1,3-Propanediol dehydrogenase from K. pneumoniae has been overexpressed in E. coli, purified and crystallized. Diffraction data have been collected to 2.7 Å resolution.
Keywords: 1,3-propanediol dehydrogenase; Klebsiella pneumoniae; glycerol metabolism; KES; opportunistic pathogens
Abstract
1,3-Propanediol dehydrogenase (1,3-PD-DH), encoded by the dhaT gene, is a key enzyme in the dissimilation process for converting glycerol to 1,3-propanediol in the human pathogen Klebsiella pneumoniae. Single colourless crystals were obtained from a recombinant preparation of 1,3-propanediol dehydrogenase overexpressed in Escherichia coli. The crystals belong to space group P21, with unit-cell parameters a = 91.9, b = 226.6, c = 232.6 Å, β = 92.9°. The crystals probably contain two decamers in the asymmetric unit, with a V M value of 3.07 Å3 Da−1 and an estimated solvent content of 59%. Diffraction data were collected to 2.7 Å resolution using synchrotron radiation at the ID14-4 beamline of the European Synchrotron Radiation Facility.
1. Introduction
Klebsiella pneumoniae belongs to the KES group of opportunistic pathogens (which also includes Enterobacter and Serratia species). It is an important cause of respiratory infections and is responsible for significant morbidity and mortality in immuno-compromised individuals (Carpenter, 1990 ▶; Podschun & Ullmann, 1998 ▶). Protein structures from this organism are being determined as part of the Structural Proteomics In Europe (SPINE) programme.
K. pneumoniae posseses an extremely active metabolism under microaerophilic conditions, being able to grow fermentatively using glycerol as the sole carbon source in a dissimilatory pathway (Huang et al., 2002 ▶). The central enzyme of glycerol metabolism is 1,3-propanediol dehydrogenase (1,3-PD-DH), which is encoded by the dhaT gene and is involved in the reductive pathway of anaerobic glycerol dissimilation, which is essential for the use of glycerol as a carbon source in the absence of an exogenous electron acceptor (Biebl et al., 1998 ▶; Johnson & Lin, 1987 ▶). The dhaT gene encodes a polypeptide of 387 amino acids with a calculated MW of 41 544 Da and a theoretical isoelectric point of 5.9. This enzyme is part of the dha regulon that encodes two parallel pathways. Through the oxidative pathway, glycerol is dehydrogenated by an NAD+-linked enzyme, glycerol dehydrogenase type I (glyDH-I), to dihydroxyacetone (DHA), which is then phosphorylated by an ATP-dependent kinase to dihydroxyacetone phosphate (DHAP; Cheng et al., 2006 ▶; Huang et al., 2002 ▶). Through the parallel reductive pathway, glycerol is dehydrated by a B12-dependent enzyme to form 3-hydroxypropionaldehyde, which is then reduced to 1,3-propanediol (1,3-PD) by the NADH-linked oxidoreductase 1,3-propanediol dehydrogenase (Cheng et al., 2004 ▶; O’Brien et al., 2004 ▶). The physiological reason for 1,3-PD formation could be the regeneration of NAD+ needed by the DHA branch of the pathway controlled by the dha regulon (Yuanyuan et al., 2004 ▶).
There are no structures available of 1,3-propanediol dehydrogenases. The most closely related structure to have been solved is that of lactaldehyde 1,2-propanediol oxidoreductase from Escherichia coli (PDB code 2bl4), which shares 44% sequence identity with 1,3-PD-DH.
2. Experimental
2.1. Protein expression and purification
The sequence of the dhaT gene coding for 1,3-propanediol dehydrogenase was amplified by the polymerase chain reaction (PCR) from K. pneumoniae genomic DNA using KOD Hot Start DNA polymerase (Novagen) and complementary gene-specific primers, to which were appended sequences to facilitate ligation-independent cloning (LIC; Aslanidis & de Jong, 1990 ▶). For LIC, the PCR-amplification products were treated with T4 DNA polymerase in the presence of dATP to generate 5′ single-stranded overhangs at both ends of the fragment through the enzyme’s combined 3′-5′ exonuclease and DNA polymerase activities. Complementary 5′ single-stranded overhangs were generated in the vector pET-YSBLIC by cleavage with the restriction endonuclease BseRI and treatment with T4 DNA polymerase in the presence of dTTP. This plasmid pET-YSBLIC is a pET28a (Novagen) derivative that has been adapted for LIC and adds an N-terminal hexahistidine tag to the cloned gene. The vector and PCR products were mixed and used to transform E. coli NovaBlue cells (Novagen). Recombinants were selected through kanamycin resistance and colony PCR using T7 promoter and gene-specific oligonucleotide primers to confirm the presence of a correctly sized insert; the DNA sequence was subsequently verified. The pET-YSBLICdhaT plasmid was transformed into E. coli BL21(DE3) for recombinant protein expression.
Six 100 ml cell cultures were grown to an OD600 of 0.6 at 310 K in Luria–Bertani broth containing 50 µg ml−1 kanamycin. Expression of the full native enzyme sequence fused directly to the amino-terminal sequence (MGSSHHHHHH) encoded by pET-YSBLIC was then induced by the addition of isopropyl β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. Cells were harvested by centrifugation and resuspended in 50 mM HEPES buffer pH 7.4 containing 1 M NaCl, 1 mM MnCl2 and 2 mM DTT (buffer A). The cells were disrupted in a French press at 131 MPa and the cell-free supernatant was collected by centrifugation at 30 000g in a Beckman JA-20 rotor. A 5 ml Hi-Trap chelating column (Amersham Biosciences) previously loaded with nickel and equilibrated with buffer A containing 20 mM imidazole was attached to an ÄKTA Explorer system (Amersham Biosciences). The soluble fraction was applied onto the column and washed with 30 ml buffer A containing 20 mM imidazole followed by 50 ml buffer A containing 50 mM imidazole. The protein was eluted with buffer A containing 500 mM imidazole and passed to a Sephacryl S-200 gel-filtration column (Amersham Pharmacia) pre-equilibrated in 50 mM HEPES pH 7.4 containing 150 mM NaCl, 1 mM MnCl2 and 2 mM DTT for size-exclusion chromatography. Fractions containing 1,3-PD-DH were identified by SDS–PAGE, pooled and concentrated by ultrafiltration to 55 mg ml−1 in the same buffer.
2.2. Crystallization
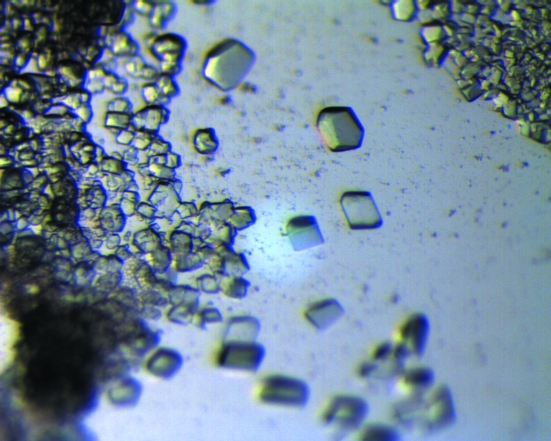
Automated crystallization screening was carried out at 295 K using sitting-drop vapour diffusion in 96-well plates (Greiner) using a Cartesian Minibee nanolitre pipetting robot (Genomic Solutions) and screens from Nextal Biotechnologies (Classics, PEGs, MbClass and MbClass II). Each crystallization drop contained 150 nl protein solution and 150 nl reservoir solution and was equilibrated against 150 µl reservoir solution. The most promising crystallization conditions were found to be 0.1 M MES pH 6.5 with 15%(w/v) PEG 20K. These conditions were optimized manually to 0.1 M MES pH 6.5 with 12%(w/v) PEG 20K. A screen of cationic additives added directly to the crystallization drops using the optimized conditions was also performed. The best results were obtained by the addition of CaCl2 to the drop to a final concentration of 10 mM (Fig. 1 ▶).
Figure 1.
1,3-PD-DH crystals grown in 0.1 M MES pH 6.5 with 12% PEG 20K, 10 mM CaCl2.
2.3. Data collection
Appropriate cryoprotecting conditions were obtained by soaking crystals for 1 min in crystallization solution with 25%(v/v) glycerol. Several data sets were collected using synchrotron radiation at DESY, Hamburg or at the ESRF, Grenoble. The highest resolution data collected to date (see statistics in Table 1 ▶) were obtained from a crystal with dimensions of 0.15 × 0.15 × 0.15 mm vitrified in liquid nitrogen and were measured at ESRF beamline ID14-4. Diffraction data were collected at 100 K using an Oxford Cryostream and an ADSC Q4 CCD detector. Diffraction images were processed with MOSFLM (Leslie, 2006 ▶) and experimental intensities were scaled with SCALA from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). The data statistics are summarized in Table 1 ▶.
Table 1. Diffraction data-processing statistics.
Values in parentheses are for the highest resolution shell.
| Source | ESRF ID14-4 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 91.44, b = 226.87, c = 232.34, β = 92.71 |
| Wavelength (Å) | 0.939 |
| No. of unique intensities | 257983 |
| Redundancy | 3.5 |
| Resolution (Å) | 80–2.7 (2.85–2.7) |
| Completeness (%) | 97.1 (92.2) |
| Rmerge† (%) | 8.3 (36.8) |
| I/σ(I) | 12.1 (3.1) |
R
merge = 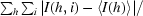
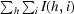 , where I(h, i) is the intensity of the ith measurement of reflection h and 〈I(h)〉 is the mean value of I(h, i) for all i measurements.
, where I(h, i) is the intensity of the ith measurement of reflection h and 〈I(h)〉 is the mean value of I(h, i) for all i measurements.
3. Results and discussion
The dhaT gene from the human pathogen K. pneumoniae was successfully cloned in E. coli and the encoded hexahistidine-tagged 1,3-PD-DH was purified to homogeneity by metal-chelating affinity chromatography. The best crystallization conditions included 12% PEG 20K as a precipitant agent and 10 mM CaCl2 as an additive. Crystals typically appeared in 8–24 h, reaching maximum dimensions of 200 × 200 × 300 µm. The direct addition of CaCl2 to the crystallization drop produced a significant reduction in crystal mosaicity.
Size-exclusion chromatography suggested that 1,3-PD-DH is a decamer in solution (data not shown). In consequence, and taking into account the presence of a peak in the self-rotation function corresponding to twofold noncrystallographic symmetry, the crystals probably contain two decamers in the asymmetric unit, with a V M value of 3.07 Å3 Da−1 and an estimated solvent content of 59%.
Structure solution by molecular replacement is currently under way, using several homologous structures present in the PDB database.
Acknowledgments
This research received funding from the European Commission under the SPINE project, contract No. QLG2-CT-2002-00988, under the RTD programme ‘Quality of Life and Management of Living Resources’. The authors would like to thank both the staff of the EMBL Outstation in Hamburg and of the ESRF for support with data collection and Dr Pedro Matias for help during data analysis. DM was funded by a Fundação para a Ciência e Tecnologia grant (SFRH/BD/13738/2003). DM and ATR were also supported by a travel grant from the SPINE project.
References
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res.18, 6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebl, H., Zeng, A. P., Menzel, K. & Deckwer, W. D. (1998). Appl. Microbiol. Biotechnol.50, 24–29. [DOI] [PubMed] [Google Scholar]
- Carpenter, J. L. (1990). Rev. Infect. Dis.12, 672–682. [DOI] [PubMed] [Google Scholar]
- Cheng, K. K., Liu, D. H., Sun, Y. & Liu, W. B. (2004). Biotechnol. Lett.26, 911–915. [DOI] [PubMed] [Google Scholar]
- Cheng, K. K., Zhang, J. A., Liu, D. H., Sun, Y., Yang, M. D. & Xu, J. M. (2006). Biotechnol. Lett.28, 1817–1821. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Huang, H., Gong, C. S. & Tsao, G. T. (2002). Appl. Biochem. Biotechnol.98–100, 687–698. [PubMed] [Google Scholar]
- Johnson, E. A. & Lin, E. C. (1987). J. Bacteriol.169, 2050–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed] [Google Scholar]
- O’Brien, J. R., Raynaud, C., Croux, C., Girbal, L., Soucaille, P. & Lanzilotta, W. N. (2004). Biochemistry, 43, 4635–4645. [DOI] [PubMed] [Google Scholar]
- Podschun, R. & Ullmann, U. (1998). Clin. Microbiol. Rev.11, 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuanyuan, Z., Yang, C. & Baishan, F. (2004). Biotechnol. Lett.26, 251–255. [DOI] [PubMed] [Google Scholar]