Crystals of MotY, a stator protein of the V. alginolyticus polar flagellar motor, have been produced and characterized by X-ray diffraction.
Keywords: MotY, sodium-driven flagellar motor, stator proteins, peptidoglycans, T rings
Abstract
The polar flagellum of Vibrio alginolyticus is rotated by the sodium motor. The stator unit of the sodium motor consists of four different proteins: PomA, PomB, MotX and MotY. MotX and MotY, which are unique components of the sodium motor, form the T-ring structure attached to the LP ring in the periplasmic space. MotY has a putative peptidoglycan-binding motif in its C-terminal region and MotX is suggested to interact with PomB. Thus, MotX and MotY are thought to be required for incorporation and stabilization of the PomA/B complex. In this study, mature MotY composed of 272 amino-acid residues and its SeMet derivative were expressed with a C-terminal hexahistidine-tag sequence, purified and crystallized. Native crystals were grown in the hexagonal space group P6122/P6522, with unit-cell parameters a = b = 104.1, c = 132.6 Å. SeMet-derivative crystals belonged to the same space group with the same unit-cell parameters as the native crystals. Anomalous difference Patterson maps of the SeMet derivative showed significant peaks in their Harker sections, indicating that the derivatives are suitable for structure determination.
1. Introduction
Many bacteria swim by rotating a locomotive organelle called the flagellum. Rotation of the flagellum is driven by the motor, which is embedded in the bacterial membranes at the base of the flagellar filament. The bacterial flagellar motor is a molecular machine that couples the influx of specific ions to mechanical rotation of the flagellar filament. While the flagella of Escherichia coli and Salmonella typhimurium are driven by proton flow, the polar flagella of marine bacteria, such as Vibrio alginolyticus, V. parahaemolyticus and V. cholera, utilize sodium ions for rotation (Yorimitsu & Homma, 2001 ▶).
The motor is composed of two distinct complex structures, the rotor and the stator, and the torque is generated between these two structures. The stator is a selective membrane ion-channel complex composed of several proteins. The stator unit of the sodium motor consists of four different proteins: PomA, PomB, MotX and MotY. PomA and PomB are cytoplasmic membrane proteins and orthologues of MotA and MotB (Asai et al., 1997 ▶), which form a proton-channel complex in the proton motor. Sato and Homma demonstrated that the purified PomA–PomB complex, which is composed of four PomA and two PomB molecules (Sato & Homma, 2000b ▶; Yorimitsu et al., 2004 ▶), has a sodium-conducting activity (Sato & Homma, 2000a ▶). Thus, the PomA–PomB complex was identified as the sodium ion channel of the sodium motor.
MotX and MotY are unique components of the sodium motor of Vibrio and are believed to form a complex associated with the outer membrane from the periplasmic space. MotX and MotY are synthesized with a signal sequence and exported to the periplasmic space through the Sec system (Okabe et al., 2002 ▶). Mature MotY consists of 272 amino-acid residues with a molecular weight of 32 000 Da. In the C-terminal region, MotY contains a putative peptidoglycan-binding motif, which is also found in MotB, PomB, outer membrane protein A (OmpA), peptidoglycan-associated lipoprotein (Pal) and RmpM (Grizot & Buchanan, 2004 ▶; Yakushi et al., 2005 ▶).
The null mutants of MotX and MotY showed nonmotile phenotypes (Okunishi et al., 1996 ▶; Okabe et al., 2001 ▶; Yagasaki et al., 2006 ▶); thus both proteins are indispensable for torque generation, but their actual roles are unclear. MotX has been found to affect the membrane localization of PomB, suggesting that MotX interacts with PomB (Okabe et al., 2005 ▶). Recently, a new ring structure called the T ring has been found on the periplasmic side of the P ring of the hook-basal body purified from V. alginolyticus and the components of the T ring have been identified as MotX and MotY (Terashima et al., 2006 ▶). In addition, PomA and PomB do not localize to the cell pole in null mutants of MotX and MotY, suggesting that MotX and MotY are involved in the incorporation of the PomA–PomB complex into the motor by forming the T-ring complex.
To elucidate the rotation and construction mechanism of the sodium motor, we crystallized MotY in order to determine its structure. Here, we report the expression, purification, crystallization and preliminary X-ray analysis of mature MotY with a C-terminal hexahistidine tag and its SeMet derivative.
2. Materials and methods
2.1. Protein expression and purification
The gene encoding MotY tagged with a C-terminal hexahistidine was transferred from pKJ503 (Okabe et al., 2002 ▶) into pJY19 (Yorimitsu et al., 2003 ▶), a derivative of plasmid pTTQ18 (Ampr; GE Healthcare Biosciences), using SalI–XbaI restriction sites and transformed into E. coli strain BL21 (Novagen). Cells were grown at 303 K in LB medium [1% polypeptone (Wako), 0.5% yeast extract, 0.5% NaCl] containing 50 mg ampicillin per litre until the culture density reached an OD660 of 1.2. Expression of MotY was induced with isopropyl β-d-thiogalactoside (IPTG) at a final concentration of 0.5 mM and culture was continued for 2 h. Cells were harvested by centrifugation at 10 000g for 10 min and the pellet was suspended in buffer (20 mM Tris–HCl pH 7.5, 100 mM NaCl, 25% sucrose, 0.5 mM PMSF). The cells were converted to spheroplasts by adding a solution containing lysozyme and DNaseI (bovine, Sigma), followed by gradual addition of a twofold volume of EDTA solution (1.5 mM EDTA, 100 mM NaCl pH 8.0; Osborn et al., 1972 ▶; Osborn & Munson, 1974 ▶). The spheroplast suspension was incubated for 20 min on ice and centrifuged at 10 000g for 10 min in order to separate the periplasmic fraction from the spheroplast cells. NaCl was then added to the supernatant to a final concentration of 500 mM and centrifuged at 100 000g for 60 min. The supernatant containing the mature MotY protein was loaded onto an Ni–NTA Superflow column (Qiagen). After washing with five column volumes of buffer (10 mM Tris–HCl pH 7.5, 500 mM NaCl, 0.5 mM PMSF, 10 mM imidazole), proteins were eluted with a stepwise gradient of imidazole from 40 to 200 mM. The eluate was loaded onto a Superdex 75 (10/300) gel-filtration column (GE Healthcare Biosciences) equilibrated and eluted with 10 mM Tris–HCl pH 7.5, 500 mM NaCl. The fractions containing MotY were concentrated to 10 mg ml−1 by ultrafiltration using AmiconUltra (Millpore). The purity of MotY was assayed by SDS–PAGE and MALDI–TOF mass spectrometry (Voyager DE/STR, Applied Biosystems).
2.2. Crystallization
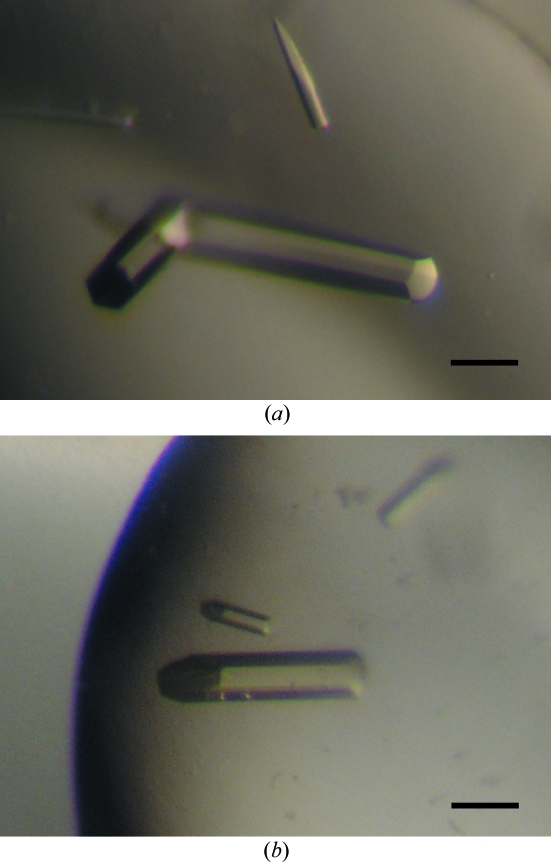
Initial crystallization screening was performed by the sitting-drop vapour-diffusion technique using screening-solution kits (Crystal Screens I and II, Hampton Research; Wizard I and II, Emerald BioSystems) at 293 K. Within a week, small hexagonal column-shaped crystals and needle-shaped crystals appeared using several conditions containing PEG or NaH2PO4/K2HPO4 as precipitant. We optimized the conditions by varying the precipitant concentration, pH, salts and additives using the sitting-drop method. The best crystals were obtained from drops prepared by mixing 1 µl protein solution with 1 µl reservoir solution containing 1.12 M NaH2PO4/K2HPO4, 5% PEG 1000 and 0.1 M acetate pH 4.5 equilibrated against 500 µl reservoir solution. The optimized hexagonal column-shaped crystals appeared at 293 K within one week and grew to typical dimensions of 0.5 × 0.1 × 0.1 mm (Fig. 1 ▶ a).
Figure 1.
Hexagonal column-shaped crystals of (a) MotY and (b) SeMet-labelled MotY. The scale bar is 0.1 mm in length.
2.3. Preparation and crystallization of SeMet-labelled MotY
Mature MotY with a hexahistidine tag contains five methionine residues in its 278-amino-acid sequence; therefore, we prepared and crystallized SeMet-labelled MotY for phasing. SeMet MotY was expressed in B834(DE3)pLysS, which is a met− autotroph (Leahy et al., 1992 ▶; Wood, 1966 ▶), carrying pJY19-motY-his6 using M9 medium containing 0.2 g MgSO4, 4 g glucose, 40 mg of all l-amino acids except Met, 40 mg seleno-l-methionine, 25 mg pyridoxine hydrochloride, 1.25 mg thiamine hydrochloride and 50 mg ampicillin per litre. Cells were cultured at 303 K until the density of the medium reached an OD660 of 0.4–0.5 and expression of MotY was induced with IPTG at a final concentration of 0.5 mM; culture was continued for 4 h. The labelled protein was then purified using the same protocol as described for nonlabelled MotY. Crystals of SeMet MotY were obtained under the same conditions as the native MotY crystals, but only diffracted to 5 Å resolution. Therefore, we tested the SeMet protein using other conditions under which small crystals of native MotY had grown in the initial screening. Finally, crystals suitable for X-ray experiments were obtained at 293 K using a reservoir solution containing 24% polyethylene glycol monoethyl ether (PEG MME) 2000, 0.36 M ammonium sulfate and 0.1 M Tris–HCl pH 8.5. They were also hexagonal column-shaped crystals and grew to maximum dimensions of 0.3 × 0.06 × 0.06 mm (Fig. 1 ▶ b). The drop size, reservoir volume and protein concentration were the same as for the native crystals.
2.4. Data collection and processing
X-ray diffraction experiments were performed at SPring-8 (Harima, Japan) beamline BL41XU. Both native and SeMet crystals were soaked in a solution containing 90%(v/v) of the respective optimized reservoir solution and 10%(v/v) MPD for a few seconds, immediately transferred into liquid nitrogen for freezing and mounted in a cryo-gas flow. The diffraction data were recorded on an ADSC Quantum 315 CCD detector (Area Detector Systems Corporation) under a 100 K nitrogen-gas flow for the native crystals and a 35 K helium-gas flow (Rigaku cryocooling device) for the SeMet crystals. The diffraction data were indexed, integrated and scaled using the programs MOSFLM (Leslie, 1992 ▶) and SCALA from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). The statistics of data collection are summarized in Table 1 ▶.
Table 1. Summary of the data statistics.
Values in parentheses are for the highest resolution shell.
| SeMet derivative | |||||
|---|---|---|---|---|---|
| Native | Peak | Inflection | Remote 1 | Remote 2 | |
| Space group | P6122/P6522 | ||||
| Unit-cell parameters | a = b = 104.1, c = 132.6 | a = b = 104.7, c = 132.3 | |||
| Wavelength (Å) | 1.1765 | 0.97913 | 0.97940 | 0.986 | 0.964 |
| Resolution range (Å) | 53.53–2.85 (3.00–2.85) | 66.08–2.9 (3.06–2.9) | 66.08–3.0 (3.16–3.0) | 66.08–3.0 (3.16–3.0) | 66.08–3.0 (3.16–3.0) |
| Observations | 40463 (5999) | 65286 (9668) | 59007 (8795) | 59073 (8787) | 59348 (8841) |
| Unique reflections | 10448 (1490) | 10022 (1410) | 9095 (1290) | 9087 (1291) | 9086 (1292) |
| Completeness (%) | 99.6 (100) | 99.9 (100) | 99.9 (100) | 99.9 (100) | 99.9 (100) |
| Anomalous completeness (%) | — | 99.5 (99.8) | 99.5 (99.5) | 99.6 (99.5) | 99.6 (99.6) |
| Redundancy | 6.5 (6.9) | 6.5 (6.9) | 6.5 (6.8) | 6.5 (6.8) | 6.5 (6.8) |
| Anomalous redundancy | — | 3.6 (3.6) | 3.6 (3.6) | 3.6 (3.6) | 3.6 (3.6) |
| I/σ(I) | 11.4 (2.7) | 16.3 (5.0) | 17.0 (5.3) | 15.4 (4.7) | 14.6 (3.8) |
| Rmerge (%) | 8.7 (38.4) | 8.0 (35.1) | 8.4 (32.4) | 8.5 (33.1) | 9.1 (44.4) |
| Ranom (%) | — | 4.9 (15.4) | 4.0 (14.1) | 3.8 (14.2) | 4.6 (19.5) |
3. Results and discussion
MotY tends to form insoluble aggregates under low ionic strength conditions (Okabe et al., 2005 ▶); hence, we kept the concentration of NaCl at 500 mM throughout the purification. Typical protein solutions used for crystallization also contained 500 mM NaCl with 10 mM Tris–HCl pH 7.5 and 10 mg ml−1 MotY.
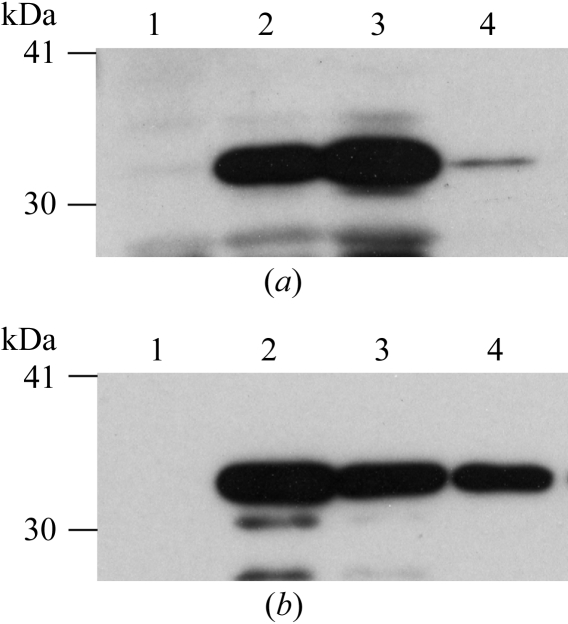
Initially, we were not able to obtain a sufficient amount of SeMet MotY for crystallization. After spheroplast formation, most MotY was precipitated with the cells. Soluble MotY was only recovered from the periplasmic fraction at a very low level (Fig. 2 ▶ a). We analysed the molecular weight of MotY from the spheroplast fraction by MALDI–TOF mass spectrometry and found that it had no signal sequence (the observed molecular weight was 32 kDa and the calculated monoisotopic weights of the His-tagged SeMet MotY with and without signal sequence were 34 400 and 32 100 Da, respectively). This observation means that the expressed protein should have been translocated to the periplasmic space through the Sec system. However, the protein was not in the periplasmic fraction (Fig. 2 ▶ a, lane 4) and remained in the spheroplast fraction (Fig. 2 ▶ a, lane 3). We then checked the spheroplast formation with a microscope and found that the cell spheroplasts were not well produced. While BL21 cells were used for expression of native MotY, B834(DE3)pLysS cells were used for expression of SeMet MotY. We thought that the T7 lysozyme expressed by the pLysS plasmid in the cells might have disturbed the formation of spheroplasts, as it is known that control of lysozyme concentration is one of the key factors in spheroplast formation. The T7 lysozyme is encoded on the pLysS plasmid, which carries the chloramphenicol-resistance gene. Therefore, we cultured the cells without chloramphenicol in order to drop the pLysS plasmid and converted them into spheroplasts. In this way, we recovered soluble SeMet MotY from the periplasmic fraction and obtained a sufficient amount of the protein for crystallization (Fig. 2 ▶ b).
Figure 2.
SDS–PAGE analysis of SeMet-labelled MotY expressed (a) with and (b) without chloramphenicol. MotY was detected by immunoblotting using an anti-MotY antibody. Lane 1, before induction; lane 2, after induction; lane 3, spheroplast fraction; lane 4, periplasmic fraction.
MotY crystals diffracted to 2.9 Å resolution. The native crystals belong to the hexagonal space group P6122/P6522, with unit-cell parameters a = b = 104.1, c = 132.6 Å. The Matthews coefficient (V M; Matthews, 1968 ▶) indicates the presence of a single MotY molecule in the asymmetric unit, with a solvent content of 62%. The self-rotation function map calculated using the program POLARRFN (Collaborative Computational Project, Number 4, 1994 ▶) showed no significant peak corresponding to local symmetry.
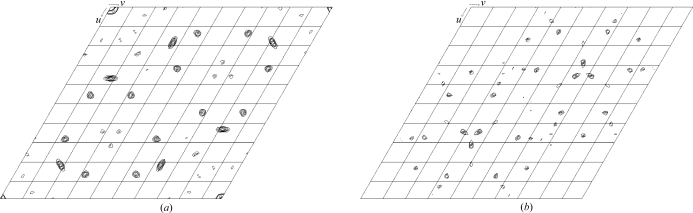
Although the crystallization conditions for the SeMet derivative are quite different from those for native MotY, the unit-cell parameters of the derivative crystals, a = b = 104.7, c = 132.3 Å, were almost the same as those of the native crystals (Table 1 ▶). The derivative crystals diffracted to 3.0 Å resolution, but were sensitive to X-ray irradiation. Therefore, we used a helium cryocooling device in order to reduce the radiation damage. Statistics of the diffraction data at four different wavelengths are summarized in Table 1 ▶. Bijvoet and dispersive difference Patterson maps of the SeMet derivative showed common clear peaks on the Harker sections (Fig. 3 ▶), suggesting the usefulness of these data for phasing by the MAD method.
Figure 3.
(a) Bijovoet and (b) dispersive difference Patterson maps (w = 0.5 Harker section) calculated using the data from the SeMet derivative at 3.5 Å resolution. The contour lines are drawn from 2.5σ to 6.5σ, with an increment of 0.5σ.
Acknowledgments
We acknowledge H. Matsunami, M. Shimada, R. Iwase and T. Tajima for assistance in data collection and N. Shimizu, M. Kawamoto and K. Hasegawa at SPring-8 for technical help in the use of beamlines BL38B1 and BL41XU. This work was supported in part by Grants-in-Aid for Scientific Research to KI (16310088 and 18074006) and KN (16087207) and the ‘National Project on Protein Structural and Functional Analyses’ (to KI and MH) from the Ministry of Education, Science and Culture of Japan and the Soft Nano-Machine Project of Japan Science and Technology Agency (to TY and MH).
References
- Asai, Y., Kojima, S., Kato, H., Nishioka, N., Kawagishi, I. & Homma, M. (1997). J. Bacteriol.179, 5104–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Grizot, S. & Buchanan, S. K. (2004). Mol. Microbiol.51, 1027–1037. [DOI] [PubMed] [Google Scholar]
- Leahy, D. J., Hendrickson, W. A., Aukhil, I. & Erickson, H. P. (1992). Science, 258, 987–991. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Okabe, M., Yakushi, T., Asai, Y. & Homma, M. (2001). J. Biochem. (Tokyo), 130, 879–884. [DOI] [PubMed] [Google Scholar]
- Okabe, M., Yakushi, T. & Homma, M. (2005). J. Biol. Chem.280, 25659–25664. [DOI] [PubMed] [Google Scholar]
- Okabe, M., Yakushi, T., Kojima, M. & Homma, M. (2002). Mol. Microbiol.46, 125–134. [DOI] [PubMed] [Google Scholar]
- Okunishi, I., Kawagishi, I. & Homma, M. (1996). J. Bacteriol.178, 2409–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn, M. J. & Munson, R. (1974). Methods Enzymol.31, 642–653. [DOI] [PubMed] [Google Scholar]
- Osborn, M. J., Gander, J. E., Parisi, E. & Carson, J. (1972). J. Biol. Chem.247, 3962–3972. [PubMed] [Google Scholar]
- Sato, K. & Homma, M. (2000a). J. Biol. Chem.275, 5718–5722. [DOI] [PubMed] [Google Scholar]
- Sato, K. & Homma, M. (2000b). J. Biol. Chem.275, 20223–20228. [DOI] [PubMed] [Google Scholar]
- Terashima, H., Fukuoka, H., Yakushi, T. & Homma, M. (2006). Mol. Microbiol.62, 1170–1180. [DOI] [PubMed] [Google Scholar]
- Wood, W. (1966). J. Mol. Biol.16, 118–133. [DOI] [PubMed] [Google Scholar]
- Yagasaki, J., Okabe, M., Kurebayashi, R., Yakushi, T. & Homma, M. (2006). J. Bacteriol.188, 5308–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushi, T., Hattori, N. & Homma, M. (2005). J. Bacteriol.187, 778–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu, T. & Homma, M. (2001). Biochim. Biophys. Acta, 1505, 82–93. [DOI] [PubMed] [Google Scholar]
- Yorimitsu, T., Kojima, M., Yakukshi, T. & Homma, M. (2004). J. Biochem. (Tokyo), 130, 879–884.
- Yorimitsu, T., Mimaki, A., Yakushi, T. & Homma, M. (2003). J. Mol. Biol.344, 567–583. [DOI] [PubMed]