DHNA synthetase from G. kaustophilus has been cloned, expressed, purified and crystallized.
Keywords: thermophilic microorganisms, vitamin K2, Geobacillus kaustophilus
Abstract
The aerobic Gram-positive bacterium Geobacillus kaustophilus is a bacillus species that was isolated from deep-sea sediment from the Mariana Trench. 1,4-Dihydroxy-2-naphthoate (DHNA) synthetase plays a vital role in the biosynthesis of menaquinone (vitamin K2) in this bacterium. DHNA synthetase from Geobacillus kaustophilus was crystallized in the orthorhombic space group C2221, with unit-cell parameters a = 77.01, b = 130.66, c = 131.69 Å. The crystal diffracted to a resolution of 2.2 Å. Preliminary studies and molecular-replacement calculations reveal the presence of three monomers in the asymmetric unit.
1. Introduction
Interest in thermophilic microorganisms that live and thrive at high temperatures of up to or above 373 K has been increasing in recent decades (Andrade et al., 1999 ▶; Singleton & Amelunxen, 1973 ▶). One thermophilic organism of immense research interest is Geobacillus kaustophilus, a bacillus species isolated from deep-sea sediment from the Mariana Trench (Takami et al., 1997 ▶). It is an aerobic endospore-forming Gram-positive bacterium that grows optimally at 333 K with an upper temperature limit of 347 K (Takami et al., 2004 ▶). Vitamin K2 (menaquinone) plays a vital role as an electron carrier in this bacterium (Bentley & Meganathan, 1982 ▶; Johnston et al., 2005 ▶). Quinones serve as a source of energy and are of fundamental importance to all life forms (Bentley & Meganathan, 1982 ▶). They occur as ubiquinone in the mitochondria of animals, plastoquinone in the chloroplasts of plants and menaquinone in bacteria. A key reaction in the biosynthesis of menaquinone involves the conversion of O-succinylbenzoyl-CoA to 1,4-dihydroxy-2-;naphthoyl-CoA (Truglio et al., 2003 ▶). 1,4-Dihydroxy-2-naphthoate synthetase (MenB), abbreviated as DHNA synthetase (EC 4.1.3.36), is believed to be responsible for cyclization in the reaction (Meganathan & Bentley, 1981 ▶; Sharma et al., 1992 ▶).
Menaquinone has been found to be an important product of metabolism in many bacterial species (Bentley & Meganathan, 1982 ▶). Inactivation of the genes encoding the menaquinone-biosynthetic enzymes has been found to produce a demand for menaquinone in the growth medium (Truglio et al., 2003 ▶). Thus, the enzymes involved in the menaquinone-biosynthesis pathway play a significant role in the metabolism of bacterial species. Understanding the three-dimensional crystal structures of enzymes from the Gram-positive bacterium G. kaustophilus will be of great significance in the field of thermophile research. Intensive research needs to be performed to study the thermostability of macromolecules and to screen new thermophiles for biotechnological processes. The recent focus has been on the study of metabolic pathways and mechanisms involved in the regulation of thermophilic proteins. Furthermore, insight into the crystal structure may shed light on the design of a potent inhibitor for the protein.
2. Materials and methods
2.1. Cloning, expression and purification of the protein
The DHNA gene (GK2873) was amplified by PCR using G. kaustophilus HTA426 genomic DNA as the template. The amplified fragment was cloned under the control of the T7 promoter of the Escherichia coli expression vector pET-HisTEV, with the tobacco etch virus (TEV) protease-recognition site, Glu-Asn-Leu-Tyr-Phe-Gln-Gly, instead of the thrombin-recognition site of the pET-15b vector (Novagen, Madison, WI, USA). The expression vector was introduced into E. coli BL21-CodonPlus(DE3)-RIL strain (Stratagene, La Jolla, CA, USA) and the recombinant strain was cultured in 2.5 l LB broth supplemented with 50 µg ml−1 ampicillin. The cells (9.6 g) were collected by centrifugation, washed with 50 ml buffer A (20 mM Tris–HCl pH 8.0) containing 0.5 M NaCl, 5 mM 2-mercaptoethanol and 1 mM phenylmethylsulfonyl fluoride and resuspended in 25 ml of the same buffer. The cells were then disrupted by sonication in a chilled water bath and the cell lysate was incubated at 343 K for 13 min. The sample was centrifuged at 15 000g for 30 min and the supernatant was applied onto a HisTrap Chelating HP5 Superdex 200 column (GE Healthcare Bioscience Corp., Piscataway, NJ, USA) pre-equilibrated with buffer A containing 0.5 M NaCl and 20 mM imidazole, which was eluted with a linear gradient of 0.02–0.5 M imidazole. The sample containing DHNA synthetase was then loaded onto a HiLoad 16/60 Superdex 200 column (GE Healthcare Bioscience Corp.) pre-equilibrated with buffer A containing 0.5 M NaCl and 20 mM imidazole. The eluted fractions containing DHNA synthetase were collected and treated with TEV protease at 303 K for 1 h. The protein sample was applied onto a HisTrap Chelating HP5-Superdex 200 column (GE Healthcare Bioscience Corp.) pre-equilibrated with buffer A containing 0.5 M NaCl and 20 mM imidazole. The flowthrough fraction was collected and the purified protein was loaded onto a HiPrep 26/10 desalting column, which was eluted with buffer A containing 0.2 M NaCl.
2.2. Crystallization experiments
The protein was concentrated using a Vivaspin 20 concentrator (10 kDa molecular-weight cutoff, Sartorius, AG, Göttingen, Germany). The protein concentration was determined by measuring the absorbance at 280 nm (Kuramitsu et al., 1990 ▶). The concentration of the purified protein was ∼10 mg ml−1 in 20 mM Tris–HCl pH 8.0 and 200 mM NaCl. Preliminary crystallization screening was carried using the Index kit (Hampton Research). Sitting-drop vapour-diffusion experiments were set up by pipetting drops consisting of 1 µl protein solution and 1 µl well solution. The well solution contained 30%(v/v) pentaerythritol ethocylate, 0.05 M ammonium sulfate and 0.05 M bis-tris pH 6.5. Crystals were obtained when 1 µl protein solution was mixed with 1 µl well solution and allowed to equilibrate against 100 µl well solution at 293 K. 50%(v/v) Paratone-N and 50%(v/v) paraffin oil were used as a cryoprotectant. Crystals (Fig. 1 ▶) appeared in around 20 d.
Figure 1.
Crystal of DHNA synthetase from G. kaustophilus (C-centred orthorhombic).
2.3. Data collection and processing
The diffraction data were collected from a single crystal (approximately 0.2 × 0.2 × 0.2 mm in size) at 100 K using the RIKEN Structural Genomics Beamline II (BL26B2) at SPring-8 (Hyogo, Japan) with a Jupiter 210 CCD detector (Rigaku MSC Co., Tokyo, Japan). The crystal-to-detector distance was maintained at 230 mm. The program DENZO (Otwinowski, 1993 ▶) was used for data processing. The crystal diffracted to 2.2 Å resolution. The details of the diffraction data statistics are given in Table 1 ▶.
Table 1. X-ray diffraction data statistics of DHNA synthetase.
Values in parentheses are for the last resolution bin.
| Wavelength (Å) | 1.0 |
| Space group | C2221 |
| Temperature (K) | 100 |
| Unit-cell parameters (Å) | a = 77.01, b = 130.66, c = 131.69 |
| Matthews coefficient (Å3 Da−1) | 1.83 (3 subunits per ASU) |
| Solvent content (%) | 32.8 |
| Resolution range (Å) | 50.0–2.20 (2.28–2.20) |
| No. of observed reflections | 1320862 |
| No. of unique reflections | 33813 (3181) |
| Completeness (%) | 98.9 (94.2) |
| Rsym† (%) | 4.1 (8.1) |
| Multiplicity | 9.8 (6.5) |
| Average I/σ(I) | 39 (14) |
R
sym = 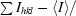
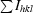 .
.
3. Results and discussion
Analysis of the diffracted intensities indicates that DHNA synthetase from G. kaustophilus belongs to the C-centred orthorhombic space group C2221, with unit-cell parameters a = 77.01, b = 130.66, c = 131.69 Å. The calculated Matthews coefficient (V M = 1.83 Å3 Da−1), corresponding to a solvent content of 32.8% (Matthews, 1968 ▶), indicates the presence of three monomers in the asymmetric unit. The X-ray diffraction data statistics are summarized in Table 1 ▶. The DHNA enzyme consists of 272 amino-acid residues with a subunit molecular weight of 30 kDa.
3.1. Molecular replacement
The crystal structure of DHNA synthetase from G. kaustophilus was solved by molecular-replacement calculations using the program CNS v.1.1 (Brünger et al., 1998 ▶). The coordinates of the search model used were those of MenB from Mycobacterium tuberculosis (PDB code 1rjn), which has 50% amino-acid sequence identity to DHNA synthetase from G. kaustophilus. Three monomers were located in the asymmetric unit and were subsequently subjected to rigid-body refinement using the program CNS v.1.1 (Brünger et al., 1998 ▶). 5% of the reflections were used for R free calculation (Brünger, 1992 ▶). The crystallographic R work and R free values of the partially refined structure were 36.6% and 38.1%, respectively.
Acknowledgments
SPK, CVR and KS thank the Bioinformatics Centre, the Interactive Graphics Based Molecular Modelling Facility and the Supercomputer Education and Research Centre. We thank Drs Satoru Watanabe and Ryoichi Arai for construction of the pET-HisTEV vector and Nagisa Takemoto for purification of the protein. We also thank Dr Koji Takio and Naoko Takahashi for the N-terminal amino-acid sequence analysis of the recombinant DHNA synthase. This work was supported by the RIKEN Structural Genomic/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Andrade, C. M. M. C., Pereira, N. Jr & Antranikian, G. (1999). Rev. Microbiol.30, 287–298.
- Bentley, R. & Meganathan, R. (1982). Microbiol. Rev.46, 241–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger, A. T. (1992). Nature (London), 355, 472–474. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Johnston, J. M., Arcus, V. L. & Baker, E. N. (2005). Acta Cryst. D61, 1199–1206. [DOI] [PubMed] [Google Scholar]
- Kuramitsu, S., Hiromi, K., Hayashi, H., Morino, Y. & Kagamiyama, H. (1990). Biochemistry, 29, 5469–5476. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Meganathan, R. & Bentley, R. (1981). Biochemistry, 20, 5336–5340. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. (1993). Proceedings of the CCP4 Study Weekend. Data Collection and Processing, edited by L. Sawyer, N. Isaacs & S. Bailey, pp. 56–62. Warrington: Daresbury Laboratory.
- Sharma, V., Suvarna, K., Meganathan, R. & Hudspeth, M. E. (1992). J. Bacteriol.174, 5057–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, R. Jr & Amelunxen, R. E. (1973). Bacteriol Rev.37, 320–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami, H., Inoue, A., Fuji, F. & Horikoshi, K. (1997). FEMS Microbiol. Lett.152, 279–285. [DOI] [PubMed] [Google Scholar]
- Takami, H., Nishi, S., Lu, J., Shimamura, S. & Takaki, Y. (2004). Extremophiles, 8, 351–356. [DOI] [PubMed] [Google Scholar]
- Truglio, J. J., Theis, K., Feng, Y., Gajda, R., Machutta, C., Tonge, P. J. & Kisker, C. (2003). J. Biol. Chem.278, 42352–42360. [DOI] [PubMed] [Google Scholar]



