To potentially gain insight into the conformational origins of substrate recognition by the enzyme from Escherichia coli, cocrystallization experiments were carried out with RNase HI–dsRNA (enzyme–inhibitor) complexes. Crystals were obtained of two complexes containing 9-mer and 10-mer RNA duplexes that diffracted X-rays to 3.5 and 4 Å resolution, respectively.
Keywords: ribonucleases, RNAse HI, substrate recognition, enzyme–inhibitor complexes
Abstract
RNase H binds RNA–DNA hybrid and double-stranded RNA (dsRNA) duplexes with similar affinity, but only cleaves the RNA in the former. To potentially gain insight into the conformational origins of substrate recognition by the enzyme from Escherichia coli, cocrystallization experiments were carried out with RNase HI–dsRNA (enzyme–inhibitor) complexes. Crystals were obtained of two complexes containing 9-mer and 10-mer RNA duplexes that diffracted X-rays to 3.5 and 4 Å resolution, respectively.
1. Introduction
Ribonucleases H (RNases H) recognize hybrid duplexes between RNA and DNA and specifically cleave the RNA strand (Hostomsky et al., 1993 ▶). The enzyme is thought to play a role in DNA replication and regulation of transcription. In Drosophila, RNase H1 is essential for development but not for proliferation (Filippov et al., 1997 ▶, 2001 ▶). The work of Filippov and coworkers represents the first loss-of-function mutation in an rnase H1 gene of a metazoan organism; none of the previously studied mutations in rnase H genes from either prokaryotes or lower eukaryotes were lethal. In addition, two genes encoding functional RNase H were determined to be essential for growth in Bacillus subtilis 168 (Itaya et al., 1999 ▶). RNase H is also believed to be an important determinant for potent antisense activity by artificial oligonucleotides (Walder & Walder, 1988 ▶; Crooke, 1998 ▶). However, most chemically modified antisense oligonucleotides (AONs) do not elicit RNase H action, with the first-generation phosphorothioate DNA (PS-DNA; Crooke, 1995 ▶) and 2′-deoxy-2′-fluoroarabinonucleic acid (2′-FANA; Damha et al., 1998 ▶) constituting exceptions. For example, AONs bearing 2′ modifications at the carbohydrate moiety bound to complementary RNA are not recognized as substrates by RNase H (Cook, 1998 ▶; Manoharan, 1999 ▶). Moreover, there are some observations based on treatment of human cell lines with antisense PS-DNAs that cast doubt on the role of RNase H as a major player in AON-mediated degradation of target mRNAs (ten Asbroek et al., 2002 ▶). Thus, RNase H activity does not simply correspond to the activity assayed in vitro, but appears to be modulated by cell-type specific factors that could, for example, affect enzyme localization.
RNase HI from Escherichia coli is the best characterized bacterial RNase H (Kanaya & Crouch, 1983 ▶). The enzyme not only binds and processes RNA–DNA hybrids, but also exhibits considerable affinity for both RNA and DNA duplexes and to a lesser extent for single-stranded oligonucleotides. Thus, RNase H binds RNA–DNA and RNA duplexes ∼60-fold more strongly than DNA duplexes and ∼300-fold more strongly than single strands (Lima & Crooke, 1997 ▶). The K d for the complex with a 17-mer dsRNA is ∼1 µM. The crystal structure of the enzyme alone was determined many years ago (Katayanagi et al., 1990 ▶; Yang, Hendrickson, Crouch et al., 1990 ▶). However, no structure of a complex between E. coli RNase HI and a substrate duplex has been reported to date, although attempts have been made in this direction (Ishikawa et al., 1991 ▶). The interactions between RNase H and heteroduplexes composed of DNA or chemically modified AONs and RNA have been the focus of numerous investigations over the years (Nakamura et al., 1991 ▶; Fedoroff et al., 1993 ▶; Lima et al., 1997 ▶, 2004 ▶; Minasov et al., 2000 ▶; Sarafianos et al., 2001 ▶; Yazbeck et al., 2002 ▶). Early models of an enzyme–substrate complex were based on the assumption that the presence of 2′-hydroxyl groups in the RNA strand and the lack thereof in the DNA would be used by the enzyme to discriminate between RNA–DNA and dsRNA (Nakamura et al., 1991 ▶). The mystery of how the enzyme can discriminate between hybrids and dsRNA is complicated by the fact that hybrid duplexes can adopt a variety of conformations, including the canonical A-form (Egli et al., 1993 ▶; Ban et al., 1994 ▶; Horton & Finzel, 1996 ▶; Sarafianos et al., 2001 ▶). An NMR investigation of a hybrid duplex in solution provided evidence that the sugars of the DNA strand adopted the O4′-endo (Eastern) pucker (Fedoroff et al., 1993 ▶). This particular conformation of the sugars results in a narrowing of the minor groove compared with dsRNA with A-form geometry. In the structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA–DNA the minor groove is indeed contracted, but 2′-deoxyriboses in the section of the duplex that is contacted by the RNase H domain adopt Southern-type puckers (Sarafianos et al., 2001 ▶). In addition, the crystal structure of the complex between a bacterial RNase H and an RNA–DNA hybrid revealed that riboses adopt the C3′-endo pucker and 2′-deoxyriboses adopt C2′-endo or C1′-exo puckers (Nowotny et al., 2005 ▶). Interestingly, the minor groove of the hybrid duplex in this structure is narrowed compared with the canonical A-form and five RNA 2′-hydroxyl groups are contacted directly by the enzyme.
A complete understanding of the substrate-specificity of RNase H will require analyses of its complexes with both RNA–DNA (substrate) and dsRNA (inhibitor). No structure of an inhibitor complex (RNase H–dsRNA) is available at present. In addition, the interpretation of the large amount of functional data with regard to the dependence of E. coli or human RNase H cleavage activity on location and nature of chemical modifications in the DNA strand (Lima et al., 2004 ▶) would greatly benefit from the three-dimensional structures of enzyme–substrate and enzyme–inhibitor complexes. Our recent investigation of the conformational preferences of DNA duplexes with incorporated 2′-FANA residues provided evidence that this analog is unable to adopt a Southern pucker (Li et al., 2006 ▶). This observation begs the following questions: (i) does the enzyme tolerate a limited range of conformations of the DNA or AON strand paired to RNA? and (ii) is the minor-groove width really the central recognition feature exploited by RNase H for substrate recognition?
The advent of RNA interference and the identification of the argonaute 2 (Ago2) enzyme that is responsible for the cleavage of mRNA targeted by miRNAs and siRNAs (Liu et al., 2004 ▶; Meister et al., 2004 ▶) provides further motivation for gaining an improved understanding of the conformational bases of substrate recognition by RNase H. The PIWI domain that forms part of the Ago2 enzyme adopts an RNase H fold (Song et al., 2004 ▶; Ma et al., 2005 ▶; Parker et al., 2005 ▶) and yet is responsible for cleavage of dsRNA. In this context, it is also interesting to mention that the recently characterized RNase HI from the thermoacidophilic archaeon Sulfolobus tokodaii possesses both dsRNase and RNase H activity (Ohtani et al., 2004 ▶).
In order to potentially gain insight into the structural origins of the ability of E. coli RNase to discriminate between RNA–DNA and dsRNA, we first directed our efforts towards the crystallization of enzyme–inhibitor (RNase HI–dsRNA) complexes. The decision to tackle the complex with RNA was partly a consequence of the notion that no particular precautions (i.e. use of inactive mutant proteins or Mg2+-free crystallization buffers) would be necessary to prevent cleavage of dsRNAs, something that would be likely to hamper crystallization of the complex between wt-RNase HI and native RNA–DNA substrate. Cocrystallization of a protein with nonspecific nucleic acid sequences has the disadvantage that a strategy that combines a recognition sequence with a variety of sequences in the flanks cannot be pursued. Thus, numerous structural studies have concentrated on proteins bound to their specific DNA and RNA sequences. Conversely, relatively few structures have been determined for complexes that do not involve sequence-specific interactions (for examples, see Luger et al., 1997 ▶; Ryter & Schultz, 1998 ▶; Viadiu et al., 2000 ▶). Here, we report the crystallization of two different E. coli RNase HI–dsRNA complexes and the results of the data collection and preliminary crystallographic analysis of these complexes.
2. Materials and methods
2.1. Cloning, protein expression and purification
To prepare large amounts of E. coli RNase HI, the corresponding cDNA was cloned into the pET-29b expression vector with a TGA STOP codon followed by the GTT codon for C-terminal valine. Overexpression of RNase HI was performed in the E. coli BL21 (DE3) strain according to a standard protocol for expression with the pET-29b vector (Novagen). Briefly, transformed E. coli cells were grown in 2×YT media to an OD600 of 1.0 at 310 K and expression was induced with 1 mM IPTG. Cells were harvested by centrifugation after 3.5 h of cultivation at 310 K and the cell pellet was either used immediately for RNase HI isolation or was stored at 253 K. RNase HI was purified using DEAE-52 and P-11 columns according to published procedures (Kanaya & Crouch, 1983 ▶; Kanaya et al., 1989 ▶; Yang, Hendrickson, Kalman et al., 1990 ▶).
2.2. RNA synthesis and purification
All RNA oligonucleotides were purchased from Dharmacon Inc. (West Lafayette, CO, USA) and were PAGE-purified, deprotected and desalted.
2.3. Crystallization experiments
Our efforts to crystallize an E. coli RNase HI–dsRNA complex were based on the assumption that it was possible to identify an RNA duplex with just the right length and sequence to trap the complex in a well packed crystal lattice. Crystallization experiments were conducted using commercially available sparse-matrix screens (Jancarik & Kim, 1991 ▶; Berger et al., 1996 ▶; Hampton Research, Aliso Viejo, CA, USA) and ‘home-made’ crystallization buffers. To date, we have included some 25 different RNA duplexes based on 15 oligoribonucleotides of various lengths and sequences in the trials (Table 1 ▶; different Y and R RNAs can be combined). Light scattering was routinely used to establish whether solutions of the complexes were monodisperse. In addition to screening crystallization conditions with the various RNase HI–dsRNA complexes, we also produced setups with protein and RNAs alone under identical conditions. Thus, crystals of wt-RNase H were grown from 20 mM HEPES pH 8.0, 16% PEG 3350 (Yang, Hendrickson, Kalman et al., 1990 ▶; Fig. 1 ▶ a). In many cases, crystals were obtained from RNA alone (Fig. 1 ▶ b) and in some cases these crystals diffracted to atomic resolution, but structure determination was not further pursued.
Table 1. RNA oligonucleotide sequences employed in the crystallization trials.
| Code | Length | 5′→3′ sequence |
|---|---|---|
| SC | 15 | GGA CUG AUC AGU CCA |
| Y | 15 | CAC UUG ACC UGG CUC |
| R | 15 | GAG CCA GGU CAA GUG |
| Y1 | 17 | G CAC UUG ACC UGG CUC G |
| R1 | 17 | C GAG CCA GGU CAA GUG C |
| Y2 | 16 | G CAC UUG ACC UGG CUC |
| R2 | 16 | C GAG CCA GGU CAA GUG |
| Y3 | 16 | CAC UUG ACC UGG CUC G |
| R3 | 16 | GAG CCA GGU CAA GUG C |
| Y4 | 9 | CCU GGC UCG |
| R4 | 9 | CGA GCC AGG |
| Y5 | 9 | CUG GCU CGC |
| R5 | 9 | GCG AGC CAG |
| Y6 | 10 | GCA CUU GAC C |
| R6 | 10 | GGU CAA GUG C |
Figure 1.
Micrographs of crystals. (a) wt-RNase HI (E. coli); (b) RNA duplex (CAC UUG ACC UGG CUC)–(GAG CCA GGU CAA GUG) (Y–R in Table 1 ▶), diffraction limit ∼2.1 Å; (c) complex between RNase HI and RNA duplex Y–R; (d) complex between RNase HI and RNA duplex (GCA CUU GAC C)–(GGU CAA GUG C) (Y6–R6 in Table 1 ▶).
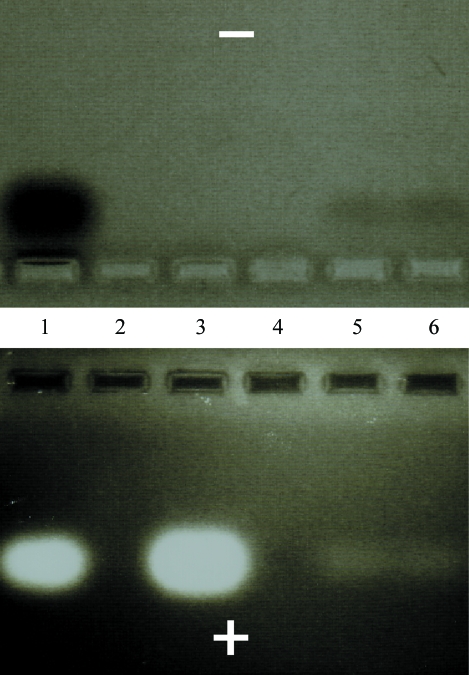
An adapted gel-electrophoretic assay (Su et al., 1994 ▶) that allows simultaneous detection of positively charged (protein) and negatively charged species (RNA) at a specific pH was employed to confirm that crystals contained both RNase HI and dsRNA. The pH of the running buffer for the single gel with central wells was kept at ∼6.7 so that RNA and protein migrated to the anode and cathode, respectively (Fig. 2 ▶). Crystals obtained from droplets containing RNase HI and dsRNA were thoroughly washed, dissolved and run on a 4.6% agarose gel that was first stained with ethidium bromide and then with Coomassie G250 (Bio-Rad) to detect RNA and protein, respectively. This particular method does not permit determination of the stoichiometry of the complex in the crystals.
Figure 2.
Gel-electrophoretic assay to establish the presence of both dsRNA (bottom panel) and E. coli RNase HI (top panel) in crystals: RNA and protein mixed in a 1:1 ratio (lane 1), RNA alone (lane 3) and two different crystals containing both RNase H and the RNA duplex Y–R (Table 1 ▶).
By screening a variety of RNA duplexes (9-mers to 17-mers with either blunt ends or overhangs; Table 1 ▶), crystals of complexes were obtained with a variety of dsRNAs. However, they often did not diffract X-rays or only diffracted to low resolution. For example, crystals of the complex with the 15-mer RNA duplex Y–R (Fig. 1 ▶ c, Table 1 ▶) diffracted to 7 Å and data of similar resolution were obtained for the complex with RNA duplex Y3–R2 (Table 1 ▶). The complex with RNA Y4–R4 diffracted to ∼10 Å. To date, the best crystals of complexes with E. coli RNase HI were obtained with RNA duplexes Y5–R5 (9-mer, complex 1) and Y6–R6 (10-mer, complex 2; Fig. 2 ▶ d). Crystals of complex 1 and complex 2 were grown using the sitting-drop vapor-diffusion method. The concentrations of the complexes were 0.11–0.15 mM. Either 1 or 2 µl of complex were combined with 1 or 2 µl 50 mM bis-tris pH 6.1, 12.5% PEG 3350, 25 mM NaCl, 2 mM MgCl2 and 1 mM TCEP. 4 µl 20 mM HEPES pH 8.0, 16% PEG 3350 was added to the drop containing complex 2 and reservoir solution.
2.4. Diffraction data collection and preliminary crystallographic analysis
Crystals were mounted in nylon loops using an initial cryoprotection protocol and screened for diffraction quality either on an in-house rotating-anode X-ray setup or at the Advanced Photon Source (APS), Argonne National Laboratory, Argonne, IL, USA (5-ID beamline, DND-CAT, Sector 5). The cryoprotection protocol for complex 1 and complex 2 was as follows. Crystals were cryoprotected with 30%(v/v) ethylene glycol, 12%(w/v) PEG 20 000, 30 mM bis-tris pH 6.1, 15 mM NaCl, 0.5 mM MgCl2 and 1 mM TCEP.
A summary of selected crystal data and diffraction data statistics for the two crystals is given in Table 2 ▶. Crystals of complexes 1 and 2 diffracted to maximum resolutions of 3.5 and 3.99 Å, respectively. Unit-cell volume considerations indicated that complex 1 and 2 crystals could contain either one or two copies of a 1:1 RNase HI–dsRNA complex per crystallographic asymmetric unit (ASU) or alternatively could contain two copies of the dsRNA and a single copy of the protein or two copies of the protein and a single copy of the RNA. The values of the Matthews coefficient V M (Matthews, 1968 ▶) for complex 1 crystals based on the above scenarios range between 6.84 Å3 Da−1 (single copy of 1:1 complex, 82.0% solvent content) and 3.42 Å3 Da−1 (two copies of 1:1 complex, 64.0% solvent content). In the case of complex 2 crystals, the corresponding values range between 6.04 Å3 Da−1 (79.6% solvent content) and 3.02 Å3 Da−1 (solvent content 59.3%).
Table 2. Selected crystal data and diffraction data statistics.
Data were collected on the 5-ID beamline at APS and were processed using the program AUTOMAR (Bartels & Klein, 2003 ▶). Values in parentheses are for the outer resolution shell.
| Complex 1 | Complex 2 | |
|---|---|---|
| Protein | E. coli RNase HI | E. coli RNase HI |
| RNA duplex | CUG GCU CGC (Y5) | GCA CUU GAC C (Y6) |
| GCG AGC CAG (R5) | GGU CAA GUG C (R6) | |
| CCD detector | MAR Mosaic 225 | MAR Mosaic 225 |
| Wavelength (Å) | 1.000 | 1.000 |
| Space group | P21 | P212121 |
| Unit-cell parameters | ||
| a (Å) | 42.13 | 68.10 |
| b (Å) | 61.42 | 78.19 |
| c (Å) | 121.28 | 106.63 |
| β (°) | 93.16 | — |
| Resolution range (Å) | 30.0–3.50 (3.62–3.50) | 30.0–3.99 (4.14–3.99) |
| Observations | 36900 | 25887 |
| Unique reflections | 7476 (739) | 5099 (490) |
| Completeness (%) | 91.3 (94.6) | 98.6 (95.4) |
| Rmerge | 0.102 (0.359) | 0.061 (0.202) |
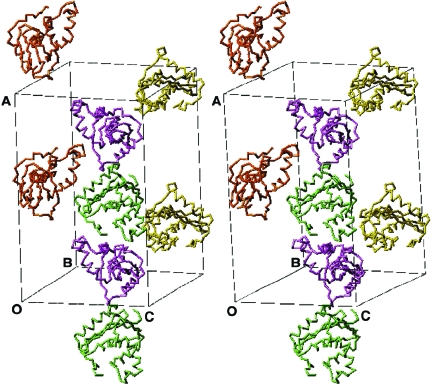
Molecular-replacement searches in CNS (Brünger et al., 1998 ▶) using the structure of wt-RNase HI from E. coli (PDB code 2rn2; Katayanagi et al., 1992 ▶) as a search model indicate the presence of two protein molecules per ASU for both complex 1 and complex 2 crystals (Fig. 3 ▶). In both cases, this leaves ample space to accommodate either one or two RNA duplexes. However, the electron density in the regions presumably occupied by the RNAs after rigid-body and positional and B-factor refinements of the protein molecules alone is relatively weak. The lack of clear density at this stage may be a sign of high mobility of the RNA duplexes.
Figure 3.
Stereo diagram of the crystallographic unit cell of complex 2 between E. coli RNase HI and RNA duplex (GCA CUU GAC C)–(GGU CAA GUG C) (Y6–R6; Table 1 ▶). Only the locations of protein molecules (two copies per crystallographic asymmetric unit, space group P212121) are shown. There is sufficient space to accommodate two RNA duplexes per asymmetric unit.
3. Conclusions
By conducting crystallization experiments with some 25 different E. coli RNase HI–dsRNA complexes, we have identified crystals of two complexes with 9-mer and 10-mer RNA duplexes that diffract X-rays to medium resolution. In both cases, the asymmetric unit appears to contain two enzyme molecules and one or perhaps two RNA duplexes. Further analysis of the crystal structures will reveal whether the resolution is sufficient to gain insight into the origins of the inability of RNase HI to process dsRNA. Inclusion of a larger number of RNA constructs in the crystallization trials can be expected to yield crystals that diffract X-rays to <3 Å.
Acknowledgments
This research was supported by US National Institutes of Health grant R01 GM55237. We thank Drs Walt Lima, Isis Pharmaceuticals Inc., Carlsbad, CA and Muthiah Manoharan, Alnylam Pharmaceuticals, Cambridge, MA for discussions and Drs Zdzislaw Wawrzak and Adriana Irimia for help with data collection and molecular replacement, respectively. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science under Contract No. W-31-109-Eng-38. The DuPont–Northwestern–Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center at the Advanced Photon Source (Sector 5) is supported by E. I. DuPont de Nemours & Co., The Dow Chemical Company, the National Science Foundation and the State of Illinois.
References
- Ban, C., Ramakrishnan, B. & Sundaralingam, M. (1994). J. Mol. Biol.236, 275–285. [DOI] [PubMed] [Google Scholar]
- Bartels, K. S. & Klein, C. (2003). AUTOMAR version 3.04-0. MAR Research GmbH, Norderstedt, Germany.
- Berger, I., Kang, C. H., Sinha, N., Wolters, M. & Rich, A. (1996). Acta Cryst. D52, 465–468. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cook, P. D. (1998). Annu. Rep. Med. Chem.33, 313–325.
- Crooke, S. T. (1995). Therapeutic Applications of Oligonucleotides, pp. 63–79. Austin, TX, USA: R. G. Landes.
- Crooke, S. T. (1998). Editor. Antisense Research and Application, pp. 1–50. Berlin: Springer.
- Damha, M. J., Wilds, C. J., Noronha, A., Brukner, I., Borkow, G., Arion, D. & Parniak, M. A. (1998). J. Am. Chem. Soc.120, 12976–12977.
- Egli, M., Usman, N. & Rich, A. (1993). Biochemistry, 32, 3221–3237. [PubMed] [Google Scholar]
- Fedoroff, O. Y., Salazar, M. & Reid, B. R. (1993). J. Mol. Biol.233, 509–523. [DOI] [PubMed] [Google Scholar]
- Filippov, V., Filippova, M. & Gill, S. S. (1997). Biochem. Biophys. Res. Commun.240, 844–849. [DOI] [PubMed] [Google Scholar]
- Filippov, V., Filippova, M. & Gill, S. S. (2001). Mol. Genet. Genomics, 265, 771–777. [DOI] [PubMed] [Google Scholar]
- Horton, N. C. & Finzel, B. C. (1996). J. Mol. Biol.264, 521–533. [DOI] [PubMed] [Google Scholar]
- Hostomsky, Z., Hostomska, Z. & Matthews, D. A. (1993). Nucleases, 2nd ed., edited by S. M. Linn, S. R. Lloyd & R. J. Roberts, pp. 341–376. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press.
- Ishikawa, M., Oda, Y., Katayanagi, K., Iwai, S., Ohtsuka, E. & Morikawa, K. (1991). Nucleic Acids Res. Symp. Ser.24, 253. [PubMed]
- Itaya, M., Omori, A., Kanaya, S., Crouch, R. J., Tanaka, T. & Kondo, K. (1999). J. Bacteriol.181, 2118–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Kanaya, S. & Crouch, R. J. (1983). J. Biol. Chem.258, 1276–1281. [PubMed] [Google Scholar]
- Kanaya, S., Kohara, A., Miyagawa, M., Matsuzaki, T., Morikawa, K. & Ikehara, M. (1989). J. Biol. Chem.264, 11546–11549. [PubMed] [Google Scholar]
- Katayanagi, K., Miyagawa, M., Matsushima, M., Ishikawa, M., Kanaya, S., Ikehara, M., Matsuzaki, T. & Morikawa, K. (1990). Nature (London), 347, 306–309. [DOI] [PubMed] [Google Scholar]
- Katayanagi, K., Miyagawa, M., Matsushima, M., Ishikawa, M., Kanaya, S., Nakamura, H., Ikehara, M., Matsuzaki, T. & Morikawa, K. (1992). J. Mol. Biol.223, 1029–1052. [DOI] [PubMed] [Google Scholar]
- Li, F., Sarkhel, S., Wilds, C. J., Wawrzak, Z., Prakash, T. P., Manoharan, M. & Egli, M. (2006). Biochemistry, 45, 4141–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, W. F. & Crooke, S. T. (1997). Biochemistry, 36, 390–398. [DOI] [PubMed] [Google Scholar]
- Lima, W. F., Mohan, V. & Crooke, S. T. (1997). J. Biol. Chem.272, 18191–18199. [DOI] [PubMed] [Google Scholar]
- Lima, W. F., Nichols, J. G., Wu, H., Prakash, T. P., Migawa, M. T., Wyrzykiewicz, T. K., Bhat, B. & Crooke, S. T. (2004). J. Biol. Chem.279, 36317–36326. [DOI] [PubMed] [Google Scholar]
- Liu, J., Carmell, M. A., Rivas, F. V., Marsden, C. G., Thomson, J. M., Song, J.-J., Hammond, S. M., Joshua-Tor, L. & Hannon, G. J. (2004). Science, 305, 1437–1441. [DOI] [PubMed] [Google Scholar]
- Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997). Nature (London), 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Ma, J.-B., Yuan, Y.-R., Meister, G., Pei, Y., Tuschl, T. & Patel, D. J. (2005). Nature (London), 434, 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoharan, M. (1999). Biochim. Biophys. Acta, 1489, 117–130. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G. & Tuschl, T. (2004). Mol. Cell, 15, 185–197. [DOI] [PubMed] [Google Scholar]
- Minasov, G., Teplova, M., Nielsen, P., Wengel, J. & Egli, M. (2000). Biochemistry, 39, 3525–3532. [DOI] [PubMed] [Google Scholar]
- Nakamura, H., Oda, Y., Iwai, S., Inoue, H., Ohtsuka, E., Kanaya, S., Kimura, S., Katsuda, C., Katayanagi, K., Morikawa, K., Miyashiro, H. & Ikehara, M. (1991). Proc. Natl Acad. Sci. USA, 88, 11535–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny, M., Gaidamakov, S. A., Crouch, R. J. & Yang, W. (2005). Cell, 121, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Ohtani, N., Yanagawa, H., Tomita, M. & Itaya, M. (2004). Nucleic Acids Res.32, 5809–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J. S., Roe, S. M. & Barford, D. (2005). Nature (London), 434, 663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter, J. M. & Schultz, S. C. (1998). EMBO J.17, 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafianos, S. G., Das, K., Tantillo, C., Clark, A. D. Jr, Ding, J., Whitcomb, J. M., Boyer, P. L., Hughes, S. H. & Arnold, E. (2001). EMBO J.20, 1449–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.-J., Smith, S. K., Hannon, G. J. & Joshua-Tor, L. (2004). Science, 305, 1434–1437. [DOI] [PubMed] [Google Scholar]
- Su, C., Wang, F., Ciolek, D. & Pan, Y.-C. E. (1994). Anal. Biochem.223, 93–98. [DOI] [PubMed] [Google Scholar]
- ten Asbroek, A. L., van Groenigen, M., Nooij, M. & Baas, F. (2002). Eur. J. Biochem.269, 583–592. [DOI] [PubMed] [Google Scholar]
- Viadiu, H., Kucera, R., Schildkraut, I. & Aggarwal, A. K. (2000). J. Struct. Biol.130, 81–85. [DOI] [PubMed] [Google Scholar]
- Walder, R. T. & Walder, J. A. (1988). Proc. Natl Acad. Sci. USA, 85, 5011–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W., Hendrickson, W. A., Crouch, R. J. & Satow, Y. (1990). Science, 249, 1398–1405. [DOI] [PubMed] [Google Scholar]
- Yang, W., Hendrickson, W. A., Kalman, E. T. & Crouch, R. J. (1990). J. Biol. Chem.265, 13553–13559. [PubMed] [Google Scholar]
- Yazbeck, D. R., Min, K.-L. & Dahma, M. J. (2002). Nucleic Acids Res.30, 3015–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]