The crystal structure of the catalytic domain of a chitinase from P. furiosus is reported.
Abstract
The crystal structure of the catalytic domain of a chitinase from the hyperthermophilic archaeon Pyrococcus furiosus (AD2PF-ChiA) has been determined at 1.5 Å resolution. This is the first structure of the catalytic domain of an archaeal chitinase. The overall structure of AD2PF-ChiA is a TIM-barrel fold with a tunnel-like active site that is a common feature of family 18 chitinases. Although the catalytic residues (Asp522, Asp524 and Glu526) are conserved, comparison of the conserved residues and structures with those of other homologous chitinases indicates that the catalytic mechanism of PF-ChiA is different from that of family 18 chitinases.
1. Introduction
Chitinases (EC 3.2.1.14) hydrolyze chitin, a polymer of β-1,4-linked N-acetylglucosamine (GlcNAc), and are classified into two families (families 18 and 19 in the CAZy database; http://afmb.cnrs-mrs.fr/CAZY/) according to amino-acid sequence similarity (Henrissat, 1991 ▶). Family 18 chitinases include glycoside hydrolases, which have been found in many organisms ranging from bacteria to humans (Henrissat & Davies, 1997 ▶).
The catalytic domains of family 18 chitinases have (β/α)8-barrel folds and are characterized by several conserved sequence motifs (Terwisscha van Scheltinga et al., 1996 ▶; Suzuki et al., 1999 ▶). Three-dimensional structures have been reported for bacterial family 18 chitinases, such as chitinases A and B from Serratia marcescens and chitinase A1 from Bacillus circulans (Perrakis et al., 1994 ▶; van Aalten et al., 2000 ▶; Matsumoto et al., 1999 ▶). In particular, the catalytic mechanism of chitinase B has been investigated through extensive structure and mutagenesis studies (van Aalten et al., 2001 ▶). However, little is known about the structure and function of archaeal chitinases.
In the genome database of the hyperthermophilic archaeon Pyrococcus furiosus, we found two adjacent genes (PF1234 and PF1233) homologous to those of the chitinase of Thermococcus kodakaraensis. On analysis of the structural gene of P. furiosus, it appeared that a nucleotide insertion (the 1006th adenine from the ORF start position) in PF1234 has caused a frame shift and separated the genes. By deletion of the inserted nucleotide in PF1234, the best match was achieved between the chitinases of T. kodakaraenesis and P. furiosus (Oku & Ishikawa, 2006 ▶). This artificial recombinant chitinase (referred to as PF-ChiA) possesses two chitin-binding domains (ChBD1PF-ChiA and ChBD2PF-ChiA; Nakamura et al., 2005 ▶) and two active (catalytic) domains (AD1PF-ChiA and AD2PF-ChiA). Both catalytic domains are classified into family 18. Interestingly, this PF-ChiA exhibits hydrolytic activity toward not only colloidal but also crystalline (β/α) chitins at high temperature (Oku & Ishikawa, 2006 ▶).
Here, we report the first crystal structure of the active (catalytic) domain (AD2PF-ChiA) of a hyperthemophilic archaeal chitinase at 1.5 Å resolution and discuss the structural comparisons with other family 18 chitinases.
2. Materials and methods
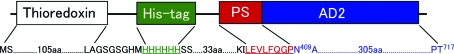
The construction of the expression plasmid has been described previously (Mine et al., 2006 ▶). In this construct, AD2PF-ChiA was expressed with an N-terminal thioredoxin-His6 tag (Fig. 1 ▶), the fusion tag being removed by proteolytic cleavage with PreScission Protease (Amersham Biosciences; Mine et al., 2006 ▶). Escherichia coli cells harbouring the expression plasmid were cultivated in a modified M9 medium containing selenomethionine and inhibitors of methionine biosynthesis (Doublié, 1997 ▶) to produce a selenomethionine derivative of AD2PF-ChiA. The selenomethionine derivative of AD2PF-ChiA was purified by the same method for the native protein (Mine et al., 2006 ▶). In brief, the fusion protein was isolated by Ni2+-chelating chromatography followed by removal of the fusion tag. The final purification step was anion-exchange chromatography using a HiTrap-Q column (Amersham Biosciences). Crystallization of the selenomethionine derivative of AD2PF-ChiA was performed using the same method as used for the native protein (Mine et al., 2006 ▶).
Figure 1.
Schematic representation of the fusion protein thioredoxin-His6tag-PS-AD2PF-ChiA. PreScission protease recognizes a subset of sequences which includes the PS sequence LEVFQGP (underlined), cleaving between the Q and G residues.
Diffraction data for the selenomethionine derivative were collected at BL38B1, SPring-8 (Harima, Japan) and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). Selenium sites were searched for with SOLVE (Terwilliger & Berendzen, 1999 ▶) and all 14 sites were found in the asymmetric unit. Initial phase sets were calculated by the MAD method and improved by density modification with RESOLVE (Terwilliger & Berendzen, 1999 ▶). The model of a single polypeptide was built in the experimental electron-density map using O (Jones et al., 1991 ▶). This initial model was used to phase a higher resolution native data set (1.5 Å; Mine et al., 2006 ▶) using MOLREP from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). The native crystal contained two polypeptides in the asymmetric unit. Refinement was performed using CNS (Brünger et al., 1998 ▶). Stereochemical analysis and structural comparisons were carried out using PROCHECK (Laskowski et al., 1993 ▶) and CHIMERA (Pettersen et al., 2004 ▶), respectively. The diffraction data statistics and the crystallographic refinement statistics are summarized in Table 1 ▶.
Table 1. Summary of the crystallographic data.
Values in parentheses are for the highest resolution shell.
| Native† | Peak | Edge | Low remote | High remote | |
|---|---|---|---|---|---|
| Data-collection statistics | |||||
| X-ray source | BL44XU, SPring-8 | BL38B1, SPring-8 | BL38B1, SPring-8 | BL38B1, SPring-8 | BL38B1, SPring-8 |
| Wavelength (Å) | 0.7 | 0.97915 | 0.97934 | 0.99506 | 0.96411 |
| Space group | P212121 | P212121 | |||
| Unit-cell parameters (Å) | a = 90.0, b = 92.8, c = 107.2 | a = 89.9, b = 92.6, c = 107.2 | |||
| Resolution range (Å) | 50–1.50 (1.55–1.50) | 50–2.00 (2.07–2.00) | 50–2.00 (2.07–2.00) | 50–2.00 (2.07–2.00) | 50–2.00 (2.07–2.00) |
| Rsym‡ (%) | 10.7 (26.6) | 8.6 (15.4) | 6.7 (14.0) | 6.7 (14.9) | 7.3 (17.6) |
| I/σ(I) | 18.0 | 20.0 | 19.2 | 19.0 | 15.8 |
| Total reflections | 1109463 | 433225 | 435356 | 415388 | 433555 |
| Unique reflections | 142710 (14039) | 58665 | 58691 | 56051 | 61306 |
| Redundancy | 7.8 (5.9) | 7.4 (6.7) | 7.4 (6.6) | 7.4 (5.8) | 7.4 (7.1) |
| Completeness (%) | 99.5 (98.8) | 96.1 (60.4) | 96.0 (59.5) | 91.6 (14.5) | 100 (100) |
| B factor of data from Wilson plot (Å2) | 14.8 | ||||
| Se atoms found/total | 14/14 | ||||
| Average figure of merit§ | 0.66 | ||||
| Refinement statistics | |||||
| Resolution range (Å) | 25.2–1.50 (1.59–1.50) | ||||
| No. of reflections | 142675 (22020) | ||||
| Rcryst¶ (%)/Rfree†† (%) | 16.8 (20.2)/18.0 (22.3) | ||||
| R.m.s.d. bond lengths (Å) | 0.005 | ||||
| R.m.s.d. angles (°) | 1.3 | ||||
| Polypeptide chains in the asymmetric unit | 2 | ||||
| Matthews coefficient (Å 3 Da−1) | 3.20 | ||||
| Protein atoms | 4768 | ||||
| Average B factor (Å2) | 14.9 | ||||
| Water molecules | 650 | ||||
| Average B factor (Å2) | 29.6 | ||||
| Sulfate ion | 1 | ||||
| Glycerol molecules | 6 | ||||
| Ramachandran plot‡‡ (%) | |||||
| Favoured | 90.8 | ||||
| Allowed | 8.8 | ||||
| Disallowed§§ | 0.4 | ||||
Data-collection statistics for the native crystal are taken from Mine et al. (2006 ▶).
R
sym = 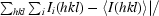
 , where I
i(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I(hkl)〉 is the average.
, where I
i(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I(hkl)〉 is the average.
Figure of merit = |F(hkl)best|/F(hkl).
R
cryst = 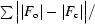
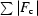 , where F
o and F
c are the observed and calculated structure-factor amplitudes, respectively.
, where F
o and F
c are the observed and calculated structure-factor amplitudes, respectively.
R free was calculated using a randomly selected 5% of the data set that was omitted through all stages of refinement.
The Ramachandran plot was obtained for all residues other than Gly and Pro.
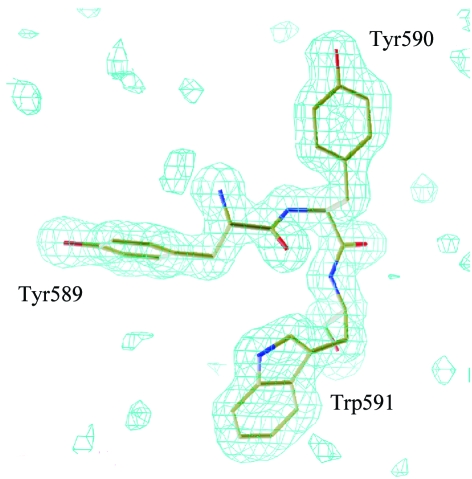
The disallowed conformation is found for Tyr590; nevertheless, the electron-density map around Tyr590 is clear and is shown in Fig. 7 ▶.
3. Results and discussion
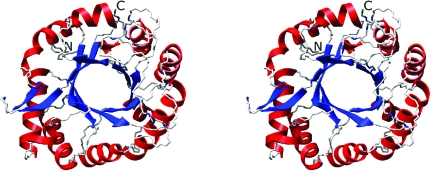
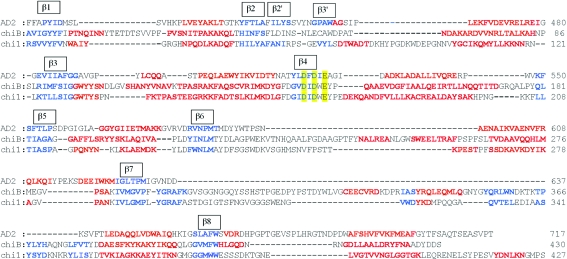
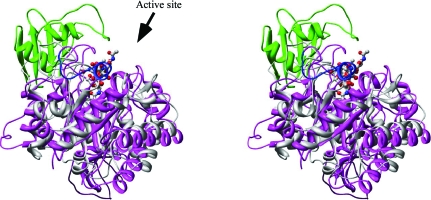
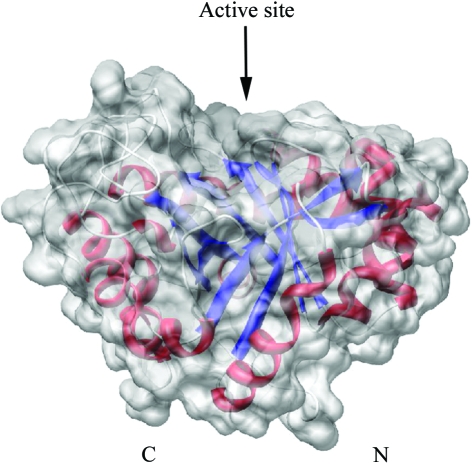
The structure of AD2PF-ChiA was solved at 1.5 Å resolution. The polypeptide chain was successfully traced from the N-terminal Asn409 to Phe706. At the C-terminus, 11 amino acids could not be traced owing to a lack of electron density. The overall structure of AD2PF-ChiA is a TIM-barrel (β/α)8-fold with a tunnel-like active site (Fig. 2 ▶) that is a common feature of exo-chitinases (Davies & Henrissat, 1995 ▶; Spezio et al., 1993 ▶; Rouvinen et al., 1990 ▶). Chitinase B from S. marcescens (PDB code 1e15) and chitinase 1 from Coccidioides immitis (PDB code 1d2k) were retrieved from the family 18 chitinases with the highest Z scores (19.8 and 18.8, respectively) using the DALI server (http://www.ebi.ac.uk/dali/; Fig. 3 ▶). AD2PF-ChiA has shorter β1 and β7 strands than chitinases B and 1. Moreover, AD2PF-ChiA lacks an insertion domain between the β7 and β8 strands. Chitinase B and chitinase 1 contain an additional small α + β insertion domain which provides one side of a deep substrate-binding cleft at the top of the catalytic (β/α)8 domain (Fig. 4 ▶). Therefore, the substrate-binding cleft of AD2PF-ChiA is not as deep as those of chitinases B and 1. Another notable deletion in AD2PF-ChiA is the ‘porch loop’ between β3 and β4. The porch loop is a barrier that is formed by the short helix and loop in chitinase B (van Aalten et al., 2001 ▶). In chitinase B, this loop prevents binding of substrate (GlcNAc) extending longer than three units from the scissile glycosidic bond (Fig. 4 ▶). The absence of the porch loop in AD2PF-ChiA indicates that the substrate-binding cleft is open on both sides of the active site (Fig. 5 ▶). Thus, the active-site cleft of AD2PF-ChiA may accommodate substrates longer than those of chitinase B (Fig. 4 ▶).
Figure 2.
Overall structure of AD2PF-ChiA. The α-helices and β-strands are shown in red and blue, respectively.
Figure 3.
Multiple alignments of the protein sequences were performed based on the three-dimensional structure. Red and blue characters indicate α-helices and β-strands, respectively. The D2 XD3 XE motif is indicated in yellow.
Figure 4.
Stereoview of the overlaid structure of AD2PF-ChiA (grey) and the complex of chitinase B with (GlcNAc)6 (magenta). The porch loop and the α/β domain of chitinase B are shown in blue and green, respectively, and GlcNAc is shown in ball-and-stick representation. The β-strands in TIM-barrel fold were used for the superposition (the r.m.s. deviation is 1.26 Å between backbone atoms for 46 residues).
Figure 5.
Surface representation of AD2PF-ChiA. The active-site cleft is indicated by an arrow. The α-helices and the β-strands are shown in red and blue, respectively. N and C indicate the N- and C-termini, respectively.
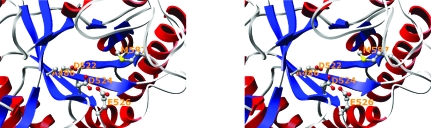
It has been suggested that the conserved amino-acid sequence (D2 XD3 XE motif) that spans the β4 strand plays an important role in the catalytic mechanism of family 18 chitinases (Fig. 6 ▶; Watanabe et al., 1993 ▶, 1994 ▶). The catalytic Glu526 is located at the end of the β4 strand and the two conserved aspartic acid residues [Asp522 (D2) and Asp524 (D3)] are linearly aligned in the active-site core (Fig. 6 ▶). Structural studies of chitinase B–substrate complexes have shown that the functionality of Asp142 (D3) during catalysis depends on the presence of Ser93 and that Tyr214 interacts with the distorted N-acetyl group of GlcNAc (van Aalten et al., 2001 ▶). However, these residues, Ser93 and Tyr214 in chitinase B, are not conserved in AD2PF-ChiA, the corresponding residues being Ala486 and Met587. These observations indicate that the chitin-recognition mechanism of AD2PF-ChiA must be different from that of chitinase B. PF-ChiA possesses a unique feature in that it catalyzes the hydrolysis of both colloidal and crystalline (β/α) chitins. This may arise from the catalytic mechanism of PF-ChiA, which is uncommon in family 18 chitinases. In addition, comparison of the AD2PF-ChiA structure with that of chitinase B suggests that AD2PF-ChiA may be an endo-type chitinase. AD2PF-ChiA has a groove-like cleft that is typical of endo-enzymes, whereas chitinase B is an exo-type chitinase with a tunnel-like cleft. To clarify this issue, we are now attempting to solve the structure of AD2PF-ChiA complexed with its substrate.
Figure 6.
Close-up view of the active site containing the conserved residues in family 18 chitinases. The side chains of Asp522, Asp524 and Glu526, which are important for catalysis, are shown. In addition, Ala486 and Met587, which correspond to Ser and Tyr in other chitinases, are also shown.
Supplementary Material
PDB reference: archaeal chitinase, 2dsk, r2dsksf
Figure 7.
The conformation around Tyr590. The electron density shows a σA-weighted F o − F c map at the 3.0σ level when three amino-acid residues (Tyr589, Tyr590 and Trp591) were removed from the structure-factor calculation.
Acknowledgments
We thank Dr H. Matsumura (Osaka University) for helpful advice on X-ray diffraction analysis. We also thank Ms C. Yoshikawa-Kageyama for performing part of the protein-purification and molecular-biological work. This work was supported by the National Project on Protein Structural and Functional Analyses. The X-ray diffraction experiments were carried out with the approval of the Japan Synchrotron Radiation Research Institute (proposal No. 2005AC05A44XU-7201-N).
References
- Aalten, D. M. van, Komander, D., Synstad, B., Gaseidnes, S., Peter, M. G. & Eijsink, V. G. (2001). Proc. Natl Acad. Sci. USA, 98, 8979–8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aalten, D. M. van, Synstad, B., Brurberg, M. B., Hough, E., Riize, B. W., Eijsink, V. G. & Wierenga, R. K. (2000). Proc. Natl Acad. Sci. USA, 97, 5842–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L, Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Davies, G. & Henrissat, B. (1995). Structure, 3, 853–859. [DOI] [PubMed] [Google Scholar]
- Doublié, S. (1997). Methods Enzymol.276, 523–530. [PubMed] [Google Scholar]
- Henrissat, B. (1991). Biochem. J.280, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat, B. & Davies, G. (1997). Curr. Opin. Struct. Biol.7, 637–644. [DOI] [PubMed] [Google Scholar]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., Moss, D. S. & Thornton, J. M. (1993). J. Mol. Biol.231, 1049–1067. [DOI] [PubMed] [Google Scholar]
- Matsumoto, T., Nonaka, T., Hashimoto, M., Watanabe, T. & Mitsui, Y. (1999). Proc. Jpn Acad.75, 269–274.
- Mine, S., Nakamura, T., Hirata, K., Ishikawa, K., Hagihara, Y. & Uegaki, K. (2006). Acta Cryst. F62, 791–793. [DOI] [PMC free article] [PubMed]
- Nakamura, T., Ishikawa, K., Hagihara, Y., Oku, T., Nakagawa, A., Inoue, T., Ataka, M. & Uegaki, K. (2005). Acta Cryst. F61, 476–478. [DOI] [PMC free article] [PubMed]
- Oku, T. & Ishikawa, K. (2006). Biosci. Biotechnol. Biochem.70, 1969–1701. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Perrakis, A., Tews, I., Dauter, Z., Oppenheim, A. B., Chet, I., Wilson, K. S. & Vorgias, C. E. (1994). Structure, 2, 1169–1180. [DOI] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem.25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Rouvinen, J., Bergfors, T., Teeri, T., Knowles, J. K. & Jones, T. A. (1990). Science, 249, 380–386. [DOI] [PubMed] [Google Scholar]
- Spezio, M., Wilson, D. B. & Karplus, P. A. (1993). Biochemistry, 32, 9906–9916. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., Taiyoji, M., Sugawara, N., Nikaidou, N., Henrissat, B. & Watanabe, T. (1999). Biochem. J.343, 587–596. [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwisscha van Scheltinga, A. C., Hennig, M. & Dijkstra, B. W. (1996). J. Mol. Biol.262, 243–257. [DOI] [PubMed] [Google Scholar]
- Watanabe, T., Kobori, K., Miyashita, K., Fujii, T., Sakai, H., Uchida, M. & Tanaka, H. (1993). J. Biol. Chem.268, 18567–18572. [PubMed] [Google Scholar]
- Watanabe, T., Uchida, M., Kobori, K. & Tanaka, H. (1994). Biosci. Biotechnol. Biochem.58, 2283–2285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: archaeal chitinase, 2dsk, r2dsksf