plot of one subunit of CmeR is shown in yellow.
The available structures of members of the TetR family of transcriptional repressors, including those of TetR (Hinrichs et al., 1994 ▶; Orth et al., 2000 ▶), QacR (Schumacher et al., 2001 ▶, 2002 ▶), CprB (Natsume et al., 2003 ▶) and EthR (Dover et al., 2004 ▶; Frenois et al., 2004 ▶), indicate that all these repressors assemble as dimers. Recently, the preliminary X-ray diffraction data of AcrR (another member of the TetR family) also suggest a dimeric assembly of the repressor (Li et al., 2006 ▶). Thus, it is likely that the structure of CmeR is also dimeric.
The MC program (Matthews, 1968 ▶, 1977 ▶) from the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶) suggests that the crystals are likely to contain one CmeR monomer in the asymmetric unit, which gives a Matthews value V M of 2.0 Å3 Da−1 and a solvent content of 36.9%. By applying the crystallographic symmetry operators, a dimeric arrangement of the protein was found. Analysis of the structure is currently in progress.
Acknowledgments
This work was supported by NIH grants DK063008 (to QZ) and GM074027 (to EWY).
References
- Akiba, M., Lin, J., Barton, Y. W. & Zhang, Q. J. (2006). J. Antimicrob. Chemother.57, 52–60. [DOI] [PubMed] [Google Scholar]
- Cagliero, C., Mouline, C., Payot, S. & Cloeckaert, A. (2005). J. Antimicrob. Chemother.56, 948–950. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Dover, L. G., Corsino, P. E., Daniels, I. R., Cocklin, S. L., Tatituri, V., Besra, G. S. & Fütterer, K. (2004). J. Mol. Biol.340, 1095–1105. [DOI] [PubMed] [Google Scholar]
- Frenois, F., Engohang-Ndong, J., Locht, C., Baulard, A. R. & Villeret, V. (2004). Mol. Cell, 16, 301–307. [DOI] [PubMed] [Google Scholar]
- Friedman, C. R., Neimann, J., Wegener, H. C. & Tauxe, R. V. (2000). Campylobacter, edited by I. Nachamkin & M. J. Blaser, pp. 121–138. Washington DC: ASM Press.
- Grkovic, S., Brown, M. H. & Skurray, R. A. (2002). Microbiol. Mol. Biol. Rev.66, 671–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs, W., Kisker, C., Duvel, M., Muller, A., Tovar, K., Hillen, W. & Saenger, W. (1994). Science, 264, 418–420. [DOI] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Li, M., Qiu, X., Su, C.-C., Long, F., Gu, R., McDermott, G. & Yu, E. W. (2006). Acta Cryst. F62, 1150–1152. [DOI] [PMC free article] [PubMed]
- Lin, J., Akiba, M., Sahin, O. & Zhang, Q. (2005). Antimicrob. Agents Chemother.49, 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J., Akiba, M. & Zhang, Q. (2005). Campylobacter: Molecular and Cell Biology, edited by J. M. Ketley & M. E. Konkel, pp. 205–218. Norwich: Horizon Bioscience.
- Lin, J., Cagliero, C., Guo, B., Barton, Y. W., Maurel, M. C., Payot, S. & Zhang, Q. (2005). J. Bacteriol.187, 7417–7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J., Michel, L. O. & Zhang, Q. (2002). Antimicrob. Agents Chemother.46, 2124–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J., Sahin, O., Michel, L. O. & Zhang, Q. (2003). Infect. Immun.71, 4250–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, N., Sahin, O., Lin, J., Michel, L. O. & Zhang, Q. (2003). Antimicrob. Agents Chemother.47, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1977). The Proteins, edited by H. Neurath & R. L. Hill, Vol. 3, pp. 468–477. New York: Academic Press.
- Natsume, R., Ohnishi, Y., Senda, T. & Horinouchi, S. (2003). J. Mol. Biol.336, 409–419. [DOI] [PubMed]
- Orth, P., Schnappinger, D., Hillen, W., Saenger, W. & Hinrichs, W. (2000). Nature Struct. Biol.7, 215–219. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, M. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Parkhill, J. et al. (2000). Nature (London), 403, 665–668. [Google Scholar]
- Pumbwe, L. & Piddock, L. J. (2002). FEMS Microbiol. Lett.206, 185–189. [DOI] [PubMed] [Google Scholar]
- Ramos, J. L., Martinez-Bueno, M., Molina-Henares, A. J., Teran, W., Watanabe, K., Zhang, X. D., Gallegos, M. T., Brennan, R. & Tobes, R. (2005). Microbiol. Mol. Biol. Rev.69, 326–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, M. A., Miller, M. C., Grkovic, S., Brown, M. H., Skurray, R. A. & Brennan, R. G. (2001). Science, 294, 2158–2163. [DOI] [PubMed] [Google Scholar]
- Schumacher, M. A., Miller, M. C., Grkovic, S., Brown, M. H., Skurray, R. A. & Brennan, R. G. (2002). EMBO J.21, 1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T. C. (2001). Acta Cryst. D57, 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks, C. M., Shah, N., Green, M. L., Miller, R. & Furey, H. (2005). Acta Cryst. A61, C152.