A novel cardiotoxin-like basic protein from Naja naja atra was crystallized and diffraction data were collected to 2.35 Å resolution.
Keywords: snake venoms, cardiotoxin-like basic protein, vascular endothelial growth factor, basic fibroblast growth factor, neurotoxins, toxicity, haemolytic activity
Abstract
A novel cardiotoxin-like basic protein was isolated from the venom of the Chinese cobra (Naja naja atra) from the south of Anhui in China. The protein inhibits the expression of vascular endothelial growth factor and basic fibroblast growth factor in human lung cancer cell line H1299 and induces the haemolysis of rabbit erythrocytes under low-lecithin conditions. After a two-step chromatographic purification, the resultant 7 kDa protein was crystallized by the hanging-drop vapour-diffusion method at room temperature. A complete data set was collected to 2.35 Å resolution using an in-house X-ray diffraction system. The crystal belongs to space group P41212, with unit-cell parameters a = b = 43.2, c = 147.9 Å. There are two molecules in the crystallographic asymmetric unit.
1. Introduction
In cobra snake venoms, there are a number of small proteins of 60–70 amino acids in length that show a broad spectrum of pharmacological activities (Endo et al., 1991 ▶). The most abundant and most investigated of these proteins are the so-called ‘three-finger toxins’. The neurotoxins and the cardiotoxins are the two main three-finger toxic components involved in the lethality of these snake venoms (Menez, 1998 ▶; Tsetlin, 1999 ▶). The cardiotoxins exhibit activities including haemolysis, cytoxicity and depolarization of muscle (Harvey, 1985 ▶; Menez et al., 1990 ▶; Dufton & Hider, 1991 ▶; Fletcher & Jiang, 1993 ▶). In addition to the main toxic factors, weak toxic constituents have also been discovered, which include the cardiotoxin-like basic proteins (CLBPs). To date, three CLBPs have been identified (Sun et al., 1997 ▶; Takechi et al., 1985 ▶; Chang et al., 2004 ▶). CLBPs are cardiotoxin homologues, but possess weak cardiotoxic activity at most and therefore are not classified as cardiotoxins (Sivaraman et al., 1997 ▶). Here, we have purified a novel CLBP with low toxicity from the venom of Naja naja atra (from the south of Anhui in China). The protein shows 85% sequence homology to the CLBP from the Formosan cobra (Takechi et al., 1985 ▶) in the 21 N-terminal residues. Because the N. naja atra was from the south of Anhui in China, we named the protein CLBPah. We found that CLBPah has haemolytic activity under low-lecithin conditions.
Vascular endothelial growth factor (VEGF) is a mitogen, primarily for vascular endothelial cells. The broad term ‘VEGF’ covers a number of proteins that result from alternate splicing of the mRNA from a single eight-exon VEGF gene. Both VEGF and basic fibroblast growth factor (bFGF) are angiogenic and have been implicated in tumour angiogenesis, which is a prerequisite for tumour progression (Brattstrom et al., 2004 ▶; Komorowski et al., 2000 ▶). Recently, Yu and coworkers found that CLBPah inhibits the expression of VEGF and bFGF in human non-small-cell lung cancer cell line H1299 in a dose-dependent manner (Yu et al., 2005 ▶).
In order to obtain insights into the new CLBP, we crystallized the protein and performed preliminary crystallographic analysis.
2. Experimental procedures
2.1. Materials
Dried crude N. naja atra venom was obtained from the southern mountain region of Anhui Province, China. CM-Sepharose and Sephadex G-50 were purchased from Pharmacia (Uppsala, Sweden). Standard protein markers were produced by the Shanghai Institute of Biochemistry (Shanghai, China). Other chemicals and reagents were of analytical grade from commercial sources. Deionized water was used in the preparation of all solutions.
2.2. Purification of CLBPah and molecular characterization
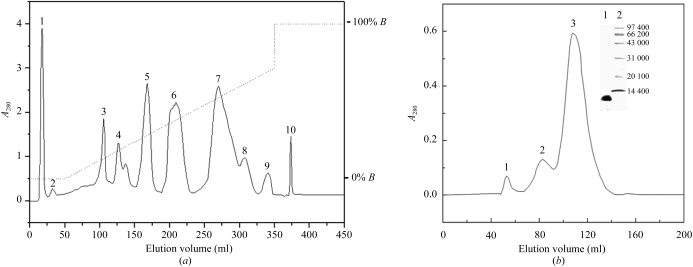
The preparation of CLBPah was carried out by a two-step chromatography procedure at room temperature (∼298 K; see Fig. 1 ▶). The purified samples were desalted and lyophilized for subsequent crystallization and characterization. Both conventional SDS–PAGE and MALDI–TOF mass spectrometry were performed in order to check for homogeneity and to estimate the molecular weight.
Figure 1.
(a) Purification of CLBPah from N. naja atra venom. 0.5 g of dried crude venom was dissolved in 10 ml buffer A (0.02 M NaH2PO4/Na2HPO4 pH 6.0), centrifuged at 10 000 rev min−1 to remove insoluble materials and then applied onto a CM-Sepharose column (1 × 20 cm) pre-equilibrated with the same buffer. The elution was performed at a flow rate of 0.5 ml min−1 with a combined gradient of pH value and salt concentration produced by mixing 200 ml buffer A with 200 ml buffer B (0.02 M Na2HPO4/NaH2PO4 pH 8.0 containing 0.5 M NaCl) followed by two column volumes of buffer B. The ninth fraction, containing CLBPah, was pooled and desalted for further isolation. The absorbance at 280 nm is indicated by a solid line and the salt gradient by a dashed line. (b) Purification of CLBPah on a gel-filtration column. Fraction 9 was applied onto a Sephadex G-50 column (1.6 × 100 cm) and eluted with 0.15 M NaCl solution. Peak 3 is CLBPah. The inset shows SDS–PAGE analysis of CLBPah. Lane 1, CLBPah. Lane 2, molecular-weight markers: rabbit phosphorylase b (97 400 Da), bovine serum albumin (66 200 Da), rabbit actin (43 000 Da), bovine carbonic anhydrase (31 000 Da), trypsin inhibitor (20 100 Da) and hen egg-white lysozyme (14 400 Da).
The lethal activity LD50 was assayed intravenously on laboratory mice according to the procedure of Aird & Kaiser (1985 ▶). Mice with body weights of 20–30 g were randomly assigned to five cages. Each cage contained five females and five males. CLBPah was dissolved in phosphate-buffered saline (made by mixing 8.0 g NaCl, 0.2 g KCl, 0.2 g KH2PO4 and 0.15 g Na2HPO4 per litre and then adjusting the pH value to 7.1 with Na2HPO4) for injection. The in vitro injections were performed through one of the two dorsolateral caudal veins. The outcome (survival or death) was recorded 24 h post-injection. All operations were carried out at room temperature.
2.3. Protein-sequence analysis
N-terminal amino-acid sequences were obtained by the Edman degradation method on a Procise Protein Sequencer (Applied Biosystems 475A sequencer). The protein sequence was subjected to a BLAST database search (http://www.ncbi.nlm.nih.gov/BLAST) to find proteins with significant sequence homology.
2.4. Haemolytic activity of CLBPah
Rabbit erythrocytes were used in the in vitro experiments. The blood was collected from one carotid of a male rabbit under urethane anaesthesia using sodium citrate as an anticoagulant. The procedure for the determination of the percentage of haemolysis was as described by Eno et al. (1998 ▶). The whole blood was centrifuged (600g, 10 min) to obtain erythrocytes. These were washed at least three times in PBS (10 mM potassium phosphate, 150 mM NaCl pH 7.4) at room temperature. Following the final centrifugation step, 3 ml of erythrocytes was added to 197 ml of the incubation medium (PBS plus 0.025% lecithin and 65 nM sodium deoxycholate). 1 ml preparations were incubated with or without CLBPah, after which 3 ml PBS was added followed by 5 min incubation at 273 K. The sample was centrifuged at 600g for 10 min and the absorbance of the supernatant was measured for released haemoglobin at 540 nm. The 100% haemolysis point was determined by incubating 1 ml of the preparation in 3 ml deionized water. Blanks containing no CLBPah were subtracted from all samples. The percentage of haemolysis was calculated using the formula
2.5. Crystallization
For crystallization trials, the protein was used at a concentration of 20 mg ml−1 in 20 mM Tris–HCl buffer pH 8.0. Crystallization was performed at room temperature using the hanging-drop vapour-diffusion method. Crystal Screens I and II (Hampton Research) were used to establish initial crystallization conditions. The drops were composed of equal volumes (2 µl) of protein solution and precipitant solution and were equilibrated against 300 µl reservoir solution. The final crystallization conditions were 21% PEG 3350 and 0.2 M ammonium sulfate (see Fig. 2 ▶).
Figure 2.
Crystals of CLBPah.
2.6. Data collection and processing
Data could be collected to a resolution of 2.35 Å from a single crystal using a rotating-anode generator (Rigaku) and a MAR345 dtb detector (MAR Research). Single crystals were soaked in cryoprotectant solution [30%(v/v) glycerol, 21% PEG 3350, 0.2 M ammonium sulfate] for 90 s and mounted in nylon cryoloops. The pre-soaked crystals were immediately subjected to flash-freezing in a cold nitrogen stream. Data were collected at 110 K with a crystal-to-detector distance of 200 mm, an oscillation range of 1° and an exposure time of 5 min per image. A total of 99 images were recorded. The diffraction data were processed with the AUTOMAR package.
3. Results and discussion
3.1. Chemical characterization
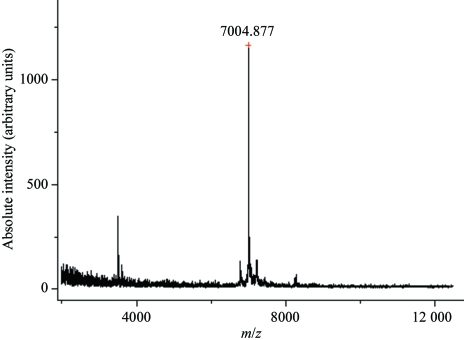
Separation of the CLBPah was carried out as described in §2. A CM-Sepharose column with a linear gradient of 0–0.5 M NaCl was used for initial isolation of the crude venom. Fraction 9 (Fig. 1 ▶ a) was further applied onto a column of Sephadex G-50 and separated into three fractions. Peak 3 (Fig. 1 ▶ b) contained the protein of interest. Its homogeneity was evident from SDS–PAGE (inset in Fig. 1 ▶ b). The 21 amino acids at the N-terminus are DCCHNTQLPFIYKTCPEGCNL, which shows it to be a cardiotoxin-like basic protein (Fig. 3 ▶). The molecular weight of the native CLBPah was determined to be 7005 Da by MALDI–TOF MS (Fig. 4 ▶).
Figure 3.
Sequence alignments of the 7005 Da protein CLBPah with other CLBPs (Corpet, 1988 ▶).
Figure 4.
MALDI–TOF mass spectrometry of CLBPah. The profile shows the molecular weight of CLBPah to be 7005 Da.
3.2. Biological characterization
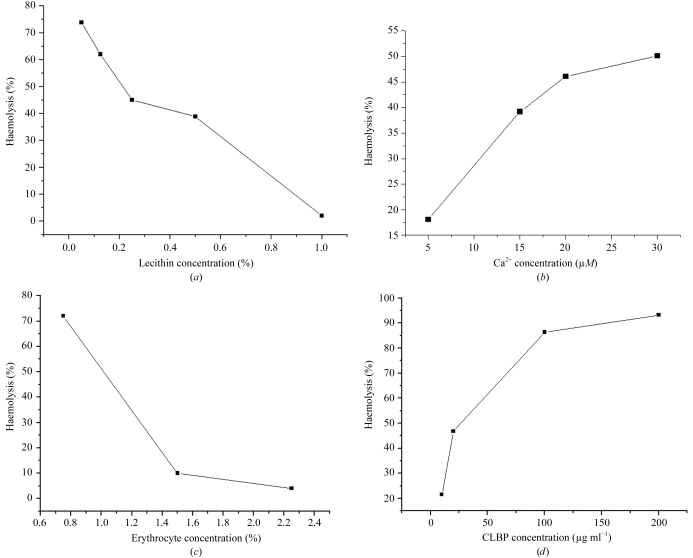
No CLBPah-mediated haemolysis was observed when the medium did not contain lecithin. Haemolytic activity was observed at low lecithin concentrations (0.005−0.1%) and decreased when the lecithin concentration was increased (Fig. 5 ▶ a). When the concentration of lecithin was increased to 1.0%, the rate of haemolysis dropped to 2.0%, which was 1/40th of that observed at a concentration of 0.005%. Fig. 5 ▶(b) shows that the percentage of haemolysis increases with increasing Ca2+ concentration over the range 5–30 µM. Fig. 5 ▶(c) shows that the percentage of haemolysis is related to the concentration of erythrocytes. When the concentration of erythrocytes is increased, the percentage of haemolysis decreases. Fig. 5 ▶(d) shows that the percentage of haemolysis increases nonlinearly with CLBPah concentration.
Figure 5.
(a) The effect of the concentration of lecithin on percentage haemolysis. The reaction medium contains 1.5%(v/v) erythrocytes, 10 µM Ca2+ and 50 µg CLBPah. The reaction was incubated at 310 K for 15 min. (b) The effect of Ca2+ on the percentage of haemolysis. The effect of Ca2+ was determined by using four Ca2+ concentrations. The medium consisted of 2.25%(v/v) erythrocytes, 0.025% lecithin and 50 µg CLBPah. The time and temperature of reaction were 15 min and 310 K, respectively. (c) The effect of erythrocytes on the percentage of haemolysis. Three concentrations of erythrocytes were examined. The medium contains 0.025% lecithin, 10 µM Ca2+ and 50 µg CLBPah. The temperature and time of reaction were 310 K and 15 min, respectively. (d) The effect of concentration of CLBPah on the percentage of haemolysis. The concentration of erythrocytes was 2.25%(v/v) and the concentration of lecithin was 0.025%. The concentration of Ca2+ was 10 µM.
In our experiments, the percentage of haemolysis increased with increasing concentrations of Ca2+, erythrocytes and CLBPah. Small quantities of lecithin are required for CLBPah-mediated haemolysis, but large amounts of lecithin inhibit haemolysis. We speculate that low concentrations of lecithin might be required for the formation of a continuous hydrophobic column by the three-finger loops in CLBPs that can penetrate the surface of erythrocytes and induce haemolysis (Sun et al., 1997 ▶). However, increasing lecithin concentrations might saturate the lipid-binding ability of CLBPs and prevent their attachment to the surface of erythrocytes.
Takechi et al. (1985 ▶) estimated that the LD50 of purified CLBP in mice after subcutaneous injection was 60 µg g−1. In contrast, the LD50 of CLBPah in mice was 9.7 µg g−1 within 24 h. The toxic activity of CLBPah is therefore greater than that of CLBP, but is lower than that of cardiotoxins and neurotoxins.
3.3. Preliminary X-ray diffraction analysis
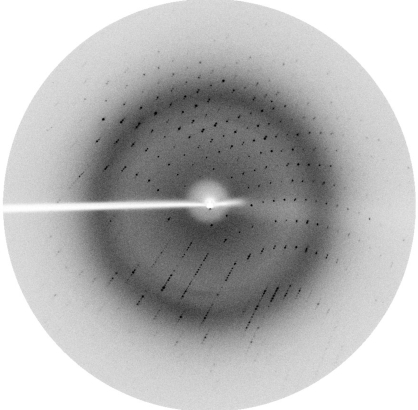
Calculation of the Matthews coefficient (Matthews, 1968 ▶) gave a volume-to-weight ratio V M of 2.746 Å3 Da−1. This suggests the presence of two monomers in the asymmetric unit. Data-collection and reduction statistics are summarized in Table 1 ▶. The diffraction pattern of CLBPah can be seen in Fig. 6 ▶.
Table 1. Diffraction data-collection and reduction statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P41212/P43212 |
| Unit-cell parameters | |
| a (Å) | 43.5 |
| b (Å) | 43.5 |
| c (Å) | 145.1 |
| No. of reflections | 38726 (3350) |
| No. of independent reflections | 6088 |
| Resolution limits (Å) | 20–2.35 (2.40–2.35) |
| Rmerge (%) | 6.6 (9.0) |
| Completeness (%) | 99.6 (95.4) |
| Redundancy | 6.4 |
| Mosaicity (°) | 0.302 |
| I/σ(I) | 9.8 (3.4) |
Figure 6.
Diffraction pattern of the crystal of CLBPah.
3.4. Molecular replacement
Molecular replacement was performed using the MOLREP software (Vagin & Teplyakov, 1997 ▶). The best result was obtained using PDB code 1kxi monomer A as an initial model. We found two monomers per asymmetric unit in the CLBPah crystal structure. The rotation and translation functions yielded a best result with a correlation coefficient and R factor of 40 and 47%, respectively. The model produced was then submitted to initial crystallographic refinement using REFMAC5 (Collaborative Computational Project, Number 4, 1994 ▶). The preliminary refinement was performed using rigid-body refinement followed by restrained refinement (maximum-likelihood method), resulting in a model with an R factor of 30% and an R free of 36.2%.
The sequencing of all amino-acid residues and complete structural refinement of CLBPah are being carried out in our laboratory, which will provide further insight into the structural features of CLBPs and may help in the study of the evolutionary relationship between cardiotoxins and CLBPs.
Acknowledgments
This work was supported by research grants from the Chinese National Natural Science Foundation (grant Nos. 30121001, 30025012 and 30571066), the ‘973’ and ‘863’ Projects of the Chinese Ministry of Science and Technology (grant Nos. 2004CB520801 and 2002BA711A13) and the Chinese Academy of Sciences (grant No. KSCX1-SW-17).
References
- Aird, S. D. & Kaiser, I. I. (1985). Toxicon, 23, 361–374. [DOI] [PubMed] [Google Scholar]
- Brattstrom, D., Bergqvist, M., Hesselius, P., Larsson, A., Wagenius, G. & Brodin, O. (2004). Lung Cancer, 43, 55–62. [DOI] [PubMed] [Google Scholar]
- Chang, L. S., Lin, S. K. & Chung, C. (2004). Biochem. Genet.42, 429–440. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Corpet, F. (1988). Nucleic Acids Res.16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufton, M. J. & Hider, R. C. (1991). Snake Toxins, edited by A. L. Harvey, pp. 259–302. New York: Pergamon Press.
- Endo, T., Tamiya, N. & Harvey, A. L. (1991). Snake Toxins, edited by A. L. Harvey, pp. 165–222. New York: Pergamon Press.
- Eno, A. E., Konya, R. S. & Ibu, J. O. (1998). Toxicon, 36, 2013–2020. [DOI] [PubMed] [Google Scholar]
- Fletcher, J. E. & Jiang, M. S. (1993). Toxicon, 31, 669–695. [DOI] [PubMed] [Google Scholar]
- Harvey, A. L. (1985). J. Toxicol. Toxin Rev.4, 41–49.
- Komorowski, J., Jankewicz, J. & Stepien, H. (2000). Cytobios, 101, 151–159. [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Menez, A. (1998). Toxicon, 36, 1557–1572. [DOI] [PubMed] [Google Scholar]
- Menez, E., Gatineau, E., Roumestand, C. & Harvey, A. L. (1990). Biochimie, 72, 575–588. [DOI] [PubMed] [Google Scholar]
- Sivaraman, T., Kumar, T. K. S., Yang, P. W. & Yu, C. (1997). Toxicon, 35, 1367–1371. [DOI] [PubMed] [Google Scholar]
- Sun, Y. J., Wu, W. G., Chiang, C. M., Hsin, A. Y. & Hsiao, C. D. (1997). Biochemistry, 36, 2403–2413. [DOI] [PubMed] [Google Scholar]
- Takechi, M., Tanaka, Y. & Hayashi, K. (1985). Biochem. Int.11, 795–802. [PubMed] [Google Scholar]
- Tsetlin, V. I. (1999). Eur. J. Biochem.264, 281–286. [DOI] [PubMed] [Google Scholar]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025. [Google Scholar]
- Yu, Q. S., Liu, X. Y., Qiu, Y. & Hang, S. (2005). Clin. Pharmacol. Exp. Ther. Asia, 2, 13–16.