In situ proteolysis with fungal protease or subtilisin is crucial for the crystallization of murine CstF-77.
Keywords: CstF-77, in situ proteolysis
Abstract
The cleavage-stimulation factor (CstF) is required for the cleavage of the 3′-end of messenger RNA precursors in eukaryotes. During structure determination of the 77 kDa subunit of the murine CstF complex (CstF-77), it was serendipitously discovered that a solution infected by a fungus was crucial for the crystallization of this protein. CstF-77 was partially proteolyzed during crystallization; this was very likely to have been catalyzed by a protease secreted by the fungus. It was found that the fungal protease can be replaced by subtilisin and this in situ proteolysis protocol produced crystals of sufficient size for structural studies. After an extensive search, it was found that 55% glucose can be used as a cryoprotectant while maintaining the diffraction quality of the crystals; most other commonly used cryoprotectants were detrimental to the diffraction quality.
1. Introduction
The cleavage-stimulation factor (CstF) plays a crucial role in the 3′-end processing of mRNA precursors (pre-mRNAs; Colgan & Manley, 1997 ▶; Zhao et al., 1999 ▶; Shatkin & Manley, 2000 ▶). This complex contains three subunits: CstF-50 (with molecular weight of 50 kDa), CstF-64 and CstF-77. CstF-64 contains an RNA-recognition motif (RRM) that helps the binding of the pre-mRNA substrate by the processing machinery (Canadillas & Varani, 2003 ▶; Deka et al., 2005 ▶). The CstF-77 subunit contains a HAT (half a TPR) domain at its N-terminus (approximately residues 1–550; Preker & Keller, 1998 ▶), followed by a proline-rich segment (approximately residues 560–630). The HAT domain may be related to the tetratricopeptide-repeat (TPR) domain (Preker & Keller, 1998 ▶; Goebl & Yanagida, 1991 ▶) and is expected to mediate protein–protein interactions (Lamb et al., 1995 ▶).
Currently, only the structure of the RRM of CstF-64 has been determined (Canadillas & Varani, 2003 ▶) and there is no structural information on the other components of the CstF complex. To help understand the molecular basis of the functional roles of these proteins, we initiated structural studies on the CstF-77 subunit. Here, we describe some of the special features of the crystallization of this protein, especially our discovery that in situ proteolysis with a fungal protease or subtilisin is essential for the crystallization of this protein. In addition, we found that the crystals were damaged by most common cryoprotectants; an extensive search identified 55% glucose as the only cryoprotectant that maintains the diffraction quality of the crystals.
2. Materials and methods
2.1. Protein expression and purification
Various segments of the HAT domain of mouse CstF-77 were subcloned into the pET28a vector (Novagen), overexpressed in Escherichia coli at 293 K and purified by nickel-agarose affinity chromatography (Ni–NTA, Qiagen) followed by gel-filtration chromatography (S300, GE Healthcare). The protein was concentrated in a buffer containing 20 mM Tris pH 8.5, 200 mM NaCl, 5%(v/v) glycerol and 5 mM DTT. The protein concentration was determined by the Bradford method. The expression constructs contain a hexahistidine tag at either the N- or the C-terminus.
2.2. Identification of a proteolysis product by mass spectrometry
The purified HAT domain of murine CstF-77 (residues 10–600) was incubated for 20 h at 277 K with subtilisin (1000:1 protein:subtilisin ratio). Sample aliquots were taken at various time points and analyzed by SDS–PAGE. The band corresponding to the stable domain obtained after overnight incubation (Fig. 1 ▶) was excised from the gel and washed for 1 h with 500 µl 100 mM NH4HCO3. The gel slice was then incubated for 30 min at 333 K with 150 µl 100 mM NH4HCO3 and 10 µl 45 mM DTT. After cooling the solution to room temperature, 10 µl 100 mM iodoacetamide was added and the mixture was incubated for 30 min in the dark at room temperature. The solvent was discarded and the gel slice was washed with 500 µl 50%(v/v) acetonitrile/100 mM NH4HCO3 for 1 h. The gel slice was then shrunk with 150 µl acetonitrile for 15 min and dried in a rotating evaporator. 10 µl (enough to cover the gel slice) of 25 mM NH4HCO3 containing modified trypsin (Promega; 10:1 protein:trypsin ratio) was added to the gel slice and the mixture was incubated overnight at 310 K. The resulting solution was analyzed by MALDI–TOF mass spectrometry on a Voyager-DE Pro instrument (PerSeptive Biosystems) using α-cyano-4-hydroxycinnamic acid as the matrix.
Figure 1.
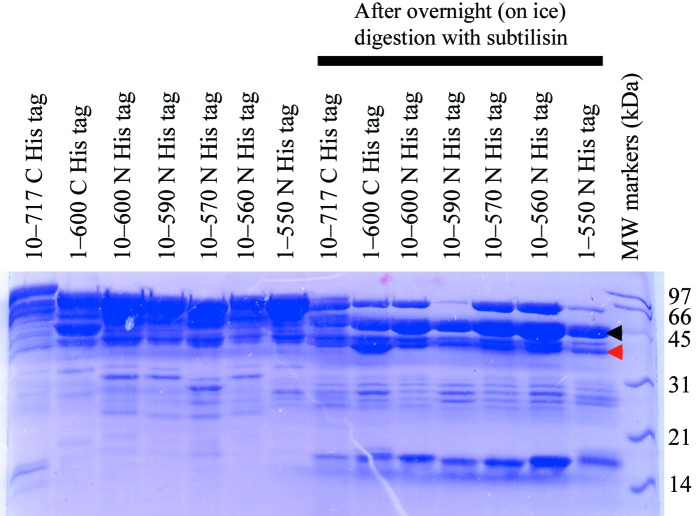
Limited proteolysis of murine CstF-77 with subtilisin. The various protein samples are labeled. After overnight incubation on ice in the presence of a 1000:1 weight ratio of subtilisin, a stable domain is observed, indicated by the black arrowhead. This band was excised for identification by mass spectrometry. The domain that ultimately crystallized is indicated by the red arrowhead.
2.3. Protein crystallization
Crystals of murine CstF-77 were obtained by the sitting-drop vapor-diffusion method. The reservoir solution contained 200 mM sodium tartrate and 20%(w/v) PEG 3350. The protein was at 6 mg ml−1 concentration and the drops also contained 10 mM EDTA and subtilisin (at 5000:1 protein:subtilisin ratio). Details of the crystallization procedures are described in §3.
The crystals were cryoprotected with 55% glucose and were then flash-frozen in liquid nitrogen for diffraction analysis and data collection at 100 K. X-ray diffraction data were collected on an ADSC CCD at the X4A beamline of the National Synchrotron Light Source.
3. Results and discussion
3.1. Protein samples containing the entire HAT domain are aggregated in solution
Based on the domain structure of CstF-77, our initial expression constructs covered the entire HAT domain (approximately residues 1–550) and various fragments of the proline-rich segment (residues 560–630) (Table 1 ▶). Sequence analysis suggests that the HAT domain probably forms a contiguous structure (Preker & Keller, 1998 ▶) and therefore we did not design any constructs that contained fragments of this domain. Most of the constructs produced soluble protein in bacteria and we carried out large-scale purifications on all of them (Table 1 ▶). Unfortunately, gel-filtration chromatography showed that they all had various degrees of aggregation in solution, with some of the protein migrating in the void volume of the gel-filtration column. Such protein samples are not suitable for crystallization and structural analysis.
Table 1. Bacterial expression constructs for murine CstF-77.
| His-tag position | Start residue (range) | End residue (range) | No. of constructs made | No. of strongly soluble constructs | No. of proteins purified | No. showing aggregation |
|---|---|---|---|---|---|---|
| Constructs covering the entire HAT domain | ||||||
| N-terminus | 110 | 540717 | 14 | 5 | 5 | 5 |
| C-terminus | 110 | 560717 | 12 | 4 | 4 | 4 |
| Constructs designed based on the mass-spectrometry data | ||||||
| N-terminus | 200 | 540650 | 5 | 3 | 2 | 0 |
| C-terminus | 200 | 540650 | 5 | 0 | ||
| N-terminus | 150 | 560600 | 3 | 1 | 0 | |
| C-terminus | 150 | 560600 | 3 | 1 | 0 | |
| Constructs designed based on the crystal structure | ||||||
| N-terminus | 241 | 550 | 1 | 1 | 1 | 1 |
3.2. A stable segment was identified by limited proteolysis and mass spectrometry
We next carried out limited proteolysis in order to search for a stable fragment, with the expectation that such a fragment would behave well in solution. Time courses were taken using chymotrypsin, trypsin and subtilisin as the protease and we found that subtilisin (1000:1 protein:subtilisin ratio) gave the best results (Fig. 1 ▶). A stable domain of approximately 45 kDa molecular weight was identified which appeared to remain as the major species even after overnight proteolysis with subtilisin (Fig. 1 ▶). Moreover, this domain could be produced using any of our purified proteins containing different amounts of extension of the HAT domain (Fig. 1 ▶). The SDS gel suggests that between 20 and 30 kDa is removed from the protein samples by the subtilisin treatment.
We next used MALDI–TOF mass spectrometry (MS) to characterize the proteolysis product in more detail. The band corresponding to the domain was excised from the gel and subjected to trypsin digest. Several tryptic peptides were identified from the MS analysis, corresponding to residues 203–207, 210–215, 253–258, 357–363, 468–485, 506–527 and 541–551 of murine CstF-77. This suggests that the domain may contain the C-terminal segment of the protein, as no peptides were found from the N-terminal segment. Based on the peptides and the molecular weight of this domain, we estimated that approximately the first 200 residues of the protein were removed by the subtilisin treatment. This was somewhat unexpected, as the cleavage site is then located in the middle of the HAT domain.
According to the MS analysis, we designed a new series of constructs covering the second half of the HAT domain. The N-terminus of these constructs started at residue 150 or 200 of CstF-77. Satisfyingly, we found that the construct covering residues 200–600, with an N-terminal His tag, produced a large amount of soluble protein (Table 1 ▶). More importantly, the protein behaved very well in solution, with no sign of aggregation.
3.3. Crystallization required in situ proteolysis
Potential crystallization conditions for the protein sample covering residues 200–600 were screened at 294 K using several commercially available kits (Hampton Research, Emerald Biosystems and others; Jancarik & Kim, 1991 ▶; Cudney et al., 1994 ▶). A few conditions from the PEG/Ion screen (Hampton Research) produced medium-sized crystals (Fig. 2 ▶).
Figure 2.
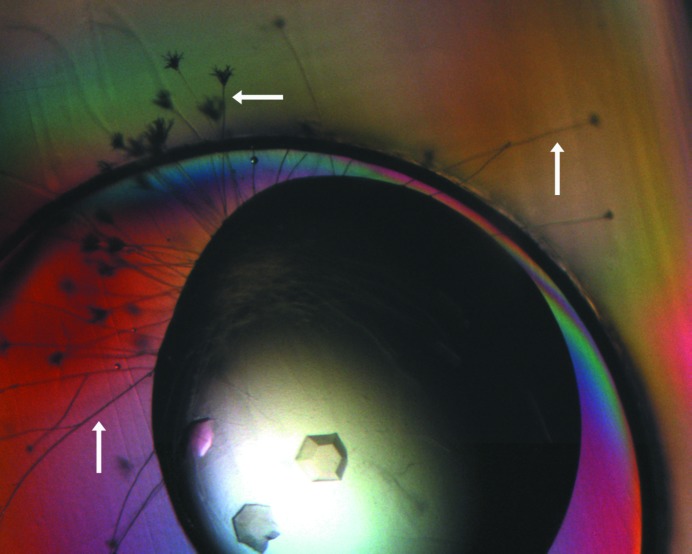
Crystals of murine CstF-77 (residues 200–600). The crystals were obtained from a commercial screen condition. Fungal growth is visible around the drop and is indicated by the arrows.
However, we were not able to reproduce these initial hits using homemade solutions. We then realised that the commercial screen solution was infected by a fungus. (This solution had been in use in the laboratory for nearly 2 y.) In fact, fungal growth could be observed around the drops that produced crystals of the protein (Fig. 2 ▶). Our previous experience with another protein (Mandel, Gebauer et al., 2006 ▶; Mandel, Kaneko et al., 2006 ▶) suggested that the CstF-77 protein sample may have been cleaved by a protease that is secreted by the fungus and that this cleavage is required for the crystallization of the protein. An SDS gel on the crystal confirmed our suspicion, showing that the protein in the crystal is smaller than the sample purified from bacteria (Fig. 3 ▶). The molecular weight of the protein in the crystal is less than 45 kDa (Fig. 3 ▶), smaller than the major stable species after subtilisin treatment, but may instead correspond to one of the minor species there (Fig. 1 ▶).
Figure 3.
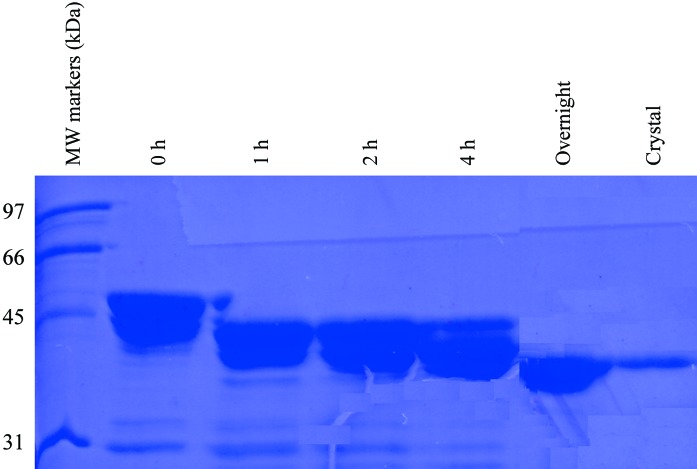
SDS gel of crystals of murine CstF-77. A subtilisin-digestion time course with the protein sample covering residues 200–600 is also shown. The lane labeled ‘Crystal’ is from crystals grown in the presence of the fungus.
These observations demonstrate that the fragment 200–600 is still larger than the stable domain and that further proteolysis is required before the protein can be crystallized. Therefore, we performed a time course with this new protein sample, again using subtilisin as the protease. Interestingly, subtilisin can produce a stable species with this sample that is essentially the same size as the fragment in the crystal (Fig. 3 ▶). This species is smaller than that generated from proteolysis of the entire HAT domain (Fig. 1 ▶), possibly because the N-terminal segment in the 200–600 fragment is no longer stabilized. We carried out a preparatory scale proteolysis experiment with subtilisin (1000:1 protein:subtilisin ratio) as the protease. However, the resulting protein appeared to have reduced solubility. While this sample could be crystallized, we only obtained showers of small crystals.
We next tried to carry out the proteolysis in situ, but we replaced the fungus with subtilisin in order to have more control over the process. We added subtilisin directly to the crystallization drop, at a ratio of 5000:1(w:w) (protein:subtilisin). With this protocol, we were able to crystallize the protein using homemade solutions. Further optimization produced crystals that were of sufficient size for crystallographic analyses. The reservoir solution contained 200 mM sodium tartrate and 20%(w/v) PEG 3350.
3.4. An extensive search for a suitable cryoprotectant
The crystals of CstF-77 have sharp faces and edges (Fig. 2 ▶) and we expected them to produce good-quality X-ray diffraction. Surprisingly, the diffraction patterns that we did obtain showed poor resolution and very high mosaicity. These crystals were cryoprotected with reservoir solution supplemented with PEG 3350. We then screened other commonly used cryoprotectants, such as ethylene glycol, PEG, PEG MME, PEG DME and Paratone. Short soaking and/or slow equilibration into the cryoprotectant solution were used. Some of these damaged the crystal and we did not observe any improvement in diffraction quality with the others.
To assess whether the poor diffraction quality is truly a feature of the crystals themselves, we mounted a crystal in a capillary and examined its X-ray diffraction at room temperature without cryoprotection. We obtained a diffraction pattern that showed good resolution and very low mosaicity, suggesting that the diffraction problem arose from the cryoprotection. Therefore, we screened a much wider range of cryoprotectants, including additional precipitants, different organic solvents, high concentrations of various salts and various sugar solutions, and finally found that our crystals could be cryoprotected by 55%(w/v) glucose (in water) without loss of diffraction quality. The crystals were soaked in this solution for 30 s and then frozen in liquid nitrogen for data collection.
3.5. Residues removed by proteolysis are incompatible with crystal packing
High-quality X-ray diffraction data were collected to 2.8 Å resolution at the X4A beamline of the National Synchrotron Light Source. The structure of this domain has been determined by the selenomethionyl single-wavelength anomalous diffraction (SAD) method (Hendrickson, 1991 ▶), the details of which will be presented elsewhere (Bai et al., 2007 ▶).
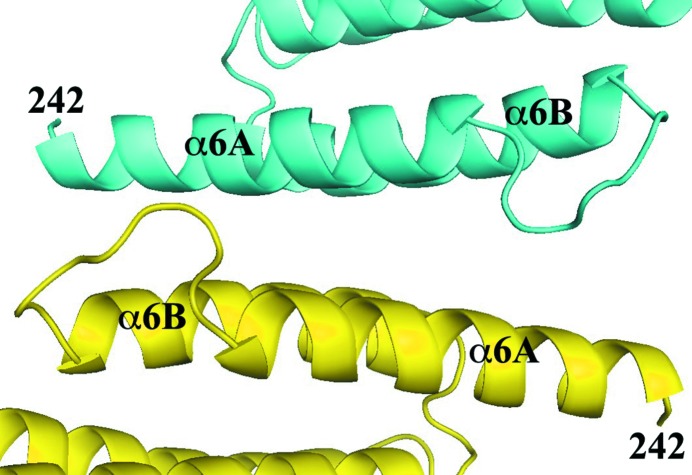
The crystallographic analysis showed that residues 200–241 and 550–600 have no electron density. Both of these segments were probably removed by subtilisin during crystallization. Residues 242–549 have a molecular weight of 36 kDa, which is consistent with the SDS gel on the crystal (Fig. 3 ▶). Moreover, residues 242–259 are involved in crystal packing, which would not allow the presence of additional residues at the N-terminus (Fig. 4 ▶). This may explain why proteolysis is required for the crystallization of this protein.
Figure 4.
The N-terminus of the protein is involved in crystal packing. Two molecules of CstF-77 related by a crystallographic twofold axis are shown in cyan and yellow, respectively.
With the identification of residues 242 and 549 as the accurate boundaries for this domain based on the structure, we created the bacterial expression construct for this domain (Table 1 ▶). Somewhat surprisingly, this construct produced only a small amount of soluble protein in E. coli. Moreover, we observed aggregation behavior for the purified protein. It is not clear why this protein sample does not behave well in solution. The presence of the N-terminal His tag, with 20 residues, may be part of the reason.
4. Conclusions
Our SDS–PAGE and MS peptide fingerprinting only produced rough boundaries for the stable domain. More accurate definition of the boundaries could be made using N-terminal sequencing coupled with MS on the undigested protein in order to obtain the accurate molecular weight. However, our experiences with the construct designed based on the final crystal structure (Table 1 ▶) suggest that, at least in this case, making the construct that corresponds to the exact boundary might not be advantageous. The additional residues outside the domain may be beneficial for the solubility and bacterial expression of the protein.
Partial proteolysis to remove flexible segments is an important component in the crystallization of many protein samples. In most cases, the proteolysis is carried out separately from the crystallization experiments and the stable domain is produced by bacterial expression from a re-engineered construct or by preparative-scale proteolysis. Our successes with CstF-77 and CPSF-100 (Mandel, Gebauer et al., 2006 ▶; Mandel, Kaneko et al., 2006 ▶) suggest that in situ proteolysis may be another useful protocol for protein crystallization. Recent reports have shown this protocol to be successful with several other proteins (Taneja et al., 2006 ▶; Johnson et al., 2006 ▶; Gaur et al., 2004 ▶).
Acknowledgments
We thank Randy Abramowitz and John Schwanof for setting up the X4A beamline at the NSLS. This research was supported in part by a grant from the NIH.
References
- Bai, Y., Auperin, T. C., Chou, C.-Y., Chang, G.-G., Manley, J. L. & Tong, L. (2007). In the press. [DOI] [PubMed]
- Canadillas, J. M. P. & Varani, G. (2003). EMBO J. 22, 2821–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan, D. F. & Manley, J. L. (1997). Genes Dev. 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- Cudney, R., Patel, S., Weisgraber, K., Newhouse, Y. & McPherson, A. (1994). Acta Cryst. D50, 414–423. [DOI] [PubMed] [Google Scholar]
- Deka, P., Rajan, P. K., Canadillas, J. M. P. & Varani, G. (2005). J. Mol. Biol. 347, 719–733. [DOI] [PubMed] [Google Scholar]
- Gaur, R. K., Kupper, M. B., Fischer, R. & Hoffmann, K. M. V. (2004). Acta Cryst. D60, 965–967. [DOI] [PubMed] [Google Scholar]
- Goebl, M. & Yanagida, M. (1991). Trends Biochem. Sci. 16, 173–177. [DOI] [PubMed] [Google Scholar]
- Hendrickson, W. A. (1991). Science, 254, 51–58. [DOI] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411. [Google Scholar]
- Johnson, S., Roversi, P., Espina, M., Deane, J. E., Birket, S., Picking, W. D., Blocker, A., Picking, W. L. & Lea, S. M. (2006). Acta Cryst. F62, 865–868. [DOI] [PMC free article] [PubMed]
- Lamb, J. R., Tugendreich, S. & Hieter, P. (1995). Trends Biochem. Sci. 20, 257–259. [DOI] [PubMed] [Google Scholar]
- Mandel, C. R., Gebauer, D., Zhang, H. & Tong, L. (2006). Acta Cryst. F62, 1041–1045. [DOI] [PMC free article] [PubMed]
- Mandel, C. R., Kaneko, S., Zhang, H., Gebauer, D., Vethantham, V., Manley, J. L. & Tong, L. (2006). Nature (London), 444, 953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker, P. J. & Keller, W. (1998). Trends Biochem. Sci. 23, 15–16. [DOI] [PubMed] [Google Scholar]
- Shatkin, A. J. & Manley, J. L. (2000). Nature Struct. Biol. 7, 838–842. [DOI] [PubMed] [Google Scholar]
- Taneja, B., Patel, A., Slesarev, A. & Mondragon, A. (2006). EMBO J. 25, 398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L. & Moore, C. L. (1999). Microbiol. Mol. Biol. Rev. 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]