A truncated form of biotin carboxyl carrier protein containing the C-terminal half fragment (BCCPΔN76) and the biotin protein ligase (BPL) with the mutation R48A (BPL*) or the double mutation R48A K111A (BPL**) were successfully cocrystallized in the presence of ATP and biotin. The BPL*–BCCPΔN76 and BPL**–BCCPΔN76 crystals belong to space group P21 and diffract X-rays to 2.7 and 2.0 Å resolution, respectively.
Keywords: biotin protein ligase, biotin carboxyl carrier protein, biotinylation, fatty-acid biosynthesis
Abstract
Biotin protein ligase (BPL) catalyses the biotinylation of the biotin carboxyl carrier protein (BCCP) subunit of acetyl-CoA carboxylase. To elucidate the exact details of the protein–protein interactions in the biotinylation function, the C-terminal half fragment of BCCP (BCCPΔN76), the R48A mutant of BPL (BPL*) and the R48A K111A double mutant of BPL (BPL**), all of which are from Pyrococcus horikoshii OT3, have been expressed, purified and successfully cocrystallized. Cocrystals of the BPL*–BCCPΔN76 and BPL**–BCCPΔN76 complexes as well as crystals of BPL*, BPL** and BCCPΔN76 were obtained by the oil-microbatch method using PEG 20 000 as a precipitant at 295 K. Complete X-ray diffraction data sets for BPL*–BCCPΔN76 and BPL**–BCCPΔN76 crystals were collected at 100 K to 2.7 and 2.0 Å resolution, respectively, using synchrotron radiation. They belong to the monoclinic space group P21, with similar unit-cell parameters a = 69.85, b = 63.12, c = 75.64 Å, β = 95.9°. Assuming two subunits of the complex per asymmetric unit gives a V M value of 2.45 Å3 Da−1 and a solvent content of 50%.
1. Introduction
Biotin is an essential coenzyme that only has biological activity when covalently attached at the active site of the biotin-dependent transcarboxylases acetyl-CoA carboxylases, a class of important enzymes for fatty-acid biosynthesis, gluconeogenesis and propionate catabolism (Knowles, 1989 ▶). Linkage of the biotin moiety to these proteins is catalyzed by biotin protein ligase (BPL) in a two-step process: synthesis of biotinyl-5′-AMP from the substrates biotin and ATP is followed by the transfer of the activated biotin to the biotin carboxyl carrier protein (BCCP) subunit of acetyl-CoA carboxylase (Beckett & Matthews, 1997 ▶; Chapman-Smith & Cronan, 1999 ▶). Because of their function of funnelling biotin into metabolism, BPL and BCCP are essential for survival.
With regard to the structural details of the first step of the biotinylation reaction performed by BPL, we have recently reported the crystal structures of BPL and of BPL with various biological ligands including the reaction intermediate biotinyl-5′-AMP (Bagautdinov et al., 2005 ▶). However, the structural details of the second step of the biotinylation reaction in which the activated biotin is transferred to a specific lysine of the BCCP subunit of acetyl-CoA carboxylase have not yet been resolved. This post-translational modification of a single lysine residue is exceptionally specific and insights into how the enzymes BPL and BCCP carry out the biotinylation process is of great interest. For the well studied Escherichia coli BPL and BCCP, the interactions of the proteins during biotinylation have been discussed by Polyak et al. (2001 ▶) using mutational analysis and a protein–protein complex model consistent with physical and biochemical studies has been suggested by Weaver et al. (2001 ▶). For Aquifex aeolicus, the isolation of a chemically cross-linked BPL–BCCP complex was performed by Clarke et al. (2003 ▶). Despite these studies, the absence of a BPL–BCCP complex structure means that there is little understanding of the structure–function relationship in the biotinylation reaction or of the unusually high reactivity towards the physiological target lysine (Streaker & Beckett, 2006 ▶).
With the aim of studying the structural basis of the BPL–BCCP interaction in the biotinylation process, we successfully cocrystallized the BPL protein and the C-terminal half fragment of BCCP, both from Pyrococcus horikoshii OT3 (PhBPL and PhBCCPΔN76, respectively), and are now resolving the three-dimensional structure of the complex. In E. coli, it has been shown that a C-terminal half fragment of BCCP functions identically to the intact protein in the biotin-transfer reaction (Nenortas & Beckett, 1996 ▶). Additionally, BCCP itself, in the absence of the other acetyl-CoA carboxylase subunits, provides an adequate model to study the interaction with BPL (Wood et al., 1980 ▶). Therefore, we assumed that PhBCCPΔN76 interacts with PhBPL in a similar way as intact PhBCCP. For the preparation of the complex crystals, we used the R48A mutant of PhBPL (PhBPL*) and the R48A K111A double mutant of PhBPL (PhBPL**). Prior to cocrystallization, we added biotin and ATP to the BPL, since the BPL species that binds the BCCP domain should be the BPL–biotinyl-5′-AMP complex rather than the uncomplexed protein. The active-site residues Arg48 and Lys111 form hydrogen bonds to biotinyl-5′-AMP in BPL–biotinyl-5′-AMP (Bagautdinov et al., 2005 ▶). Previous structure analyses validated that these replacements did not affect the overall BPL structure (PDB codes 2dzc and 2e64 for PhBPL* and PhBPL**, respectively; Bagautdinov et al., unpublished results). We attempted to cocrystallize the PhBCCPΔN76 and wild-type PhBPL proteins, but for unknown reasons failed to produce crystals. Although structural data on the wild-type BPL and BCCP complex will be important, the structural information from the present mutants may provide the first approximation to understanding the biotin-transfer reaction.
In this paper, we report the expression and purification of protein PhBCCPΔN76 and the crystallization and preliminary crystallographic analysis of PhBCCPΔN76, PhBPL*, PhBPL** and the complexes PhBPL*–PhBCCPΔN76 and PhBPL**–PhBCCPΔN76. Knowledge of the three-dimensional structures of PhBPL*–PhBCCPΔN76 and PhBPL**–PhBCCPΔN76 may clarify the biotinylation function that is essential to initiate the first step of fatty-acid biosynthesis catalysed by acetyl-CoA carboxylase. The structural details of the biotinylation reaction are also important in order to develop its novel applications; for example, simple and efficient proteomic approaches for protein purification (Boer et al., 2003 ▶) and quantum dots targeting (Howarth et al., 2005 ▶).
2. Experimental
2.1. Protein expression and purification
The C-terminal half fragment of BCCP from P. horikoshii OT3 (PhBCCPΔN76) that was used in this study has a molecular weight of 7.98 kDa and consists of 73 amino-acid residues. The plasmid encoding PhBCCPΔN76 (residues 77–149) was digested with NdeI and BglII and the fragment was inserted into the expression vector pET-11a (Novagen) linearized with NdeI and BamHI. E. coli BL21 Codon Plus (DE3)-RIL cells were transformed with the recombinant plasmid and grown at 310 K in Luria–Bertani medium containing 50 µg ml−1 ampicillin for 20 h. The cells were harvested by centrifugation at 4500g for 5 min at 277 K, suspended in 20 mM Tris–HCl pH 8.0 containing 0.5 M NaCl, 5 mM 2-mercaptoethanol and 1 mM phenylmethylsulfonyl fluoride and finally disrupted by sonication and heated at 363 K for 10 min. The cell debris and heat-denaturated proteins were removed by centrifugation at 20 000g for 30 min. The supernatant solution containing PhBCCPΔN76 was used as the crude extract for purification. The crude extract was desalted on a HiPrep 26/10 desalting column (Amersham Biosciences) and applied onto a Super Q Toyopearl 650M (Tosoh) column equilibrated with 20 mM Tris–HCl pH 8.0 (buffer A). After elution with a linear gradient of 0–0.3 M NaCl, the fraction containing PhBCCPΔN76 was desalted with a HiPrep 26/10 desalting column with 10 mM potassium phosphate pH 7.0. The sample was then applied onto a Bio-Scale CHT-20-I column (Bio-Rad) equilibrated with 10 mM potassium phosphate pH 7.0 and eluted with a linear gradient of 10–300 mM potassium phosphate pH 7.0. The sample was concentrated by ultrafiltration (VivaSpin, 5 kDa cutoff) and loaded onto a HiLoad 16/60 Superdex 200 prep-grade column (Amersham Biosciences) equilibrated with buffer A containing 0.2 M NaCl. The homogeneity and identity of the purified sample were assessed by SDS–PAGE (Laemmli, 1970 ▶) and N-terminal sequence analysis. Finally, the purified PhBCCPΔN76 was concentrated to 2.4 mg ml−1 by ultrafiltration and stored at 203 K. Analysis by liquid-chromatography electrospray ionization ion-trap mass spectroscopy (LC-ESI-IT-MS) gave the molecular weight of the protein as 7909 ± 131 Da, which is within experimental error of the predicted value of 7975 Da. bpl mutant genes encoding the R48A and K111A mutations were produced using a QuikChange II XL site-directed mutagenesis kit (Stratagene) and the expression and purification took place in the same fashion as described for the wild-type protein (Bagautdinov et al., 2005 ▶). Purified PhBPL* and PhBPL** were concentrated to 7.5 mg ml−1.
2.2. Crystallization
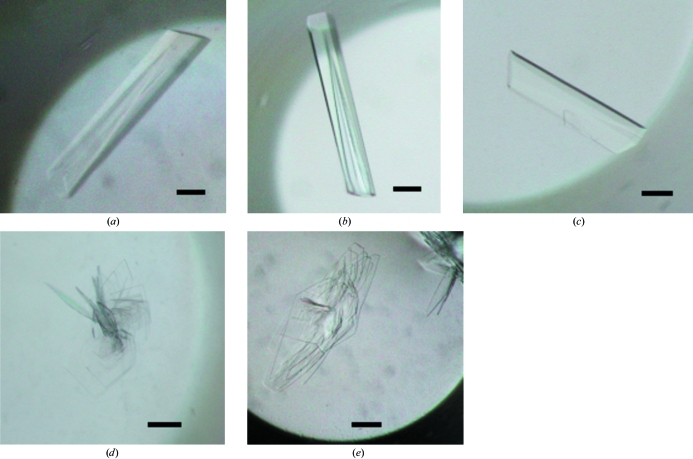
We screened the crystallization conditions for PhBCCPΔN76, BPL* and BPL** and discovered that their crystals grew under the same crystallization condition as used for wild-type PhBPL and described in Bagautdinov et al. (2005 ▶). With the aim of forming complexes with biotin and ATP, a 50 µl PhBPL* (or PhBPL**) protein aliquot comprising 7.5 mg ml−1 protein in 20 mM Tris–HCl pH 8.0 was incubated for 5 min at room temperature with 5 mM biotin and ATP. PhBCCPΔN76 was then added to the PhBPL* (or PhBPL**) in an equimolar ratio and incubation was continued for another 10 min. These mixtures were immediately used to set up crystallization. A typical SDS–PAGE pattern of the protein samples used for crystallization is presented in Fig. 1 ▶. Crystallizations of PhBCCPΔN76, PhBPL* and PhBPL** and of PhBPL*–PhBCCPΔN76 and PhBPL**–PhBCCPΔN76 mixtures were carried out by the oil-microbatch method (Chayen et al., 1990 ▶) at 295 K using Nunc HLA plates (Nalge Nunc International). The crystallization drop was made up of 1.0 µl protein solution and an equal amount of precipitant solution comprising 10.5% PEG 20 000, 0.1 M acetate–NaOH pH 5.2. The crystallization drop was overlaid with a 1:1 mixture of silicone and paraffin oil, allowing slow evaporation of water in the drop, and was stored at 295 K. For PhBPL*, PhBPL** and PhBCCPΔN76, colourless rod-shaped crystals grew to a suitable size for diffraction experiments within a week and the crystals diffracted X-rays to 1.45, 1.50 and 1.55 Å resolution, respectively. Complex crystals grow predominately as sheaves of thin colourless plates which appeared within three to four weeks (Fig. 2 ▶). Detached plates from the sheaves of PhBPL*–PhBCCPΔN76 and PhBPL**–PhBCCPΔN76 complex crystals yielded diffraction to 2.7 and 2.0 Å resolution, respectively.
Figure 1.
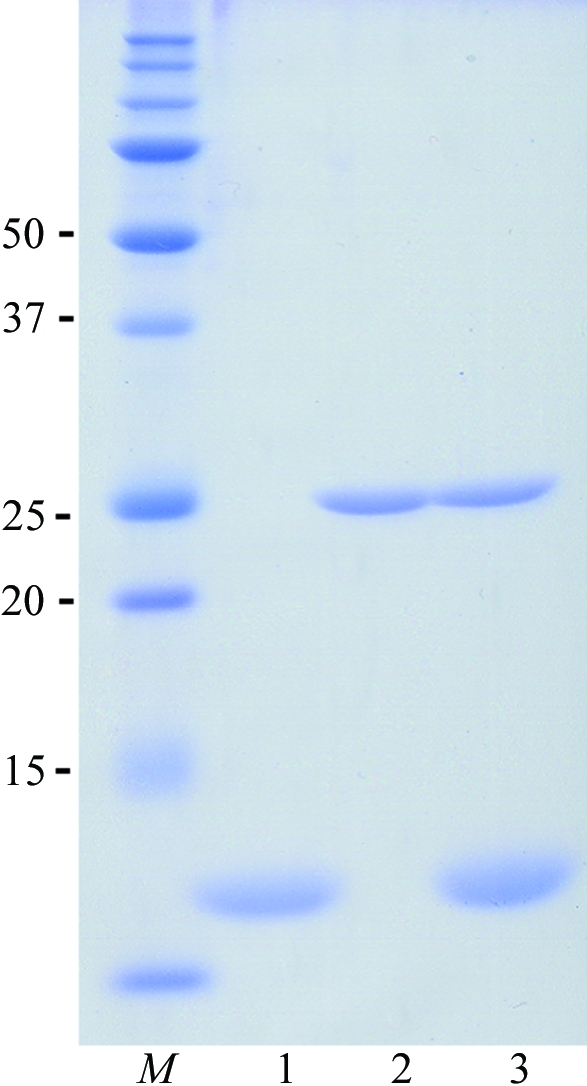
SDS–PAGE of purified samples (lane 1, PhBCCPΔN76; lane 2, PhBPL**; lane 3, PhBPL*–PhBCCPΔN76; lane M, molecular-weight markers labelled in kDa). Proteins were denatured in SDS–PAGE sample buffer, separated on a 15% polyacrylamide gel and stained with Coomassie Brilliant Blue. An analogous SDS–PAGE was observed for PhBPL* and PhBPL*–PhBCCPΔN76.
Figure 2.
Crystals of (a) PhBPL*, (b) PhBPL**, (c) PhBCCPΔN76, (d) PhBPL*–PhBCCPΔN76 and (e) PhBPL**–PhBCCPΔN76. The scale bars are 0.1 mm in length. The single plates of the PhBPL*–PhBCCPΔN76 and PhBPL**–PhBCCPΔN76 crystals used for the X-ray diffraction study had approximate dimensions of 0.5 × 0.6 × 0.02 and 0.4 × 0.6 × 0.03 mm, respectively.
2.3. Data collection
Data sets were collected from the PhBPL*, PhBPL**, PhBCCPΔN76, PhBPL*–PhBCCPΔN76 and PhBPL**–PhBCCPΔN76 crystals. The crystals were transferred from the crystallization drop using a nylon loop (Hampton Research) into a cryoprotectant solution comprising 10.5% PEG 20 000, 0.1 M acetate–NaOH pH 5.2 and 20%(v/v) glycerol. After soaking in this solution for a few seconds, the crystals were flash-cooled in a nitrogen-gas stream at 100 K. X-ray diffraction data were collected at 100 K using synchrotron radiation on a Jupiter 210 charge-coupled device detector at beamline BL26B1 of SPring-8, Japan. For the PhBPL*, PhBPL** and PhBCCPΔN76 crystals, the crystal-to-detector distance was 150 mm. Each of a total of 180 frames was exposed for 25 s with 1° oscillation. For the PhBPL*–PhBCCPΔN76 and PhBPL**–PhBCCPΔN76 crystals, the collection conditions were the same except that the crystal-to-detector distance was set to 220 mm with an exposure time of 20 s. Data were processed and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results
We have established the expression, purification and crystallization of PhBCCPΔN76, which lacks 76 residues from the N-terminus, and have successfully cocrystallized it with PhBPL* or PhBPL**. Crystals appeared about three to four weeks after crystallization setup and grew to approximate dimensions of 0.50 × 0.60 × 0.02 mm and 0.40 × 0.60 × 0.03 mm, respectively (Fig. 2 ▶). Data-collection statistics are summarized in Table 1 ▶. The PhBPL*–PhBCCPΔN76 and PhBPL**–PhBCCPΔN76 complex crystals diffracted to better than 2.7 and 2.0 Å resolution, respectively, and systematic absences revealed that they belonged to space group P21. Assuming a 2:2 complex of PhBPL–PhBCCPΔN76 in the asymmetric unit gives a crystal volume per protein weight (V M) of 2.14 Å3 Da−1 and a solvent content of 50% (Matthews, 1968 ▶). The crystal structures of PhBPL*, PhBPL** and PhBCCPΔN76 are available in the PDB (PDB codes 2dzc, 2e64 and 2d5d, respectively; Bagautdinov et al., unpublished results). Only BCCPΔN76 displayed essentially different crystal packing, belonging to space group P212121. These structures were used for molecular-replacement calculations using the program MOLREP (Vagin & Teplyakov, 1997 ▶). The calculations yielded obvious solutions and the current model building indicates that we have for the first time captured the PhBPL*–PhBCCPΔN76 and PhBPL**–PhBCCPΔN76 complex states containing biotin and partly disordered ATP. These structures will be a good foothold to obtain valuable information regarding molecular recognition and interaction in the BPL–BCCP system.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| BPL* | BPL** | BCCPΔN76 | BPL*–BCCPΔN76 | BPL**–BCCPΔN76 | |
|---|---|---|---|---|---|
| Space group | P21 | P21 | P212121 | P21 | P21 |
| Unit-cell parameters (Å) | |||||
| a (Å) | 38.36 | 38.30 | 39.69 | 69.57 | 69.85 |
| b (Å) | 82.77 | 82.88 | 39.77 | 63.48 | 63.12 |
| c (Å) | 72.71 | 72.58 | 88.42 | 75.72 | 75.64 |
| β (°) | 103.2 | 103.5 | 90.0 | 93.7 | 95.9 |
| VM (Å3 Da−1) | 2.16 | 2.15 | 2.18 | 2.45 | 2.45 |
| Wavelength (Å) | 1.00000 | 1.00000 | 1.00000 | 1.00000 | 1.00000 |
| Resolution range (Å) | 50–1.45 (1.50–1.45) | 50–1.50 (1.55–1.50) | 50–1.55 (1.61–1.55) | 50–2.70 (2.80–2.70) | 50–2.00 (2.07–2.00) |
| Total observations | 224041 | 244726 | 123723 | 47075 | 133241 |
| Unique reflections | 72231 | 69164 | 20755 | 15564 | 42152 |
| Multiplicity | 3.2 (2.9) | 3.6 (3.3) | 6.0 (3.4) | 3.1 (2.8) | 3.2 (3.0) |
| Completeness (%) | 92.4 (97.2) | 98.1 (95.9) | 98.5 (98.0) | 95.1 (86.6) | 95.8 (90.0) |
| Mean I/σ(I) | 15.5 (3.1) | 12.8 (2.2) | 27.7 (3.1) | 6.9 (2.0) | 9.0 (2.8) |
| Rmerge† (%) | 4.8 (37.9) | 4.2 (27.4) | 4.5 (29.2) | 10.7 (32.0) | 8.7 (31.3) |
R
merge = 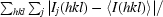
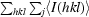 , where Ij(hkl) and 〈I(hkl)〉 are the observed intensity of measurement j and the mean intensity of the reflection with indices hkl, respectively.
, where Ij(hkl) and 〈I(hkl)〉 are the observed intensity of measurement j and the mean intensity of the reflection with indices hkl, respectively.
Acknowledgments
The authors thank the staff of RIKEN Genomic Science Center for providing plasmids and Dr M. Miyano for providing PH1284. We also thank the staff of beamline BL26B1 of SPring-8 for assistance during X-ray experiments. This work (PH0147/HTPF10023 and PH1284/HTPF12746) was supported by the National Project on Protein Structural and Functional Analyses funded by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
References
- Bagautdinov, B., Kuroishi, C., Sugahara, M. & Kunishima, N. (2005). J. Mol. Biol.353, 322–333. [DOI] [PubMed] [Google Scholar]
- Beckett, D. & Matthews, B. W. (1997). Methods Enzymol.279, 362–375. [DOI] [PubMed] [Google Scholar]
- Boer, E. de, Rodriguez, P., Bonte, E., Krijgsveld, J., Katsantoni, E., Heck, A., Grosveld, F. & Strouboulis, J. (2003). Proc. Natl Acad. Sci. USA, 100, 7480–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman-Smith, A. & Cronan, J. E. (1999). Trends Biochem. Sci.24, 359–363. [DOI] [PubMed] [Google Scholar]
- Chayen, N. E., Shaw Stewart, P. D., Maeder, D. L. & Blow, D. M. (1990). J. Appl. Cryst.23, 297–302. [Google Scholar]
- Clarke, D. J., Coulson, J., Baillie, R. & Campopiano, D. J. (2003). Eur. J. Biochem.270, 1277–1287. [DOI] [PubMed] [Google Scholar]
- Howarth, M., Takao, K., Hayashi, Y. & Ting, A. Y. (2005). Proc. Natl Acad. Sci. USA, 102, 7583–7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, J. R. (1989). Annu. Rev. Biochem.58, 195–221. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Nenortas, E. & Beckett, D. (1996). J. Biol. Chem.271, 7559–7567. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Polyak, S. W., Chapman-Smith, A., Mulhern, T. D., Cronan, J. E. Jr & Wallace, J. C. (2001). J. Biol. Chem.276, 3037–3045. [DOI] [PubMed] [Google Scholar]
- Streaker, E. D. & Beckett, D. (2006). Protein Sci.15, 1928–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025. [Google Scholar]
- Weaver, L. H., Kwon, K., Beckett, D. & Matthews, B. W. (2001). Protein Sci.10, 2618–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, H. G., Harmon, F. R., Wuhr, B., Hubner, K. & Lynen, F. (1980). J. Biol. Chem.255, 7397–7409. [PubMed] [Google Scholar]