Crystals of the RNA-dependent RNA polymerase catalytic domain from the dengue virus NS5 protein have been obtained using a strategy that included expression screening of naturally occurring serotype variants of the protein, the addition of divalent metal ions and crystal dehydration. These crystals diffract to 1.85 Å resolution and are thus suitable for a structure-based drug-design program.
Keywords: dengue, flaviviruses, RNA-dependent RNA polymerases
Abstract
Dengue virus, a member of the Flaviviridae genus, causes dengue fever, an important emerging disease with several million infections occurring annually for which no effective therapy exists. The viral RNA-dependent RNA polymerase NS5 plays an important role in virus replication and represents an interesting target for the development of specific antiviral compounds. Crystals that diffract to 1.85 Å resolution that are suitable for three-dimensional structure determination and thus for a structure-based drug-design program have been obtained using a strategy that included expression screening of naturally occurring serotype variants of the protein, the addition of divalent metal ions and crystal dehydration.
1. Introduction
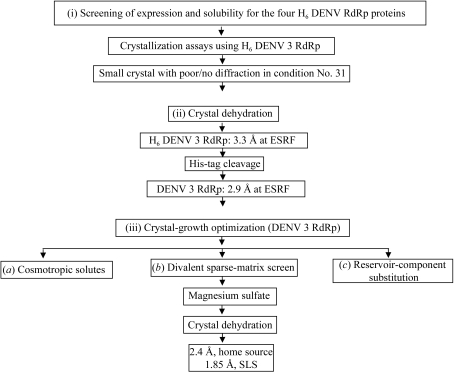
Several positive-strand RNA viruses from the Flaviviridae genus are important human pathogens, including dengue virus (DENV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), tick-borne encephalitis virus (TBEV) and West Nile virus (WNV). DENV affects ∼50–100 million individuals worldwide per year, resulting in approximately 500 000 cases of more severe pathological forms such as dengue haemorrhagic fever and dengue shock syndrome and ∼20 000 deaths, mainly in children (Henchal & Putnak, 1990 ▶). Based on serological studies, dengue viruses are subclassified into four distinct serotypes: DENV 1–4 (Blok, 1985 ▶). The NS5 protein has a molecular weight of 104 kDa and is the most conserved viral protein, with a minimum of 67% amino-acid sequence identity across the four DENV serotypes. The NS5 protein possesses three functional domains: an N-terminal S-adenosylmethionine methyltransferase (SAM) domain (Koonin & Ilyina, 1993 ▶), a nuclear-localization sequence (NLS) spanning residues 320–405 (Forwood et al., 1999 ▶) and a C-terminal RNA-dependent RNA polymerase domain (RdRp) responsible for synthesizing a transient double-stranded replication RNA intermediate (Bartholomeusz & Thompson, 1999 ▶). Viral polymerases represent attractive drug targets for the development of specific drugs, as host cells are devoid of this enzymatic activity (Wu et al., 2005 ▶). With a view to accelerating the search for antiviral compounds that are active against DENV, we expressed and purified catalytically active DENV RdRp enzymatic domains in order to determine their three-dimensional structure using X-ray crystallography. Here, we report the multi-step strategy used which led to a high-resolution structure of the DENV 3 polymerase catalytic domain suitable for further structure-based drug design. The strategy, outlined in Fig. 1 ▶, involved (i) expression screening of naturally occurring serotype variants of the RdRp domain, (ii) a crystal dehydration/cryoprotection protocol and (iii) optimization of crystal-growth conditions.
Figure 1.
Flowchart depicting the strategy followed to obtain crystals of the DENV RdRp catalytic domain that diffract to high resolution. The approach consisted of the following steps. (i) The four serotypes of the target protein (containing a hexahistidine tag) were screened for expression levels and solubility, with the RdRp from serotype 3 giving the best results (see Table 1 ▶). (ii) A major improvement of the diffraction quality for small crystals (obtained using condition No. 31) was obtained by air dehydration. Removal of the N-terminal His6 tag resulted in better diffraction. (iii) Crystals of DENV 3 RdRp (with no His6 tag) were further optimized using magnesium sulfate or magnesium acetate as additives.
2. Materials and methods
2.1. Protein expression and purification
DNA fragments spanning residues 273–900 encoding the catalytic domains of the DENV RdRp from the four serotypes (DENV 1–4) were amplified by PCR and cloned into pET-15b (Novagen, Madison, USA) using the forward and reverse primers listed in Table 1 ▶. Transformed Escherichia coli BL21 (DE3) cells (RIL) (Stratagene) were grown at 310 K in LB medium containing 100 µg ml−1 ampicillin and 50 µg ml−1 chloramphenicol until an OD600 of 0.4–0.6 was attained. Protein expression was induced at 289 K by adding isopropyl β-d-thiogalactopyranoside to a final concentration of 0.4 mM. After overnight growth, cells were harvested by centrifugation at 8000g for 10 min at 277 K. Pellets were resuspended in buffer A (20 mM Tris–HCl pH 7.5, 0.5 M NaCl, 10 mM β-mercaptoethanol, 10% glycerol) supplemented with an EDTA-free protease-inhibitor tablet (Roche, Switzerland). This was followed by sonication and centrifugation at 30 000g for 30 min at 277 K. The lysate supernatants were first purified by metal affinity using a HisTrap HP column (GE Healthcare, Sweden) equilibrated with buffer A. Unbound proteins were washed sequentially with five column volumes of buffer A supplemented with imidazole at concentrations of 25 and 125 mM. Proteins were eluted using a linear gradient of imidazole from 125 to 500 mM. Fractions containing the protein were pooled and dialyzed overnight against 50 mM MES pH 6.5, 0.15 M NaCl, 1 mM EDTA, 5 mM β-mercaptoethanol. After dialysis, DENV 3 RdRp protein retaining the N-terminal His6 tag (hereafter named H6 DENV 3 RdRp) was subjected to cation-exchange chromatography using a Source 15S column (GE Healthcare, Sweden) with a volume of 20 ml. Tagged proteins were eluted using a linear gradient from 0.15 to 1.5 M NaCl in buffer B (50 mM MES pH 6.2, 0.05 M NaCl, 5 mM β-mercaptoethanol, 1 mM EDTA). In the case of H6 DENV 3 RdRp, two distinct fractions (labelled F1 and F2) were eluted from cation-exchange chromatography (Fig. 2 ▶) and were separately purified by gel-filtration chromatography (see below). Alternatively, after elution from the HisTrap HP column the H6 DENV 3 RdRp protein was treated with thrombin overnight at 277 K in order to remove its N-terminal hexahistidine (His6) tag using 1 U thrombin per 100 µg of protein. After treatment with thrombin, the DENV 3 RdRp protein retains residues GSHMLDN at its N-terminus which are derived from the vector used for cloning. Finally, both the tagged and untagged proteins were concentrated by ultrafiltration with a molecular-weight cutoff of 30 kDa (Millipore, Volketswil, Switzerland), a final gel-filtration chromatography step (HiPrep 16/26, Superdex 200) was carried out in buffer C [20 mM Tris–HCl pH 6.8, 0.25 M NaCl, 1 mM EDTA, 2 mM β-mercaptoethanol and 0.1%(w/v) CHAPS] and both the tagged and untagged proteins were concentrated to ∼11 mg ml−1 and used in crystallization assays.
Table 1. Cloning, expression, solubility and protein yield for RdRp from DENV serotypes 1–4.
The four serotypes of DENV RdRp have ∼77% protein similarity. Genebank accession codes for the RdRps from DENV serotypes 1–4 used in this study are AAN06981, AAK67712, AY662691 and AY648301, respectively.
| DENV RdRp serotype | Expression | Solubility (%) | Crystallization | Protein yield (µg per litre of culture) | Primers (forward/reverse) |
|---|---|---|---|---|---|
| DENV 1 | + | 15–25 | Not tested | 500 | 5′-CGAAGGCTCGAGAACCTAGATATCATTGGCCAG-3′/5′-GGACGTAGATCTCTACCAGAGTGCCCCTTCGAGATC-3′ |
| DENV 2 | + | 10–20 | Not tested | 300 | 5′-CGAAGGCTCGAGAATTTAGACATAATTGGGAAAAGA-3′/5′-CGCGGATCCCTACCACAAGACTCCTGCCTCTTCC-3′ |
| DENV 3 | + | 60–80 | Yes | 3000–5000 | 5′-CCACGCGTCGACAACATGGATGTCATTGGGGAAAG-3′/5′-CCGGAATTCCTACCAAATGGCTCCCTCCGACTCCTC-3′ |
| DENV 4 | + | 5–15 | Not tested | 100–300 | 5′-CGAAGGCTCGAGGACATGACAATCATTGGGAGAAG-3′/5′-CCGGAATTCCTACAGAACTCCTTCACTCTCGAAAG-3′ |
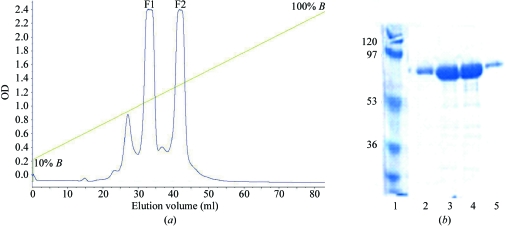
Figure 2.
(a) Elution profile from the cation-exchange purification step for the H6 DENV 3 RdRp protein. The linear gradient ranges from 0.15 M NaCl (corresponding to the inclusion of 10% buffer B) to 1.5 M NaCl (100% buffer B). Two major fractions F1 and F2 were then subjected to gel filtration and initial crystallization trials (see text). (b) SDS–PAGE analysis. Lane 1, molecular-weight markers (kDa). Lanes 2 and 5, protein DENV 3 RdRp from the minor fraction that elutes first. Lanes 3 and 4, fractions F1 and F2, respectively, both of which contain DENV 3 RdRp.
2.2. Crystallization and data collection
Approximately 300 different crystallization conditions were screened for the H6 DENV 3 RdRp protein at 293 and 277 K in 24-well Limbro plates using the hanging-drop vapour-diffusion method with the following four commercially available sparse-matrix screens: Index, SaltRx, Crystal Screen Lite (from Hampton Research, CA, USA) and the PEG suite (Qiagen/Nextal). Equal volumes (0.75 µl) of protein solution at a concentration of 11 mg ml−1 and precipitating solution were mixed over 500 µl reservoir solution. Using condition No. 31 of the Index Screen from Hampton Research [0.1 M Tris–HCl pH 8.5, 0.8 M potassium/sodium tartrate, 0.5%(w/v) polyethylene glycol monomethyl ether 5000], lens-shaped crystals were obtained at 277 K after three to four weeks, but diffracted very poorly. This crystallization condition is referred to in the following as condition No. 31. A multi-step strategy was subsequently employed in order to improve crystal diffraction. Firstly, crystal dehydration was carried out based on previously described protocols (Heras & Martin, 2005 ▶). Briefly, crystals were serially transferred to drops containing a 5 µl volume of the precipitating solution supplemented with increasing concentrations of various dehydration agents at 277 K (Table 2 ▶). Crystals were then mounted in a cryoloop and rapidly cooled to 100 K in liquid nitrogen. Other strategies adopted included the use of ‘cosmotropic solutions’ to stabilize the protein (Jeruzalmi & Steitz, 1997 ▶), screening of the effects of divalent ions using the sparse-matrix cofactors screen (Boddupalli et al., 1992 ▶; Doudna et al., 1993 ▶) and substitutions of individual components within the reservoir solution. Diffraction intensities were recorded using 1° oscillations on CCD detectors (ADSC and MAR225) on beamline ID14-4 at the ESRF (Grenoble, France) or X10SA at the Swiss Light Source (Villigen, Switzerland) using attenuated beams. Evaluation of crystal quality was performed with an R-AXIS IV++ imaging-plate detector mounted on a Rigaku/MSC FR-E X-ray generator. Integration, scaling and merging of the intensities were carried out using the programs MOSFLM (Leslie, 1999 ▶) and SCALA from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶).
Table 2. Summary of dehydration/cryoprotection assays on DENV 3 RdRp crystals.
| Dehydration agent | Concentrations† | Resolution (Å) | Radiation source |
|---|---|---|---|
| No dehydration | — | ∼20 | Home source |
| PEG 200 | 10, 15, 35%(w/v) | ∼7 | Home source |
| PEG 300 | 10, 15, 25%(w/v) | ∼10 | Home source |
| PEG 400 | 10, 15, 25%(w/v) | ∼5 | Home source |
| PEG 600 | 10, 15, 25%(w/v) | ∼10 | Home source |
| Potassium/sodium tartrate‡ | 0.84, 0.9, 1.0 M | ∼15 | Home source |
| MPD | (Dissolves crystals) | — | Home source |
| Glycerol | 10, 15, 30%(w/v) | ∼8 | Home source |
| PEG 3350§ | 15, 25, 30%(w/v) | 4.5¶ | Home source |
| 2.9¶ | Synchrotron | ||
| 2.4†† | Home source | ||
| 1.85†† | Synchrotron |
Air dehydration involved the stepwise transfer of crystals into a 5 µl drop for 15, 15 and 30 min, respectively, of the solutions listed. Each drop contains the original precipitating solution condition No. 31 (see §2) with the addition of the dehydration agents listed. Longer durations of dehydration yielded no improvement in crystal diffraction (e.g. stepwise transfers for a total time of 24 h). Very brief soaking times (∼10 s) did not improve the diffraction.
In this case, the cryoprotectant is a solution containing 30%(w/v) glycerol after the crystals had been dehydrated.
Optimal dehydration/cryoprotection used PEG 3350 for DENV 3 RdRp, leading to high-resolution diffraction.
The resolution of the diffraction pattern that was obtained for small crystals of DENV 3 RdRp after dehydration (see Fig. 3 ▶ a).
The resolution of the diffraction pattern that was obtained for large crystals after dehydration (see Fig. 3 ▶ b) and after optimization using the sparse-matrix cofactor screen.
3. Results and discussion
3.1. Protein expression
In order to increase the likelihood of obtaining crystals, we screened protein expression for the NS5 RdRp catalytic domain from the four DENV serotypes (DENV 1–4; Table 1 ▶). These constructs contain the RdRp catalytic domain, which is located at the C-terminal end of the NS5 full-length protein. The best results in terms of yield and protein solubility were obtained for a DENV 3 RdRp truncation construct spanning amino acids 273–900 of the corresponding full-length NS5 protein (Table 1 ▶), indicating that inclusion of the NLS region was essential to obtain a stable protein. This protein could be expressed in high quantity (about 3–5 mg per litre of bacterial culture) and was purified to >95% homogeneity. The study of catalytic domains from other serotypes was discontinued because of low protein-expression levels or solubility problems and only the DENV 3 RdRp was subjected to further crystallization studies. As demonstrated by other workers in the field of structural genomics, an approach that consists of cloning different strains or serotypes from an infectious agent representing naturally occurring variants of a target protein (e.g. from a bacterium or a virus) greatly increases the likelihood of obtaining a protein sample amenable to crystallization compared with a single serotype or variant (Au et al., 2006 ▶).
3.2. Crystallization
The H6 DENV 3 RdRp protein was subjected to initial crystallization assays using the F1 and F2 fractions eluted from the cation-exchange chromatography step separately (Fig. 2 ▶). Examination of the initial crystallization assays led to the observation that trials performed at 293 K gave protein precipitates in almost all conditions investigated. Therefore, subsequent crystallization assays were performed at 277 K. Using the F1 fraction of the H6 DENV 3 RdRp and condition No. 31, small crystals (Fig. 3 ▶ a) with approximate dimensions of up to of 0.05 × 0.02 × 0.02 mm were obtained that diffracted X-rays very weakly. Interestingly, no crystals could be obtained using the same crystallization conditions with the F2 fraction or pools of fractions F1 and F2. Since DENV 3 RdRp protein devoid of the His6 tag eluted as a single peak after ion-exchange chromatography, the fractions F1 and F2 at pH 6.0 might be a consequence of the presence of the N-terminal His6, resulting in the formation of two conformational states and two major elution peaks (Fig. 2 ▶). We suspected that the poor diffraction was caused by extensive dynamic properties of the enzyme introducing disorder in the crystal lattice, as observed previously for crystals of HIV-1 reverse transcriptase (HIV-1 RT; Esnouf et al., 1998 ▶). We therefore established a careful air-dehydration procedure in the hope that the reduction of solvent content thus produced might provide a more closely packed and better ordered lattice. In contrast to HIV-1 RT, we were not able to detect changes triggered by crystal dehydration in the unit-cell parameters of H6 DENV 3 RdRp crystals, since the initial diffraction was too poor to determine the unit-cell parameters. Various PEG solutions with average molecular weights ranging from 200 to 3350 were tested as potential dehydration/cryoprotection agents, as listed in Table 2 ▶. From this preliminary screen, PEG 200, PEG 400 and PEG 3350 were the most promising as they did not produce any visible damage to the crystals. A summary of the conditions tested and the resulting improvements in diffraction obtained is given in Table 2 ▶. The optimal dehydration/cryoprotection protocol involved the serial transfer of crystals at 277 K in 5 µl volumes of the precipitating solution supplemented with increasing concentrations of PEG 3350 [15, 25 and 30%(w/v)] with incubation times of 15, 15 and 30 min, respectively. In order to achieve a final concentration of 30%(w/v) PEG 3350 for the final dehydration step, the concentration of sodium/potassium tartrate was lowered from 0.8 to 0.3 M in order to avoid solubility problems. Crystals of H6 DENV 3 RdRp subjected to this procedure diffracted to ∼4 Å on a home source and to 3.3 Å resolution at the ESRF and belong to space group C2221, with average unit-cell parameters a = 160.28, b = 178.77, c = 58.05 Å and only slight variations depending on the dehydration agent. Crystals of the DENV 3 RdRp protein (with the His6 tag removed by thrombin cleavage) could also be obtained using condition No. 31. These crystals, which were subjected to the dehydration/cryoprotection procedure outlined above, diffracted better (to 2.9 Å at the ESRF ID14-4 beamline) than those originally obtained with the H6 DENV 3 RdRp protein (to 3.3 Å at ESRF; Table 2 ▶ and Fig. 1 ▶). Thus, further crystal optimizations made use of the DENV 3 RdRp protein.
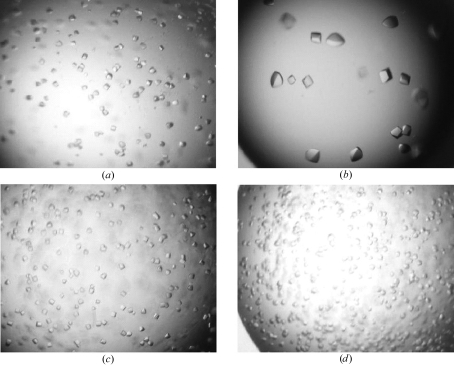
Figure 3.
Typical crystals obtained during crystal-optimization assays. The same magnification of ×20 is used throughout. (a) Outcome I. Small crystals from the original precipitating solution with linear dimensions of <0.05 mm appearing in ∼3–4 weeks. (b) Outcome II. Crystals obtained with the addition of divalent ions. Significantly larger crystals appear in 3–4 weeks (see text). (c) Outcome III. Substitution with PEG 1500 and PEG 2000 in the reservoir leads to more rapid growth of crystals without increasing their final size, which is attained in ∼5–6 days. Substitution with PEG 200 and PEG 400 produced crystals similar to those shown in (b). (d) Outcome IV. Increased rate of nucleation of crystals (approximately one week), leading to slightly smaller crystals (outcome I).
3.3. Further crystal optimization
In order to improve the diffraction properties further, several strategies were iteratively investigated, including the use of ‘cosmotropic’ solutes, the addition of cofactors and the substitution of individual components within the reservoir solution (Fig. 3 ▶ and Table 3 ▶). All crystal-optimization assays were conducted using condition No. 31 as a starting basis with the agents listed in Table 3 ▶. The effects of each factor are summarized below and typical crystals obtained using the various procedures are shown in Fig. 3 ▶.
Table 3. Summary of further crystal-optimization assays for DENV 3 RdRp crystals.
See text for a discussion of crystal-growth optimization. The crystals obtained are shown in Fig. 3 ▶.
| Crystal optimization† | Observation |
|---|---|
| Condition No. 31 (original precipitating solution) | Outcome I |
| Cosmotropic screen (Jeruzalmi & Steitz, 1997 ▶) | |
| Ethylene glycol | |
| 5% | Precipitation |
| 10% | Outcome I |
| 15% | Deters crystal growth |
| Sucrose | |
| 5, 10, 15% | Outcome IV |
| Glycerol | |
| 5, 10% | Outcome IV |
| 15% | Deters crystal growth |
| 2-Propanol | |
| 5, 10% | Deters crystal growth |
| 15% | Precipitation |
| Glucose | |
| 5, 10% | Outcome IV |
| 15% | Outcome I |
| Sparse-matrix cofactor screen (divalent ions; Boddupalli et al., 1992 ▶; Doudna et al., 1993 ▶) | |
| 5 mM CaCl2, 2 mM MgCl2 | Outcome IV |
| 0.2 M MgCl2, 2 mM MgCl2 or 2 mM MnCl2 | Deters crystal growth |
| 5 mM MgCl2, 2 mM MnCl2 or 2 mM MnCl2, 2 mM MgCl2 | Deters crystal growth |
| 5 mM MgSO4, 40 mM MgSO4, 0.2 M MgSO4 or 0.2 M magnesium acetate | Outcome II (optimal conditions) |
| PEG substitution in mother reservoir (crystallization drop using condition No. 31) | |
| PEG 1.5K and 2K [1–2.5%(w/v)] | Outcome III |
| PEG 200 and 400 [2.5–5%(w/v)] | Outcome II (crystal formation after >10 weeks) |
Crystal optimization was conducted with condition No. 31 as a starting basis, supplemented with the reagents listed above.
3.3.1. Cosmotropic solutes
Cosmotropic solutes such as glycerol or glucose, are agents that preserve protein structure against thermal denaturation or reduce conformational heterogeneity. This approach has been successfully applied to generate crystals suitable for X-ray diffraction studies in several cases, including T7 RNA polymerase (Sousa et al., 1994 ▶) and a human fibronectin fragment (Pechik et al., 1993 ▶), where the addition of glycerol proved crucial in improving crystal quality. Several cosmotropic solutes were tested and the results are summarized in Table 3 ▶ and Fig. 3 ▶. In our case, these additives either prevented crystal growth (e.g. 2-propanol) or accelerated the rate of nucleation and crystal growth, producing smaller crystals, as shown in Fig. 3 ▶(d).
3.3.2. Role of divalent ions
DENV RdRp requires divalent ions (manganese or magnesium) for its enzymatic activity. Several salts of divalent ions (as listed in Table 3 ▶) were used as additives in order to improve crystal quality. Surprisingly, crystallization condition No. 31 supplemented with 2 mM manganese chloride, magnesium chloride or 5 mM calcium chloride proved detrimental to crystal formation (Table 3 ▶ and Fig. 3 ▶ d). In contrast, addition of either magnesium acetate or magnesium sulfate at various concentrations resulted in strikingly improved crystal quality (Table 3 ▶, Figs. 3 ▶ and 4 ▶). Explanation of this effect might be found by reference to previous studies carried out on lysozyme solubility and crystal growth using various ions (Riès-Kautt & Ducruix, 1989 ▶). The best conditions found made use of 5–40 mM magnesium sulfate as an additive to condition No. 31, leading to crystals that diffracted to a resolution of 1.85 Å at SLS (Fig. 4 ▶) and 2.4 Å (Table 2 ▶) on a rotating-anode generator following the procedure outlined in Fig. 1 ▶ and described above. These crystals were used for structure determination and refinement.
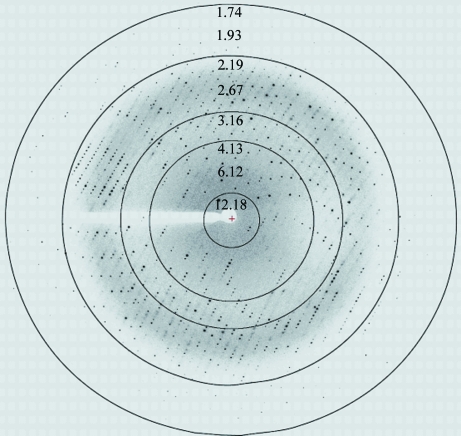
Figure 4.
Typical diffraction image obtained from crystals of the DENV 3 RdRp catalytic domain (Fig. 3 ▶ b) collected at a synchrotron-radiation source (beamline X10SA, PXII, SLS, Villigen). Diffraction extends to 1.85 Å resolution.
3.3.3. Replacement of individual component within the reservoir
Substitution of PEG 5000 monomethyl ether by various other PEGs (while retaining the other components of the crystallization solution in the reservoir; Table 3 ▶) produced two distinct results. Solutions containing PEG 200 or PEG 400 [with concentrations of 2.5–5%(w/v)] produced large crystals comparable to those displayed in Fig. 3 ▶(b), but that grew more slowly in approximately ten weeks. With reservoirs containing PEG 1500 or PEG 2000 [1–2.5%(w/v)], crystal formation occurs within 5–6 d (Fig. 3 ▶ b), with a size and morphology very similar to those of crystals obtained using the original conditions. These results show that PEGs of lower molecular weight (PEG 200 or PEG 400) slow nucleation compared with PEG 1500 or PEG 2000, which accelerate crystal growth.
3.4. Data collection and structure solution
A summary of the data-collection statistics is shown in Table 4 ▶ and a typical diffraction image is displayed in Fig. 4 ▶. Methods and procedures for measurement of the RdRp activity of the crystallized construct are detailed in a separate manuscript (Yap et al., 2007 ▶).
Table 4. Data-collection statistics.
Values in parentheses are for the last (highest) resolution shell.
| Native data (with Mg2+) | |
|---|---|
| Wavelength (Å) | 1.0000 |
| Space group | C2221 |
| Unit-cell parameters (Å) | a = 160.15, b = 180.47, c = 57. 98 |
| Resolution range (Å) | 48–1.85 (1.93–1.85) |
| No. of observed reflections | 355151 |
| No. of unique reflections | 72065 (8212) |
| Completeness (%) | 99.9 (100) |
| Multiplicity | 4.9 (4.9) |
| Rmerge† | 0.090 (0.540) |
| Rmeas‡ | 0.066 (0.584) |
| I/σ(I) | 14.62 (2.98) |
| Solvent content (%) | 58.9 |
R
merge = 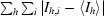 /
/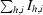 , where I
h,i is the ith observation of reflection h and 〈Ih〉 is its mean intensity.
, where I
h,i is the ith observation of reflection h and 〈Ih〉 is its mean intensity.
R
meas = 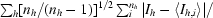
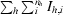 , where I
h,i is the ith observation of reflection h and nh is the multiplicity.
, where I
h,i is the ith observation of reflection h and nh is the multiplicity.
The structure was readily solved using molecular replacement with the coordinates from the West Nile virus RdRp protein (PDB code 2hfz) as a search model. After model building and refinement (Yap et al., 2007 ▶), the coordinates were deposited in the PDB with accession code 2j7u.
4. Conclusion
In this report, we described the strategy used to obtain crystals of the DENV 3 RdRp protein that diffract to high resolution. Firstly, the cloning of different serotypes of the target protein increased the likelihood of obtaining proteins that were amenable to crystallization. Secondly, small and initially poorly diffracting crystals that were not suitable for structure solution could be improved via an air-dehydration procedure, resulting in a markedly improved diffraction pattern. Thirdly, crystal quality could be further improved by the addition of salts of metal ions, leading to a high-resolution structure of the DENV RdRp catalytic domain.
Acknowledgments
We are grateful to the team of Dr Bruno Canard at AFMB Marseille, France for very useful discussions regarding construct design for the RdRp catalytic domain. Financial support via grants from NTU (RG 29/05, RG119/05) and the Singapore Biomedical Research Council (05/1/22/19/405 and 03/1/22/17/220) to the laboratory of JL are acknowledged as well as the provision of excellent beam-time by the ESRF (Grenoble, France). We thank Dr Stephanie Monaco for her support at the synchrotron beamline and Drs Clemens Scheufler, Sandra Jacob and Hans Widmer for help at beamline X10SA (PXII) at SLS, PSI, Villigen, Switzerland.
References
- Au, K. et al. (2006). Acta Cryst. D62, 1267–1275. [DOI] [PubMed] [Google Scholar]
- Bartholomeusz, A. & Thompson, P. (1999). J. Viral Hepat.6, 261–270. [DOI] [PubMed] [Google Scholar]
- Blok, J. (1985). J. Gen. Virol.66, 1323–1325. [DOI] [PubMed] [Google Scholar]
- Boddupalli, S. S., Hasemann, C. A., Ravichandran, K. G., Lu, J. Y., Goldsmith, E. J., Deisenhofer, J. & Peterson, J. A. (1992). Proc. Natl Acad. Sci. USA, 89, 5567–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Doudna, J. A., Grosshans, C., Gooding, A. & Kundrot, C. E. (1993). Proc. Natl Acad. Sci. USA, 90, 7829–7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf, R. M., Ren, J., Garman, E. F., Somers, D. O., Ross, C. K., Jones, E. Y., Stammers, D. K. & Stuart, D. I. (1998). Acta Cryst. D54, 938–953. [DOI] [PubMed] [Google Scholar]
- Forwood, J. K., Brooks, A., Briggs, L. J., Xiao, C. Y., Jans, D. A. & Vasudevan, S. G. (1999). Biochem. Biophys. Res. Commun.257, 731–737. [DOI] [PubMed] [Google Scholar]
- Henchal, E. A. & Putnak, J. R. (1990). Clin. Microbiol. Rev.3, 376–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras, B. & Martin, J. L. (2005). Acta Cryst. D61, 1173–1180. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi, D. & Steitz, T. A. (1997). J. Mol. Biol.274, 748–756. [DOI] [PubMed] [Google Scholar]
- Koonin, E. V. & Ilyina, T. V. (1993). Biosystems, 30, 241–268. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1999). Acta Cryst. D55, 1696–1702. [DOI] [PubMed] [Google Scholar]
- Pechik, I., Nachman, J., Ingham, K. & Gilliland, G. L. (1993). Proteins, 16, 43–47. [DOI] [PubMed] [Google Scholar]
- Riès-Kautt, M. M. & Ducruix, A. F. (1989). J. Biol. Chem.264, 745–748. [PubMed] [Google Scholar]
- Sousa, R., Rose, J. & Wang, B.-C. (1994). J. Mol. Biol.244, 6–12. [DOI] [PubMed] [Google Scholar]
- Wu, J. Z., Yao, N., Walker, M. & Hong, Z. (2005). Mini Rev. Med. Chem.5, 1103–1112. [DOI] [PubMed] [Google Scholar]
- Yap, T. L., Xu, T., Chen, Y. L., Malet, H., Egloff, M. P., Canard, B., Vasudevan, S. G. & Lescar, J. (2007). Submitted. [DOI] [PMC free article] [PubMed]