A crystal structure of ligand-free human glutamate carboxypeptidase II refined to 1.65 Å resolution is reported. The structure provides insight into the active-site of the enzyme in its unliganded state.
Keywords: prostate-specific membrane antigens, metallopeptidase, folate hydrolases, NAALADase
Abstract
Human glutamate carboxypeptidase II (GCPII; EC 3.4.17.21) is an established marker for prostate-cancer diagnosis as well as a candidate therapeutic target for the treatment of diverse pathologies that involve glutamatergic transmission. Structural data on GCPII are thus valuable for the design and optimization of GCPII-specific inhibitors and diagnostic probes. The currently available structure of ligand-free GCPII was refined to a resolution of 3.5 Å. This work reports the structure of the protein refined to 1.65 Å resolution, with crystallographic values of R = 0.207 and R free = 0.228. The new structure extends the resolution appreciably and the new model based on this data shows significant differences when compared with the previously published model.
1. Introduction
Human glutamate carboxypeptidase II (GCPII) is a type II membrane glycoprotein with an approximate molecular weight of 90–100 kDa. The mature full-length protein consists of 750 amino-acid residues. A short N-terminal fragment (residues 1–19), located in the cytoplasm, is followed by a single transmembrane segment and the 707-amino-acid ectodomain, which faces the extracellular milieu. GCPII is a homodimeric metallopeptidase that utilizes two zinc ions to cleave a C-terminal residue from peptidic substrates (Rawlings & Barrett, 1997 ▶). The only recognized natural substrates of GCPII to date are N-acetyl-aspartyl-glutamate (NAAG) and folyl-poly-γ-glutamates; i.e. peptides harboring glutamate at the C-terminus (Pinto et al., 1996 ▶; Robinson et al., 1987 ▶).
In healthy individuals, GCPII is expressed predominantly in the central nervous system and on the brush-border membrane of the proximal small intestine (Šácha et al., 2007 ▶; Troyer et al., 1995 ▶). In the latter compartment GCPII participates in the absorption of dietary folates (Halsted et al., 1998 ▶), while the neural form of the enzyme modulates neurotransmission by NAAG hydrolysis and the concomitant glutamate release (Robinson et al., 1987 ▶). Inhibition of GCPII peptidase activity showed efficacy under pathological conditions associated with dysregulated glutamatergic transmission, such as in experimental models of pain, brain injury or amyotrophic lateral sclerosis (for a recent review, see Neale et al., 2005 ▶ and references therein). Since GCPII expression is highly upregulated in the neovasculature of solid tumors as well as in prostate cancer (Bostwick et al., 1998 ▶), the enzyme is an obvious target for the treatment and imaging of these malignancies (Gao et al., 1997 ▶). Consequently, potent and selective small-molecule inhibitors or functionalized ligands of the enzyme are being developed and tested in preclinical settings (Zhou et al., 2005 ▶ and references therein).
The recently reported crystal structures of the human GCPII ectodomain in complex with glutamate, phosphate and GPI-18431 (PDB codes 2c6g, 2c6p and 2c6c, respectively) have revealed the presence of three GCPII subdomains, protease-like (residues 57–116 and 352–590), apical (residues 117–351) and C-terminal (residues 591–750), and it was noted that cooperation between all three subdomains is required for efficient substrate binding and hydrolysis. Furthermore, detailed analysis of the structures led to the hypothesis of an induced-fit mechanism of substrate recognition by GCPII, in which the β15/β16 hairpin (residues 692–704; termed the ‘glutarate sensor’) probes the S1′ pocket for the presence or absence of glutamate (Mesters et al., 2006 ▶). In addition to the above-mentioned moderate-resolution X-ray structures of GCPII complexes, Davis and coworkers also published the X-ray structure of ligand-free GCPII (GCPIIold; PDB code 1z8l) refined to 3.5 Å resolution (Davis et al., 2005 ▶).
Therefore, a ligand-free structure of recombinant human GCPII (rhGCPIInew) refined at a resolution of 1.65 Å and reported here represents a marked improvement in the quality of the model and allows the accurate description of the GCPII active site in the absence of ligands. This information will be significant in the elucidation of the mechanism of substrate hydrolysis by human GCPII and will permit the description of the conformational changes accompanying substrate/inhibitor binding.
2. Materials and methods
2.1. Expression and purification of rhGCPII
An extracellular part of human GCPII (rhGCPIInew; amino acids 44–750) was heterologously overexpressed in Drosophila Schneider S2 cells and purified to homogeneity as described previously (Barinka et al., 2002 ▶). Briefly, the recombinant protein was purified from conditioned medium by a succession of ion-exchange, affinity and size-exclusion chromatographies. The final protein preparation was >97% pure, as determined by Coomassie-staining SDS–PAGE (data not shown). The protein was extensively dialyzed against 100 mM NaCl, 20 mM Tris–HCl pH 8.0, concentrated to 10 mg ml−1 and stored at 193 K.
2.2. Crystallization and diffraction data collection
Initial crystallization screening was performed using a Hydra II robot (Apogent Discoveries, Hudson, USA) and a sitting-drop vapor-diffusion setup, with the rhGCPII at concentration of 10 mg ml−1. The final crystallization solution was composed of 33%(v/v) pentaerythritol propoxylate PO/OH 5/4, 1–3%(w/v) PEG 3350 and 100 mM Tris–HCl pH 8.0. For data collection, rhGCPII crystals were grown in hanging drops formed by mixing equal volumes of the protein and crystallization solutions. 2 µl drops were equilibrated against 1 ml reservoir solution at 293 K and crystals of approximate dimensions 0.5 × 0.5 × 0.2 mm grew within several weeks. Prior to X-ray exposure, crystals taken directly from the drop were cryopreserved in liquid nitrogen. The diffraction pattern was collected from a single crystal at the SER-CAT beamline (sector 22-BM) at the Advanced Photon Source (Argonne, Illinois, USA) equipped with a MAR Research CCD detector using a 1.000 Å wavelength. The measurements were conducted at 100 K. All data were integrated and scaled with the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶) and the appropriate X-ray data statistics are summarized in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data-collection statistics | |
| Wavelength (Å) | 1.000 |
| Space group | I222 |
| Unit-cell parameters (Å) | a = 101.76, b = 130.13, c = 158.87 |
| Resolution limits (Å) | 30.0–1.65 (1.71–1.65) |
| No. of unique reflections | 126082 (11591) |
| Redundancy | 9.8 (6.5) |
| Completeness (%) | 98.7 (91.6) |
| I/σ(I) | 19.7 (2.6) |
| Rmerge | 0.082 (0.598) |
| Refinement statistics | |
| Resolution limits (Å) | 30.0–1.65 (1.69–1.65) |
| Total No. of reflections | 124511 (8917) |
| No. of reflections in working set | 118244 (8477) |
| No. of reflections in test set | 6267 (440) |
| R | 0.207 (0.293) |
| Rfree | 0.228 (0.311) |
| Total No. of non-H atoms | 6511 |
| No. of non-H protein atoms | 5946 |
| No. of ions | 4 |
| No. of water molecules | 561 |
| Average B factor for all atoms (Å2) | 29.3 |
| Average B factor for protein atoms (Å2) | 27.9 |
| Ramachandran plot (%) | |
| Most favored | 89.2 |
| Additionally allowed | 10.3 |
| Generously allowed | 0.3 |
| Disallowed | 0.2 [Lys207] |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.019 |
| Bond angles (°) | 1.79 |
| Planarity (Å) | 0.009 |
| Chiral centers (Å3) | 0.12 |
2.3. Structure determination and refinement
Because of the identical symmetry and very similar unit-cell parameters of the rhGCPII–glutamate (PDB code 2c6g) and rhGCPIInew crystals, the structure solution was limited to rigid-body refinement of the rhGCPII–glutamate model (without water molecules and the S1′-bound glutamate) against the rhGCPIInew diffraction intensities. This procedure, which was conducted at 2.5 Å resolution, was followed by the refinement of atomic coordinates and B factors with a gradual extension of the resolution to the limit of the experimental data (1.65 Å). Calculations were performed with the program REFMAC v.5.1 (Murshudov et al., 1999 ▶). The refinement protocol was interspersed with manual corrections to the model using the computer-graphics program O (Jones et al., 1991 ▶). During the refinement, a randomly selected 5% of the reflections were kept aside for cross-validation (R free). The final values of the R factor and R free were 0.207 and 0.228, respectively, for data in the resolution range 30–1.65 Å. Detailed statistics for the refined structure are given in Table 1 ▶.
3. Results and discussion
3.1. Screening and crystallization
The main problem limiting the resolution of the previously reported structures of GCPII was the lack of suitable conditions for freezing the crystals prior to X-ray exposure. To address this issue, we revised the crystallization conditions for rhGCPII using several crystallization screens from commercial sources. The final selection of a mother liquor composed of 33%(v/v) pentaerythritol propoxylate (5/4 PO/OH; Hampton Research) and 100 mM Tris–HCl pH 8.0 was suggested by the results of crystallization from reagent No. 56 in Index Screen (Hampton Research). The addition of 1–3%(w/v) PEG 3350 increased the probability of crystal nucleation and accelerated the rate of crystal growth. Under the newly established conditions, rhGCPIInew crystallizes in the orthorhombic space group I222, with one molecule per asymmetric unit and unit-cell parameters a = 101.76, b = 130.13, c = 158.87 Å. This crystal form is virtually identical to the previously reported crystal forms of the GCPII complexes (Mesters et al., 2006 ▶), but the presence of 5/4 PO/OH as the primary crystallization and cryoprotectant agent (Gulick et al., 2002 ▶) allowed the cryopreservation of rhGCPIInew crystals for experiments without a visible loss in diffraction quality. It should be noted that the recently published structure of ligand-free GCPIIold was crystallized in space group P21, which is different from both the crystal form presented here and the crystal forms of the above-mentioned rhGCPII complexes.
3.2. Overall structure
The 1.65 Å model of ligand-free rhGCPIInew consists of 5946 non-H protein atoms (686 amino-acid residues), 561 water O atoms, one Ca2+ ion, one Cl− ion and two Zn2+ ions. Most of the protein residues are well defined in the electron-density peaks. The exceptions are the first 13 N-terminal residues and a flexible loop (541–547), which could not be modeled owing to the lack of corresponding electron-density data. Since these fragments do not interact with neighboring molecules, their high flexibility is probably a property native to rhGCPII. The only other fragment not modeled in the rhGCPIInew structure is the short (654–656) loop that directly interacts with an equivalent fragment from a symmetry-related molecule. Despite strong positive F o − F c peaks, we were unable to deconvolve the alternate conformations for this fragment. Two alternate conformations were modeled for the side chains of 34 residues, based on the F o − F c electron-density peaks. Unless the intensities of these peaks clearly indicated otherwise, equal occupancies were assumed for the alternate conformations. Additionally, two partially occupied main-chain conformations were evident for the protein segment 692–704. The conformational adaptability of this fragment has been suggested as being important for substrate recognition and/or binding (Mesters et al., 2006 ▶). The Ramachandran analysis of the final model using the program PROCHECK (Laskowski et al., 1993 ▶) classifies 89.2% of the residues as having the most favorable conformations, with 10.6% of the residues adopting allowed conformations. Despite being clearly defined by the electron-density maps, Lys207 adopts a conformation that is disallowed by Ramachandran analysis.
A comparison of the resulting structure with the previously reported 3.5 Å structure of ligand-free GCPIIold reveals the structures to be fairly similar, with an r.m.s. deviation between the 685 equivalent Cα atoms of 1.23 Å. Likewise, the overall fold of ligand-free rhGCPIInew is almost indistinguishable from the structures of rhGCPII complexed with reaction products/inhibitors.
3.3. The active site
Two zinc ions located in the active site of rhGCPIInew are bridged by the β-carboxylate of Asp387 and are further coordinated by the side chains of His377, Asp453, Glu425 and His553. In the structure of rhGCPIInew, an activated water molecule (or OH−) is located symmetrically between the two Zn2+ ions at distances of 2.03 and 2.01 Å. This water molecule plays a central role in the hydrolysis of a substrate and is hydrogen bonded to the conserved Glu424 residue (O...OE1, 2.92 Å) that functions as a proton shuttle during scissile bond cleavage. A similar arrangement of the active site has been reported previously for the ligand-free GCPIIold, although the interatomic distances between the activated water and Glu424 γ-carboxylate were too long (∼3.8 Å) for efficient proton transfer (Davis et al., 2005 ▶).
3.4. The substrate-binding cavity
The S1′ pocket of GCPII is defined by residues Phe209, Arg210, Asn257, Gly427, Leu428, Gly518, Lys699 and Tyr700. The latter two are part of a flexible ‘glutarate sensor’ (Mesters et al., 2006 ▶). The positions of most of the residues lining the S1′ pocket are similar in the GCPIIold and rhGCPIInew structures. The exceptions are the amino acids from the glutarate sensor (Lys699 and Tyr700) and the side chain of Phe209. The Phe209 side chain is rotated by ∼120° between rhGCPIInew and GCPIIold, resulting in a shift of ∼8 Å between the CZ atoms of the two models. The different conformation of Phe209 in GCPIIold might reflect a natural positional flexibility of this residue; however, Phe209 adopts an invariable conformation (as observed in rhGCPIInew) in all other reported structures of GCPII.
The S1′ pocket in rhGCPIInew is occupied by a network of six water molecules. Four of these molecules (one in two alternate positions) occupy the approximate positions of the N, OXT, CB, OE1 and OE2 atoms of a glutamate ligand in the rhGCPII–glutamate complex (Mesters et al., 2006 ▶), while the fifth water is conserved in both the complex and rhGCPIInew. The sixth water forms hydrogen bonds to Tyr552 OH (2.62 Å) and His553 NE2 (3.09 Å) and coordinates the Zn2+ ion at 2.48 Å.
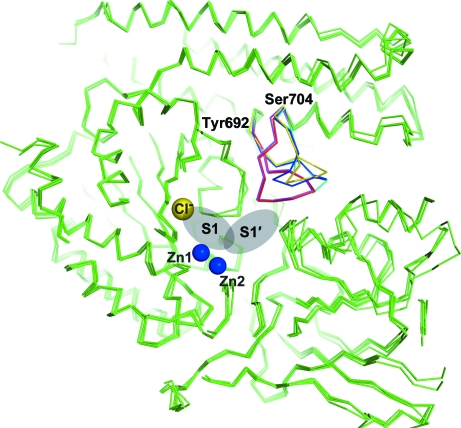
The bottom of the S1′ pocket is closed by a flexible loop formed by residues 692–704 (the ‘glutarate sensor’). Two residues from this loop (Lys699 and Tyr700) interact directly with a C-terminal residue of a substrate in the GCPII complexes and the glutarate sensor can adopt various conformations, largely depending on the occupancy of the S1′ pocket (Mesters et al., 2006 ▶; Davis et al., 2005 ▶). Two conformations of the glutarate sensor were modeled in the rhGCPIInew structure, with a maximal positional difference of ∼6 Å (Cα atoms of Gly702). An ensemble of the variable conformations of the glutarate sensor is shown in Fig. 1 ▶, where the published structures of GCPII are superimposed on the model of rhGCPIInew.
Figure 1.
Ensemble of conformations of the ‘glutarate sensor’. Two residues in this segment, Lys699 and Tyr700, interact directly with the ligands of the S1′ site. The GCPII structures available from the PDB were superimposed on the structure of rhGCPIInew. The Cα traces are shown in ribbon representation, the active-site Zn+2 ions are shown as blue spheres and the yellow sphere represents the S1-bound Cl− ion. The ‘glutarate sensor’ is shown in yellow for PDB entry 1z8l, in magenta for 2c6g, in blue for 2c6p and in red and cyan for the two conformations in rhGCPIInew. The S1 and S1′ sites are highlighted by gray ellipses.
The putative S1 pocket is defined by residues Ser454, Glu457, Asp465, Asn519, Gly548, Tyr549 and Tyr552, as well as by a stack of three arginines (Arg463, Arg534 and Arg536). This positively charged arginine stack, located 6.5–12.7 Å from the nearest Zn2+ ion, is believed to be responsible for the enzyme’s preference for negatively charged residues in the penultimate (i.e. P1) position of a substrate (Barinka et al., 2002 ▶; Mesters et al., 2006 ▶). The invariant positions of the guanidinium groups in Arg534 and Arg463 are maintained by interaction with a Cl− ion and a strong hydrogen bond (2.59 Å) to the γ-carboxylate of Glu457, respectively. The side chain of Arg536 adopts two conformations. In the slightly more abundant conformation, the guanidinium group of Arg536 forms two hydrogen bonds to Ser454 OG (NH1...OG, 3.21 Å; NH2...OG, 3.24 Å) and one to Ser454 O (3.03 Å), Glu457 OE1 (2.85 Å) and Cl− (3.26 Å) (Fig. 2 ▶). Thus, the role of the Cl− ion is in stabilizing the positions of Arg534 and Arg536 and in neutralizing the combined positive charge of their guanidinium groups. It has been suggested that the chloride ion is essential for GCPII hydrolytic activity (Robinson et al., 1987 ▶). In the less occupied conformation, the stabilization of the guanidinium group of Arg536 provided by stacking between Arg463 and Arg534 is further stabilized by ionic interactions with the negatively charged carboxylate of Asp465.
Figure 2.
Organization of the positively charged arginine stack in the putative S1 pocket. Arg463, Arg534 and Arg536 are located within the antiparallel β-strands (β14, Ser532–Thr538; β13, Thr461–Cys466). In the rhGCPIInew structure, Arg463 and Arg534 are present in single conformations, while Arg536 adopts two alternate conformations. The S1-bound Cl− ion (shown as a yellow sphere) stabilizes the invariant conformation of Arg534 as well as neutralizing the positive charge contributed by the Arg534 and Arg536 guanidinium groups. The side chains of Ser454, Asp465 and Arg536 are shown in two alternate conformations and the active-site Zn+2 ions are represented as blue spheres.
4. Conclusions
In summary, our newly established crystallization conditions allowed us to determine and refine the structure of ligand-free rhGCPIInew at a resolution of 1.65 Å. A comparison with the GCPIIold model reveals several marked differences in the organization of the substrate-binding cavity of this enzyme. Thus, the present structure could serve as a more accurate model for ensuing biochemical/structural studies.
Supplementary Material
PDB reference: human glutamate carboxypeptidase II, 2oot, r2ootsf
Acknowledgments
We acknowledge the use of beamline 22-BM of the Southeast Regional Collaborative Access Team (SER-CAT), located at the Advanced Photon Source (APS), Argonne National Laboratory, Argonne, Illinois, USA. Use of the APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38. The project was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. JK and JS were supported in part by the Ministry of Education of the Czech Republic (Research Centre for New Antivirals and Antineoplastics, 1M0508).
References
- Barinka, C., Rinnova, M., Sacha, P., Rojas, C., Majer, P., Slusher, B. S. & Konvalinka, J. (2002). J. Neurochem.80, 477–487. [DOI] [PubMed] [Google Scholar]
- Bostwick, D. G., Pacelli, A., Blute, M., Roche, P. & Murphy, G. P. (1998). Cancer, 82, 2256–2261. [DOI] [PubMed] [Google Scholar]
- Davis, M. I., Bennett, M. J., Thomas, L. M. & Bjorkman, P. J. (2005). Proc. Natl Acad. Sci. USA, 102, 5981–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X., Porter, A. T., Grignon, D. J., Pontes, J. E. & Honn, K. V. (1997). Prostate, 31, 264–281. [DOI] [PubMed] [Google Scholar]
- Gulick, A. M., Horswill, A. R., Thoden, J. B., Escalante-Semerena, J. C. & Rayment, I. (2002). Acta Cryst. D58, 306–309. [DOI] [PubMed] [Google Scholar]
- Halsted, C. H., Ling, E. H., Luthi-Carter, R., Villanueva, J. A., Gardner, J. M. & Coyle, J. T. (1998). J. Biol. Chem.273, 20417–20424. [DOI] [PubMed] [Google Scholar]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291. [Google Scholar]
- Mesters, J. R., Barinka, C., Li, W., Tsukamoto, T., Majer, P., Slusher, B. S., Konvalinka, J. & Hilgenfeld, R. (2006). EMBO J.25, 1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. (1999). Acta Cryst. D55, 247–255. [DOI] [PubMed] [Google Scholar]
- Neale, J. H., Olszewski, R. T., Gehl, L. M., Wroblewska, B. & Bzdega, T. (2005). Trends Pharmacol. Sci.26, 477–484. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pinto, J. T., Suffoletto, B. P., Berzin, T. M., Qiao, C. H., Lin, S., Tong, W. P., May, F., Mukherjee, B. & Heston, W. D. (1996). Clin. Cancer Res.2, 1445–1451. [PubMed] [Google Scholar]
- Rawlings, N. D. & Barrett, A. J. (1997). Biochim. Biophys. Acta, 1339, 247–252. [DOI] [PubMed] [Google Scholar]
- Robinson, M. B., Blakely, R. D., Couto, R. & Coyle, J. T. (1987). J. Biol. Chem.262, 14498–14506. [PubMed] [Google Scholar]
- Šácha, P., Zámečník, J., Bařinka, C., Hlouchová, K., Vícha, A., Mlčochová, P., Hilgert, I., Eckschlager, T. & Konvalinka, J. (2007). Neuroscience, 144, 1361–1372. [DOI] [PubMed] [Google Scholar]
- Troyer, J. K., Beckett, M. L. & Wright, G. L. Jr (1995). Int. J. Cancer, 62, 552–558. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Neale, J. H., Pomper, M. G. & Kozikowski, A. P. (2005). Nature Rev. Drug Discov.4, 1015–1026. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human glutamate carboxypeptidase II, 2oot, r2ootsf