Diisopropyl fluorophosphatase (DFPase) effectively hydrolyzes a number of organophosphorus nerve agents, including sarin, cyclohexylsarin, soman and tabun. Neutron diffraction data have been collected from DFPase crystals to 2.2 Å resolution in an effort to gain further insight into the mechanism of this enzyme.
Keywords: neutron diffraction, DFPase, time-of-flight, phosphotriesterase
Abstract
The enzyme diisopropyl fluorophosphatase (DFPase) from Loligo vulgaris is capable of decontaminating a wide variety of toxic organophosphorus nerve agents. DFPase is structurally related to a number of enzymes, such as the medically important paraoxonase (PON). In order to investigate the reaction mechanism of this phosphotriesterase and to elucidate the protonation state of the active-site residues, large-sized crystals of DFPase have been prepared for neutron diffraction studies. Available H atoms have been exchanged through vapour diffusion against D2O-containing mother liquor in the capillary. A neutron data set has been collected to 2.2 Å resolution on a relatively small (0.43 mm3) crystal at the spallation source in Los Alamos. The sample size and asymmetric unit requirements for the feasibility of neutron diffraction studies are summarized.
1. Introduction
Diisopropyl fluorophosphatase (DFPase; EC 3.1.8.2; 314 amino acids; 35 kDa), a Ca2+-dependent phosphotriesterase, is capable of efficiently detoxifying a wide variety of organophosphorus nerve agents, such as sarin, cyclohexylsarin, soman and tabun (Hartleib & Rüterjans, 2001 ▶). As such, it is a prime candidate for the enzymatic decontamination of existing nerve-agent stocks. On the basis of structural and biochemical experiments, two reaction mechanisms have been proposed for DFPase. Initially, a mechanism was proposed in which residue His287 acts as a general base (Scharff et al., 2001 ▶). However, mutants of the candidate residue His287 have shown little to no loss of activity and eliminated the histidine as the general base in the reaction (Katsemi et al., 2005 ▶). More recently, the catalytic calcium-coordinating residue Asp229 was identified as the nucleophile on the basis of structural, kinetic and isotope-labelling experiments and an alternative mechanism involving a phosphoenzyme intermediate was proposed (Blum et al., 2006 ▶). A water molecule then attacks the carboxyl C atom of Asp229 to generate the product. This proposed mechanism may be common to a number of structurally related proteins, such as the high-density lipoprotein (HDL) component paraoxonase (PON), that share similar active-site environments (Harel et al., 2004 ▶) and also show activity against organophosphorus compounds.
To better understand the reaction mechanism of DFPase as well as other phosphotriesterases, it is crucial to determine the protonation states of the active-site residues and important to clearly visualize the hydrogen-bonding pattern in the vicinity of the catalytic residues. Neutron diffraction, which was first utilized on protein crystals in the late 1960s (Schoenborn, 1969 ▶), can provide this information. Unlike X-rays, neutrons are readily scattered by H atoms and the coherent neutron scattering lengths of the atoms in proteins (C, N, O, S and D) are similar in magnitude. Furthermore, by taking advantage of the opposite coherent scattering lengths of H and D atoms, one can straightforwardly and accurately locate H atoms even in moderate-resolution neutron structures (2.0–2.5 Å). As such, neutron diffraction is an invaluable tool for understanding enzyme-reaction mechanisms (Kossiakoff & Spencer, 1981 ▶).
Compared with X-ray synchrotron sources, neutron sources are low-flux, so very large sample sizes are required for successful experiments, normally >1 mm3. DFPase crystals typically grow to >1 mm in one dimension and diffract X-rays to atomic resolution. A number of high-resolution and atomic resolution structures have been reported for DFPase (Scharff et al., 2001 ▶; Koepke et al., 2003 ▶). However, despite the 0.85 Å data available, only a limited number of H atoms could be discerned. The aim of this study was to prepare suitable crystals of DFPase for neutron diffraction studies in order to determine the protonation states of the catalytic residues, as well as to precisely define the hydrogen-bonding network in the vicinity of the active site. We report the collection of neutron diffraction data from DFPase crystals. We furthermore discuss the relationship between the crystal size, the size of the asymmetric unit and the feasibility of neutron data collection.
2. Crystallization and data collection
The protein was prepared as reported previously (Hartleib & Rüterjans, 2001 ▶). Briefly, the overexpressed protein was initially purified by Ni–NTA affinity chromatography. The His tag was cleaved by the addition of thrombin and after rechromatography with Ni–NTA to remove the His tag, the protein was purified to homogeneity by ion-exchange chromatography using Q-Sepharose. Crystals of DFPase were grown by hanging-drop vapour diffusion at room temperature with 1–2 mM DFPase solubilized in 10 mM Tris pH 7.5, 2 mM CaCl2 and mixed with an equal or near-equal volume of well buffer (7–11% PEG 4000, 0.1 M MES pH 6.5). The drop size ranged from 10 to 20 µl. Some crystals appeared overnight; however, the largest single crystals appeared over a period of approximately one month. Initially, capillary-mounted DFPase crystals were shipped to Los Alamos for screening. Although the crystals diffracted neutrons strongly, no diffraction was observed beyond 6 Å. Inspection of the crystals showed that they had been damaged during the shipping process. Concurrently, perdeuterated DFPase crystals were also screened. These perdeuterated crystals were of a very small size (<0.1 mm3) and diffracted poorly; they were thus unsuitable for data collection.
In order to eliminate the possibility of crystal damage during shipping, subsequent crystallization trials were performed at Los Alamos. Of the approximately 200 individual hanging-drop crystal trials screened, nine crystals were found to be of a suitable size and habit for further investigation. Of these, several candidate crystals were successfully mounted in quartz capillaries and the exchange of available H atoms for D atoms through vapour diffusion was initiated by applying ∼20 µl of 9% PEG 4000, 0.1 M MES pD 6.5 (all stock solutions prepared in D2O) on either side of the crystal. One week later, the crystals were screened at the Protein Crystallography Station (PCS) at the Los Alamos Neutron Science Center (LANSCE) spallation neutron source (Schoenborn & Pitcher, 1996 ▶; Langan et al., 2004 ▶). A 24 h test exposure was taken on the largest DFPase crystal (2.4 × 0.5 × 0.36 mm, 0.43 mm3), yielding diffraction to 2.2 Å resolution (Fig. 1 ▶). No further exchanges for deuterated mother liquor were performed, as the diffraction quality and signal-to-noise ratio were deemed to be suitable for collection of a full data set.
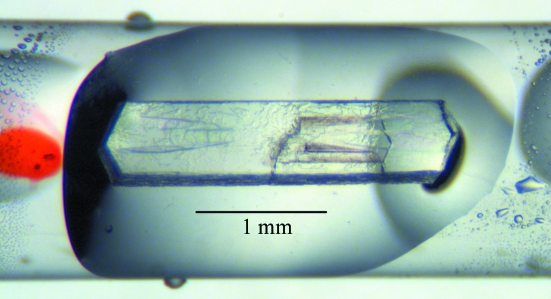
Figure 1.
Crystal of DFPase following data collection. The crystal measured 2.4 × 0.5 × 0.36 mm in size and was mounted in a 2 mm diameter thin-walled quartz capillary.
Time-of-flight wavelength-resolved Laue images were collected at 37 usable settings on a single crystal, with approximately 24 h exposure per setting (Fig. 2 ▶). The crystal-to-detector distance was 70 cm. Data were processed using a version of d*TREK (Pflugrath, 1999 ▶) modified for wavelength-resolved Laue neutron protein crystallography (Langan & Greene, 2004 ▶), wavelength-normalized using LAUENORM (Helliwell et al., 1989 ▶) and then merged using SCALA (Collaborative Computational Project, Number 4, 1994 ▶; Diederichs & Karplus, 1997 ▶; Weiss & Hilgenfeld, 1997 ▶; Weiss, 2001 ▶). The ‘tails’ of the wavelength range were cut off, with a restricted range of 0.8–4.5 Å, from the original 0.6–6 Å wavelength distribution of the neutrons. The overall completeness was 81.8% to 2.2 Å, with reasonable merging statistics and redundancy (Table 1 ▶).
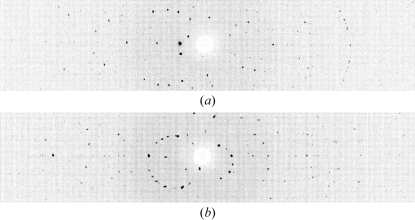
Figure 2.
Neutron Laue diffraction pattern for DFPase in two different crystal settings. In each crystal setting the data were projected in time-of-flight, thus generating a conventional Laue pattern.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Source | PCS, Los Alamos |
| Settings | 37 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 43.4, b = 83.3, c = 87.5 |
| Resolution (Å) | 10.0–2.2 (2.32–2.20) |
| Reflections (measured/unique) | 64711/10999 |
| Redundancy | 5.9 |
| Completeness (%) | 81.8 (72.9) |
| Rsym | 0.199 (0.397) |
| Wavelength range (Å) | 0.8–4.5 |
| 〈I/σ(I)〉 | 1.9 (1.8) |
| Mn(I)/sd | 4.0 (2.2) |
Refinement of the structure is in progress using SHELX (Sheldrick, 1998 ▶) and utilizing a version of CNS (Brünger et al., 1998 ▶) modified for joint X-ray and neutron refinement. A 1.86 Å room-temperature X-ray data set of DFPase (structure factors and PDB code 2gvw; Blum et al., 2006 ▶) is being used together with the recently collected neutron data. Visual inspection of the initial maps show clear signature features for neutron maps. The ‘cancellation’ effect of the opposite coherent scattering lengths of H and C atoms is seen in the absence of nuclear density around hydrocarbon groups on amino-acid side chains. The terminal amide groups of lysine residues are visible as very strong nuclear density. Further structural features will be elucidated during the course of the refinement.
3. Sample-size and asymmetric unit limitations for neutron structures
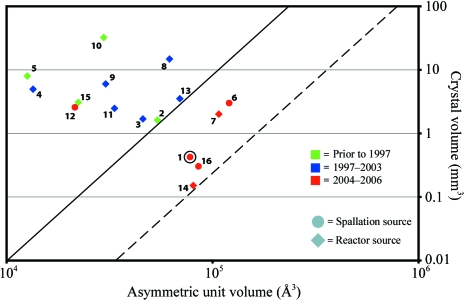
DFPase, at 35 kDa, is one of the larger proteins and asymmetric units (77 626 Å3) to be studied using neutron crystallography, although the crystal size used in the diffraction experiment reported here is among the smallest (0.43 mm3). There are around 16 unique protein structures that have been studied by neutron crystallography, including several structures that have not yet been deposited in the PDB. We have plotted the relationship between asymmetric unit volume and the size of the crystal used in successful neutron diffraction experiments, similar to that published in 1997 (Habash et al., 1997 ▶; Table 2 ▶, Fig. 3 ▶). The solid line in Fig. 3 ▶ represents the empirical limit of neutron protein crystallography as of 2003. Since then, this work, as well as other structures published using data collected at the spallation source in Los Alamos (Schoenborn & Pitcher, 1996 ▶; Langan et al., 2004 ▶) and at reactor sources such as the Institut Laue–Langevin (ILL; Cipriani et al., 1996 ▶; Myles et al., 1998 ▶), has pushed the limits of neutron diffraction with regard to the asymmetric unit volume and crystal size utilized for successful experiments. Neutron diffraction data collected from crystals <1 mm3 in size and with asymmetric units exceeding 60 000 Å3, once considered to be unreasonable, have become possible over the last 3 y and may become more routine in the future. On the basis of these recent structures and the work presented here, a dashed line has been drawn in Fig. 3 ▶ that may serve as a useful guide to the feasibility of diffraction studies at current neutron sources. The current development of a number of new neutron sources and instrument upgrades will make neutron diffraction studies of protein crystals in this size range significantly easier within the next decade.
Table 2. Sample volume (V c), unit-cell volume (V u), asymmetric unit volume (V a), molecular weight (MW) and year of publication of available neutron structures.
| Space group | Vu (Å3) | Va (Å3) | Vc (mm3) | MW (Da) | Reference | |
|---|---|---|---|---|---|---|
| 1. DFPase | P212121 | 310503 | 77626 | 0.43 | 35000 | This study |
| 2. Trypsin | P212121 | 216761 | 54190 | 1.62 | 23300 | Kossiakoff & Spencer (1981 ▶) |
| 3. DsrD | P212121 | 183143 | 45786 | 1.70 | 8840 | Chatake et al. (2003 ▶) |
| 4. Rubredoxin | P212121 | 53600 | 13400 | 5.00 | 5900 | Kurihara et al. (2004 ▶) |
| 5. BPTI | P212121 | 50111 | 12528 | 8.00 | 6530 | Wlodawer et al. (1984 ▶) |
| 6. D-Xylose isomerase† | I222 | 962934 | 120367 | 3.00 | 160000 | Katz et al. (2006 ▶) |
| 7. Rasburicase† | I222 | 814080 | 101760 | 1.80 | 135000 | Budayova-Spano et al. (2006 ▶) |
| 8. Concanavalin A | I222 | 493697 | 61712 | 15.00 | 25600 | Blakeley et al. (2004 ▶) |
| 9. Lysozyme | P43212 | 242213 | 30277 | 6.00 | 14300 | Niimura et al. (1997 ▶) |
| 10. Ribonuclease A | P21 | 59444 | 29722 | 30.00 | 13700 | Wlodawer & Sjölin (1981 ▶); Wlodawer (1980 ▶) |
| 11. Myoglobin‡ | P21 | 66735 | 33368 | 2.50 | 17200 | Shu et al. (2000 ▶) |
| 12. Amicyanin | P21 | 43127 | 21564 | 2.60 | 11500 | Sukumar et al. (2005 ▶) |
| 13. Endothiapepsin | P21 | 138925 | 69463 | 3.52 | 35000 | Coates et al. (2001 ▶) |
| 14. Aldose reductase‡ | P21 | 161663 | 80832 | 0.15 | 36000 | Hazemann et al. (2005 ▶) |
| 15. Insulin | H3 | 200409 | 22268 | 3.00 | 5790 | Wlodawer et al. (1989 ▶) |
| 16. DHFR§ | P61 | 516990 | 86165 | 0.30 | 36100 | Bennett et al. (2006 ▶) |
Tetramer.
Perdeuterated.
Dimer.
Figure 3.
Scatter plot of asymmetric unit volume versus crystal size, based on published neutron data (see Table 2 ▶). The solid line on the left represents the range of asymmetric unit volume and sample size for structures solved up to 2003. The dashed line on the right is a new guideline for current and future neutron structures.
In addition, this work describes an efficient sample-preparation and data-collection process, with a total of approximately five months between sample preparation and completion of data collection. This represents a considerably expedited ‘turnaround’ period for neutron structures, which have typically been of the order of years. This relative speed, combined with the comparatively small crystal size used in this study, potentially opens neutron diffraction as a more widely utilizable technique for the study of protein structures and enzyme mechanisms. In order to fully exploit the advantages of replacing hydrogen by deuterium in neutron crystallography, we have also prepared perdeuterated DFPase. However, despite the reduced sample-size requirement for perdeuterated protein, we have so far not been able to obtain suitable crystals for full data collection.
Together with our recent study on DFPase, the neutron structure will provide critical insight into the phosphotriesterase mechanism. Furthermore, insights from the refined neutron structure will serve as a framework for future protein-engineering efforts aimed at broadening the range of substrates that can be hydrolyzed by DFPase.
Acknowledgments
We thank Drs Leighton Coates and Marat Mustyakimov for helpful discussions and we thank Mary Jo Waltman and Sean Seaver for technical assistance. This work was funded by Fraunhofer Grants E590/3Z023/M5137 and E590/6Z004/4F170 and the Hessisches Ministerium für Wissenschaft und Kultur. A travel grant to Los Alamos for MMB and JC-HC was kindly provided by the GlaxoSmithKline Foundation. The PCS is funded by the Office of Science and the Office of Biological and Environmental Research of the US Department of Energy.
References
- Bennett, B., Langan, P., Coates, L., Mustyakimov, M., Schoenborn, B. P., Howell, E. E. & Dealwis, C. (2006). Proc. Natl Acad. Sci. USA, 103, 18493–18498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeley, M. P., Kalb, A. J., Helliwell, J. R. & Myles, D. A. A. (2004). Proc. Natl Acad. Sci. USA, 101, 16405–16410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, M.-M., Löhr, F., Richardt, A., Rüterjans, H. & Chen, J. C.-H. (2006). J. Am. Chem. Soc.128, 12750–12757. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Budayova-Spano, M., Bonneté, F., Ferté, N., El Hajji, M., Meilleur, F., Blakeley, M. P. & Castro, B. (2006). Acta Cryst. F62, 306–309. [DOI] [PMC free article] [PubMed]
- Chatake, T., Mizuno, N., Voordouw, G., Higuchi, Y., Arai, S., Tanaka, I. & Niimura, N. (2003). Acta Cryst. D59, 2306–2309. [DOI] [PubMed] [Google Scholar]
- Cipriani, F., Castagna, J. C., Wilkinson, C., Oleinek, P. & Lehmann, M. S. (1996). J. Neutron Res.4, 79–85.
- Coates, L., Erskine, P. T., Wood, S. P., Myles, D. A. A. & Cooper, J. B. (2001). Biochemistry, 40, 13149–13157. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed] [Google Scholar]
- Habash, J., Raftery, J., Weisgerber, S., Cassetta, A., Lehmann, M. S., Hoghoj, P., Wilkinson, C., Campbell, J. W. & Helliwell, J. R. (1997). J. Chem. Soc. Faraday Trans.93, 4313–4317.
- Harel, M., Aharoni, A., Gaidukov, L., Brumshtein, B., Khersonsky, O., Meged, R., Dvir, H., Ravelli, R. B. G., McCarthy, A., Toker, L., Silman, I., Sussman, J. L. & Tawfik, D. S. (2004). Nature Struct. Mol. Biol.11, 412–419. [DOI] [PubMed]
- Hartleib, J. & Rüterjans, H. (2001). Protein Expr. Purif.21, 210–219. [DOI] [PubMed] [Google Scholar]
- Hazemann, I., Dauvergne, M. T., Blakeley, M. P., Meilleur, F., Haertlein, M., Van Dorsselaer, A., Mitschler, A., Myles, D. A. A. & Podjarny, A. (2005). Acta Cryst. D61, 1413–1417. [DOI] [PubMed] [Google Scholar]
- Helliwell, J. R., Habash, J., Cruickshank, D. W. J., Harding, M. M., Greenhough, T. J., Campbell, J. W., Clifton, I. J., Elder, M., Machin, P. A., Papiz, M. Z. & Zurek, S. (1989). J. Appl. Cryst.22, 483–497. [Google Scholar]
- Katsemi, V., Lücke, C., Koepke, J., Löhr, F., Maurer, S., Fritzsch, G. & Rüterjans, H. (2005). Biochemistry, 44, 9022–9033. [DOI] [PubMed] [Google Scholar]
- Katz, A. K., Xinmin, L., Carrell, H. L., Hanson, B. L., Langan, P., Coates, L., Schoenborn, B. P., Glusker, J. P. & Bunick, G. J. (2006). Proc. Natl Acad. Sci. USA, 103, 8342–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepke, J., Scharff, E. I., Lücke, C., Rüterjans, H. & Fritzsch, G. (2003). Acta Cryst. D59, 1744–1754. [DOI] [PubMed] [Google Scholar]
- Kossiakoff, A. A. & Spencer, S. A. (1981). Biochemistry, 20, 6462–6474. [DOI] [PubMed] [Google Scholar]
- Kurihara, K., Tanaka, I., Chatake, T., Adams, M. W. W., Jenney, F. E., Moiseeva, N., Bau, R. & Niimura, N. (2004). Proc. Natl Acad. Sci. USA, 101, 11215–11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan, P. & Greene, G. (2004). J. Appl. Cryst.37, 253–257. [Google Scholar]
- Langan, P., Greene, G. & Schoenborn, B. P. (2004). J. Appl. Cryst.37, 24–31. [Google Scholar]
- Myles, D. A. A., Bon, C., Langan, P., Cipriani, F., Castagna, J. C., Lehmann, M. S. & Wilkinson, C. (1998). Physica B, 241, 1122–1130.
- Niimura, N., Minezaki, Y., Nonaka, T., Castagna, J. C., Cipriani, F., Hoghoj, P., Lehamann, M. S. & Wilkinson, C. (1997). Nature Struct. Biol.4, 909–914. [DOI] [PubMed] [Google Scholar]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- Scharff, E. I., Koepke, J., Fritzsch, G., Lücke, C. & Rüterjans, H. (2001). Structure, 9, 493–502. [DOI] [PubMed] [Google Scholar]
- Schoenborn, B. P. (1969). Nature (London), 224, 143–146. [DOI] [PubMed] [Google Scholar]
- Schoenborn, B. P. & Pitcher, E. (1996). In Neutrons in Biology, edited by B. P. Schoenborn & R. B. Knott. New York: Plenum.
- Sheldrick, G. M. (1998). In Crystallographic Computing 7, edited by K. Watenpaugh & P. E. Bourne. Oxford University Press.
- Shu, F., Ramakrishnan, V. & Schoenborn, B. P. (2000). Proc. Natl Acad. Sci. USA, 97, 3872–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar, N., Langan, P., Mathews, F. S., Jones, L. H., Thiyagarajan, P., Schoenborn, B. P. & Davidson, V. L. (2005). Acta Cryst. D61, 640–642. [DOI] [PubMed] [Google Scholar]
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135. [Google Scholar]
- Weiss, M. S. & Hilgenfeld, R. (1997). J. Appl. Cryst.30, 203–205. [Google Scholar]
- Wlodawer, A. (1980). Acta Cryst. B36, 1826–1831. [Google Scholar]
- Wlodawer, A., Savage, H. & Dodson, G. (1989). Acta Cryst. B45, 99–107. [DOI] [PubMed] [Google Scholar]
- Wlodawer, A. & Sjölin, L. (1981). Proc. Natl Acad. Sci. USA, 78, 2853–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer, A., Walter, J., Huber, R. & Sjölin, L. (1984). J. Mol. Biol.180, 301–329. [DOI] [PubMed] [Google Scholar]