The enzyme p-coumaric acid decarboxylase (PDC) from L. plantarum has been recombinantly expressed, purified and crystallized. The structure has been solved at 2.04 Å resolution by the molecular-replacement method.
Keywords: p-coumaric acid decarboxylase, Lactobacillus plantarum
Abstract
The substrate-inducible p-coumaric acid decarboxylase (PDC) from Lactobacillus plantarum has been overexpressed in Escherichia coli, purified and confirmed to possess decarboxylase activity. The recombinant His6-tagged enzyme was crystallized using the hanging-drop vapour-diffusion method from a solution containing 20%(w/v) PEG 4000, 12%(w/v) 2-propanol, 0.2 M sodium acetate, 0.1 M Tris–HCl pH 8.0 with 0.1 M barium chloride as an additive. Diffraction data were collected in-house to 2.04 Å resolution. Crystals belonged to the tetragonal space group P43, with unit-cell parameters a = b = 43.15, c = 231.86 Å. The estimated Matthews coefficient was 2.36 Å3 Da−1, corresponding to 48% solvent content, which is consistent with the presence of two protein molecules in the asymmetric unit. The structure of PDC has been determined by the molecular-replacement method. Currently, the structure of PDC complexed with substrate analogues is in progress, with the aim of elucidating the structural basis of the catalytic mechanism.
1. Introduction
Phenolic acids, mainly p-coumaric and ferulic acids, are naturally abundant plant compounds present in the cell wall that contribute to its mechanical properties (de Vries et al., 1998 ▶). They can be released from this structure by cinnamoyl esterases, which are expressed by various microorganisms (Christov & Prior, 1993 ▶; de Vries et al., 1997 ▶; McSweeney et al., 1999 ▶). Once released, phenolic acids become substrates of phenolic acid decarboxylase (PAD) enzymes, which catalyse the formation of the corresponding 4-vinyl derivatives (Cavin et al., 1993 ▶). These enzymes are produced by lactic acid bacteria, including Lactobacillus plantarum (Cavin, Barthelmebs & Diviès, 1997 ▶), a microorganism that is frequently used as a starter in the production of wine and many vegetable fermentations (Olsen et al., 1991 ▶; Cavin, Barthelmebs & Diviès, 1997 ▶). The 4-vinyl derivatives produced by PAD enzymes are volatile compounds that have important consequences in the aroma of wine (Etiévant et al., 1989 ▶) and other fermented foods and beverages and are therefore approved as food additives (flavouring substances; JECFA, 2001 ▶). Despite the importance of lactic acid bacteria in fermentative processes, little is known about the metabolism of phenolic acids in these bacteria. L. plantarum produces two inducible phenolic acid decarboxylases, one of which is specific for p-coumaric and caffeic acids and is called p-coumaric acid decarboxylase (PDC; Cavin, Barthelmebs, Guzzo et al., 1997 ▶). Considering the importance of volatile phenols in the biotechnology industry, it is obvious that a deeper understanding of the structural basis of the catalytic mechanism of PDC enzymes will be useful for the design of ad hoc recombinant microorganisms with selected substrate specificities.
Phenolic acid decarboxylase activity can be considered to be a biological response of lactic acid bacteria to the chemical stress induced by phenolic acids at low pH, mainly p-coumaric, ferulic and caffeic acids (Lambert et al., 1997 ▶; Zaldívar & Ingram, 1999 ▶; Barthelmebs, Diviès et al., 2000 ▶; Gury et al., 2004 ▶). In fact, the disruption of the pdc gene from L. plantarum makes this bacteria unable to grow in the presence of p-coumaric acid at low pH (Barthelmebs, Lecomte et al., 2000 ▶).
In this work, we report the cloning of the pdc gene from L. plantarum and the overexpression, purification, crystallization and preliminary X-ray crystallographic analysis of the p-coumaric acid decarboxylase. The structure of PDC from L. plantarum has been solved by the molecular-replacement method using the atomic coordinates of a recombinant variant of PDC from L. plantarum without fusion tags (PDB code 2gc9) as the search model.
2. Materials and methods
2.1. Cloning and overexpression
The pdc gene (coding for p-coumaric acid decarboxylase) from L. plantarum CECT 748T (ATCC 14917T) was PCR-amplified by Hot Start Turbo Pfu DNA polymerase using the primers 274 (5′-CATCATGGTGACGATGACGATAAGatgacaaaaacttttaaaacacttg) and 275 (5′-AAGCTTAGTTAGCTATTATGCGTAttacttatttaaacgatggtagttt; the nucleotides pairing the expression-vector sequence are indicated in italics and the nucleotides pairing the pdc gene sequence are written in lower case). The 0.5 kbp purified PCR product was inserted into the pURI3 vector by using the restriction-enzyme-free and ligation-free cloning strategy described by Geiser et al. (2001 ▶). Expression vector pURI3 was constructed based on the commercial expression vector pT7-7 (USB) but containing the leader sequence MGGSHHHHHHGDDDDKM consisting of an N-terminal methionine followed by three spacer amino acids, a six-histidine affinity tag, a spacer glycine residue and the five-amino-acid enterokinase-recognition site (unpublished results). Thus, the final recombinant PDC should possess 194 amino-acid residues with a molecular weight of 22.8 kDa. Escherichia coli DH5α cells were transformed, recombinant plasmids were isolated and those containing the correct insert were identified by restriction-enzyme analysis, verified by DNA sequencing and then transformed into E. coli JM109(DE3) (pLysS) cells for expression.
Cells carrying the recombinant plasmid pURI3-PDC were grown at 310 K in Luria–Bertani media containing ampicillin (100 µg ml−1) and chloramphenicol (34 µg ml−1) until they reached an optical density at 600 nm of 0.4, when they were induced by adding IPTG (0.4 mM final concentration). After induction, the cells were grown at 295 K for 20 h and were collected by centrifugation.
2.2. Purification
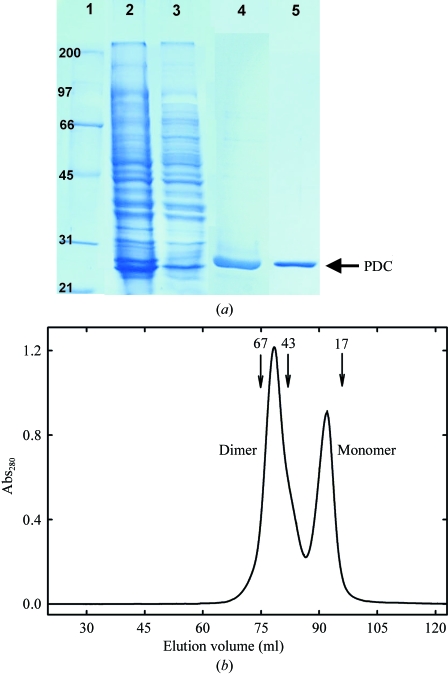
Cells were resuspended in 25 ml 20 mM Tris–HCl pH 8.0 containing 0.1 M NaCl and disrupted with a French Press. The lysate was centrifuged in an SS34 rotor at 20 000 rev min−1 using a Sorvall centrifuge for 30 min at 277 K. After filtration through a 0.45 µm filter (Millipore), the supernatant was loaded onto a His-Trap FF Ni-affinity column (GE Healthcare). The recombinant His6-tagged protein was eluted with a 10–500 mM imidazole gradient using an AKTA Prime system (GE Healthcare). Fractions containing His6-tagged PDC were pooled and dialysed overnight at 277 K against 10 mM Tris–HCl pH 8.0 containing 10 mM NaCl. The protein solution was then loaded onto a HiTrap Q HP column (GE Healthcare) and elution was performed with a 10–500 mM NaCl gradient. Pooled fractions containing PDC were dialysed overnight at 277 K against 10 mM Tris–HCl pH 8.0 containing 100 mM NaCl and further concentrated by ultrafiltration with YM-10 membranes (Amicon). The protein purity was checked by SDS–PAGE at the various stages of the purification process (Fig. 1 ▶ a). Although electrophoretically homogeneous (lane 5, Fig. 1 ▶ a), PDC exhibited an extra maximum at 357 nm in the UV–Vis absorbance spectrum (not shown). Thus, an additional chromatographic step was carried out on Superdex 200 prep grade (GE Healthcare) with a BioRad BioLogic DuoFlow FPLC to try to eliminate the accompanying molecule. The results indicated that PDC behaves as an associative monomer–dimer equilibrium (Fig. 1 ▶ b), both forms having identical spectroscopic properties. Fractions containing both dimeric and monomeric PDC were pooled and concentrated by ultrafiltration with YM-10 membranes (Amicon). The PDC solution used for crystallization was finally prepared in 20 mM Tris pH 8.0 containing 0.1 M NaCl. Protein concentration was determined by UV–Vis absorbance measurements with a Nanodrop ND-1000 spectrophotometer, using an extinction coefficient of 2.23 (1 mg ml−1, 1 cm, 280 nm).
Figure 1.
(a) SDS–PAGE of PDC samples at various steps of the purification process. Lane 1, broad-range (Bio-Rad) molecular-weight markers; lane 2, soluble fraction from E. coli JM109(DE3) cells carrying the plasmid pURI3-PDC after cell disruption; lane 3, flowthrough from the His-Trap FF Ni-affinity column; lane 4, pooled fractions containing PDC eluted from the His-Trap FF Ni-affinity column; lane 5, pooled fractions containing PDC eluted from the HiTrap Q HP column. (b) Gel-filtration chromatography of purified PDC on Superdex 200 prep grade. Two peaks consistent with the molecular weights of monomeric (22.8 kDa) and dimeric (45.6 kDa) PDC species are observed. The positions of molecular-weight standards are shown for comparison.
2.3. Crystallization
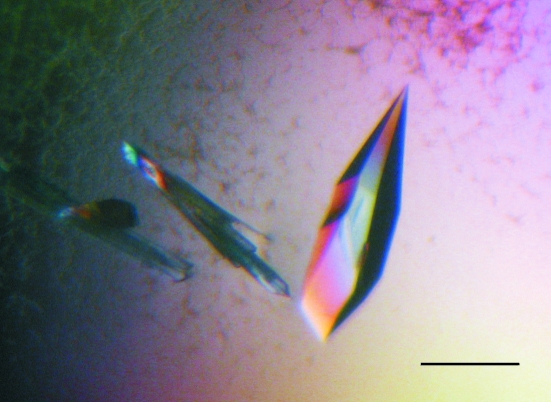
Initial screening for crystallization conditions was carried out at 291 K by the hanging-drop vapour-diffusion method using the sparse-matrix method (Jancarik & Kim, 1991 ▶) with Crystal Screen I (Hampton Research). Each drop contained 1 µl protein solution (6–8 mg ml−1) in Tris–HCl buffer (10 mM Tris–HCl pH 8.0 containing 0.1 M NaCl) and 1 µl reservoir solution. Drops were equilibrated against 500 µl reservoir solution. Similar procedures were used in the optimization process. Small and clustered crystalline needles were obtained in condition No. 9 from Crystal Screen I (Hampton Research), which contained 30%(w/v) PEG 4000, 100 mM sodium citrate pH 4.6, 200 mM ammonium acetate. As a first optimization step we used the PEG 4000-based JBScreen Classic 3 (Jena Bioscience). Three-dimensional needles were obtained in condition D2, which contained 25%(w/v) PEG 4000, 8%(w/v) 2-propanol, 100 mM sodium acetate (protein:precipitant volume ratio 1:1; total volume 2 µl). Final optimization employing the Jena Bioscience Solubility kit (Jena Bioscience) and Additive Screens I and II (Hampton Research) gave three-dimensional crystals (0.5–0.6 mm in the longest dimension; Fig. 2 ▶) in 20%(w/v) PEG 4000, 12%(w/v) 2-propanol, 0.2 M sodium acetate and 0.1 M Tris–HCl pH 8.0 with 0.1 M barium chloride as an additive (protein:precipitant:additive volume ratio 2:2:1; total volume 5 µl).
Figure 2.
Crystals of recombinant PDC from L. plantarum grown at 291 K in 20%(w/v) PEG 4000, 12%(w/v) 2-propanol, 0.2 M sodium acetate and 0.1 M Tris–HCl pH 8.0 with 0.1 M barium chloride. The bar indicates 0.4 mm.
2.4. X-ray diffraction analysis and processing
For X-ray diffraction measurements at 120 K, the crystals were cryoprotected by a quick soak (∼10 s) in optimized reservoir solution containing 15%(v/v) MPD and immediately flash-cooled in a stream of liquid nitrogen controlled by a Cryostream Controller 700 (Oxford Cryosystems). Diffraction data were collected in-house at 120 K on a Kappa 2000 Brucker–Nonius CCD detector using Cu Kα X-rays generated by an FR591 Brucker–Nonius rotating-anode generator equipped with a double-mirror focusing system and operated at 45 kV and 100 mA. The crystal-to-detector distance was kept at 200 mm. The images were processed and scaled using DENZO and SCALEPACK from the HKL-2000 suite (Otwinowski & Minor, 1997 ▶). Intensities were converted to structure-factor amplitudes using TRUNCATE (Collaborative Computational Project, Number 4, 1994 ▶). Data-collection statistics are given in Table 1 ▶.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the outermost shell.
| Temperature (K) | 120 |
| Space group | P43 |
| Unit-cell parameters (Å) | |
| a | 43.15 |
| b | 43.15 |
| c | 231.86 |
| VM (Å3 Da−1) | 2.36 |
| Solvent content (%) | 48 |
| Resolution range (Å) | 43.15–2.04 (2.11–2.04) |
| No. of measured reflections | 181494 |
| No. of unique reflections | 26502 (2641) |
| Completeness (%) | 98.5 (99.1) |
| Multiplicity | 6.8 (4.0) |
| Average I/σ(I) | 12.9 (4.2) |
| Rmerge(I)† (%) | 10.5 (24.7) |
R
merge(I) = 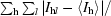
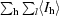 , where I
l is the lth observation of reflection h and 〈I
h〉 is the weighted average intensity for all observations l of reflection h.
, where I
l is the lth observation of reflection h and 〈I
h〉 is the weighted average intensity for all observations l of reflection h.
The crystal structure of PDC was determined by the molecular-replacement method with MOLREP (Vagin & Teplyakov, 1997 ▶) from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). The atomic coordinates of PDC from L. plantarum (PDB code 2gc9) were used as the search model.
3. Results and discussion
The pdc gene from L. plantarum CECT 748T (ATCC 14917T) was cloned and the protein was overexpressed in E. coli JM109(DE3) (pLysS) cells. The biochemical characterization of the His6-tagged PDC carried out as described previously (Cavin, Barthelmebs, Guzzo et al., 1997 ▶) revealed that this recombinant protein possesses decarboxylase activity against p-coumaric and caffeic acids (data not shown).
The purified protein behaves as a monomer–dimer associative equilibrium in solution as observed by gel filtration on Superdex 200 (Fig. 1 ▶ b). The crystals of PDC from L. plantarum grew from precipitant solution containing 20%(w/v) PEG 4000, 12%(w/v) 2-propanol, 0.2 M sodium acetate and 0.1 M Tris–HCl pH 8.0 with 0.1 M barium chloride as an additive in 5 µl drops with a 2:2:1 volume ratio of protein:precipitant:additive and reached their full size (0.5–0.6 mm in the longest dimension) in 4 d (Fig. 2 ▶). The crystals belong to the tetragonal space group P43, with unit-cell parameters a = b = 43.15, c = 231.86 Å and a unit-cell volume of 431 593 Å3. The programs DENZO and SCALEPACK from the HKL-2000 suite (Otwinowski & Minor, 1997 ▶) were used for integration and scaling of the data taken from 2825 collected images. The completeness of the data was 98.5% (99.1% in the outer shell) and the R merge was 10.5 (24.7 in the outer shell). The number of molecules in the asymmetric unit was estimated with the Matthews probability calculator, with the resolution as an additional parameter (Kantardjieff & Rupp, 2003 ▶). The highest probability (0.99) was obtained with two molecules in the asymmetric unit, resulting in a Matthews coefficient (V M; Matthews, 1968 ▶) of 2.36 Å3 Da−1 and a solvent content of 48%. Data-collection and processing statistics are shown in Table 1 ▶.
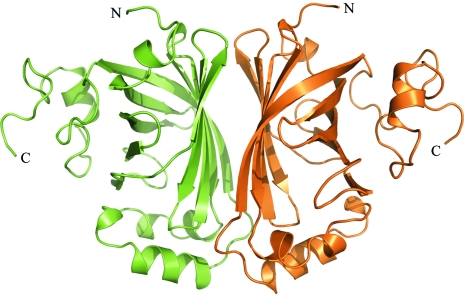
The structure of the enzyme p-coumaric acid decarboxylase from L. plantarum was determined using the molecular-replacement method. The atomic coordinates of a recombinant PDC variant from L. plantarum with no fusion tag at the N-terminus end (179 residues; PDB code 2gc9) were used as the search model. Details of the molecular-replacement solutions are given in Table 2 ▶. Two closely packed PDC monomers are present in the asymmetric unit (Fig. 3 ▶), which fully agrees with the estimated V M. In this sense, it is remarkable that although previous biochemical studies reported PDC from L. plantarum to be a homotetramer (Cavin, Barthelmebs, Guzzo et al., 1997 ▶), no tetrameric species are observed in either the tetragonal crystal reported here or in a trigonal crystal prepared under different experimental conditions (PDB code 2gc9). In fact, in these two crystal forms the asymmetric unit is made up of two closely packed monomers arranged identically, which may suggest this structure to be responsible for the high-molecular-weight species present in solution (Fig. 1 ▶ b). As the physiological oligomeric state of PDC is not known, it is obvious that further structural analyses are required. Currently, the structure refinement of PDC is in progress, together with the crystallization of PDC complexed with substrate analogues, with the aim of analysing the structural basis of the catalytic mechanism of phenolic acid decarboxylation.
Table 2. Details of the molecular-replacement solutions.
The resolution range used for the rotation function and translation was 10.0–3.0 Å.
| Eulerian angles | Translations (fractional) | |||||||
|---|---|---|---|---|---|---|---|---|
| Solution | α (°) | β (°) | γ (°) | Tx | Ty | Tz | R | Cor. |
| Molecular-replacement solutions | ||||||||
| 1 | 43.76 | 107.97 | 152.67 | 0.828 | 0.476 | 0.000 | 53.0 | 31.1 |
| 2 | 44.79 | 71.40 | 334.07 | 0.975 | 0.326 | 0.000 | 54.1 | 28.2 |
| 3 | 85.67 | 137.99 | 202.89 | 0.867 | 0.437 | 0.000 | 60.5 | 11.1 |
| Monomer 1 fixed: search for the second monomer | ||||||||
| 1 | 44.79 | 71.40 | 334.07 | 0.975 | 0.326 | 0.387 | 42.9 | 54.7 |
| 2 | 43.76 | 107.97 | 152.67 | 0.828 | 0.476 | 0.372 | 63.4 | 23.8 |
| 3 | 85.67 | 137.99 | 202.89 | 0.366 | 0.938 | 0.136 | 56.5 | 22.1 |
Figure 3.
Asymmetric unit of the PDC tetragonal crystal. Two closely packed monomers, shown as ribbon models, make up the asymmetric unit of the PDC tetragonal crystals. The figure was prepared with PyMOL (DeLano, 2002 ▶).
Acknowledgments
JMM thanks the Ministerio de Educación y Ciencia for a research grant (BFU2004-01554/BMC; DGCYT) in support of his research. This work was also supported by grants AGL2005-00470 (CICYT) and S-0505/AGR000153 (CAM). HR is a recipient of a I3P predoctoral fellowship from the CSIC.
References
- Barthelmebs, L., Diviès, C. & Cavin, J.-F. (2000). Appl. Environ. Microbiol.66, 3368–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelmebs, L., Lecomte, B., Diviès, C. & Cavin, J.-F. (2000). J. Bacteriol.182, 6724–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavin, J.-F., Andioc, V., Etiévant, P. X. & Diviès, C. (1993). Am. J. Enol. Vitic.44, 76–80.
- Cavin, J.-F., Barthelmebs, L. & Diviès, C. (1997). Appl. Environ. Microbiol.63, 1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavin, J.-F., Barthelmebs, L., Guzzo, J., Van Beeumen, J., Bart, S., Travers, J. F. & Diviès, C. (1997). FEMS Microbiol. Lett.147, 291–295.
- Christov, L. P. & Prior, B. A. (1993). Enzyme Microb. Biotechnol.15, 460–475. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. http://www.pymol.org.
- Etiévant, P. X., Issanchou, S. N., Marie, S., Ducruet, V. & Flanzy, C. (1989). Sci. Aliments, 9, 19–33.
- Geiser, M., Cèbe, R., Drewello, D. & Schemitz, R. (2001). Biotechniques, 31, 88–92. [DOI] [PubMed] [Google Scholar]
- Gury, J., Barthelmebs, L., Tran, N.-P., Diviès, C. & Cavin, J.-F. (2004). Appl. Environ. Microbiol.70, 2146–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- JECFA (2001). Evaluation of Certain Food Additives and Contaminants. Fifty-fifth Report of the Joint WHO/FAO Expert Committee on Food Additives. WHO Technical Report Series No. 901. Geneva: World Health Organization.
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci.12, 1865–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, L. A., Abshire, K., Blankenhorn, D. & Slonczewski, J. L. (1997). J. Bacteriol.179, 7595–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSweeney, C., Dulieu, A., Webb, R. I., del Dot, T. & Blackall, L. L. (1999). Arch. Microbiol.172, 139–149. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Olsen, E. B., Russel, J. B. & Henick-Kling, T. (1991). J. Bacteriol.173, 6199–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025. [Google Scholar]
- Vries, R. P. de, Michelsen, B., Poulsen, C. H., Kroon, P. A., van den Heuvel, R. H. H., Faulds, C. B., Williamson, G., van den Homberg, J. P. T. W. & Visser, J. (1997). Appl. Environ. Micobiol.63, 4638–4644. [DOI] [PMC free article] [PubMed]
- Vries, R. P. de, Poulsen, C. H., Madrid, S. & Visser, J. (1998). J. Bacteriol.180, 243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaldívar, J. & Ingram, L. O. (1999). Biotechnol. Bioeng.66, 203–210. [DOI] [PubMed] [Google Scholar]