The genes encoding XMT and DXMT, the enzymes from Coffea canephora (robusta) that catalyse the three independent N-methyl transfer reactions in the caffeine-biosynthesis pathway, have been cloned and the proteins have been expressed in Escherichia coli. Both proteins have been crystallized in the presence of the demethylated cofactor S-adenosyl-l-cysteine (SAH) and substrate (xanthosine for XMT and theobromine for DXMT).
Keywords: caffeine, SAM, N-methyltransferases
Abstract
Caffeine is a secondary metabolite produced by a variety of plants including Coffea canephora (robusta) and there is growing evidence that caffeine is part of a chemical defence strategy protecting young leaves and seeds from potential predators. The genes encoding XMT and DXMT, the enzymes from Coffea canephora (robusta) that catalyse the three independent N-methyl transfer reactions in the caffeine-biosynthesis pathway, have been cloned and the proteins have been expressed in Escherichia coli. Both proteins have been crystallized in the presence of the demethylated cofactor S-adenosyl-l-cysteine (SAH) and substrate (xanthosine for XMT and theobromine for DXMT). The crystals are orthorhombic, with space group P212121 for XMT and C2221 for DXMT. X-ray diffraction to 2.8 Å for XMT and to 2.5 Å for DXMT have been collected on beamline ID23-1 at the ESRF.
1. Introduction
Caffeine (1,3,7-trimethylxanthine) is synthesized by a small group of plants, including coffee, tea and cocoa (Ashihara & Suzuki, 2004 ▶). While caffeine is an important component in beverages made from such plants, it may also protect these plants from predators in nature (Frischknecht, 1985 ▶; Hollingsworth et al., 2002 ▶; Uefuji et al., 2005 ▶). The caffeine-biosynthetic pathway (xanthosine→7-methylxanthosine→7-methylxanthine→theobromine→caffeine) is composed of three N-methyl transfer reactions and a nucleosidase reaction. The three N-methyltransferases from Coffea arabica have recently been isolated and characterized (Ogawa et al., 2001 ▶; Uefuji et al., 2003 ▶). These proteins have been designated xanthosine methyltransferase (XMT), 7-methylxanthine methyltransferase (MXMT) and 3,7-dimethylxanthine methyltransferase (DXMT) (Uefuji et al., 2003 ▶). While the N-methyltransferases involved in caffeine biosynthesis exhibit a high degree of sequence homology (>80%) to each other, they also show remarkable substrate specificity (Uefuji et al., 2003 ▶).
The N-methyltransferases from the caffeine-biosynthesis pathway belong to the plant-specific motif B′ methyltransferases (Kato & Mizuno, 2004 ▶), a subgroup of the larger S-adenosyl-l-methione (SAM) dependent methyltransferase family (Martin & McMillan, 2002 ▶). They share ∼40% sequence homology with other motif B′ methyltransferases (Kato & Mizuno, 2004 ▶), enzymes that are involved in the biosynthesis of small-molecule esters, some of whom have been implicated in plant–plant and plant–insect communications (Shulaev et al., 1997 ▶). Only one member of this family, salicyclic acid O-methyltransferase (SAMT), has been structurally characterized to date (Zubieta et al., 2003 ▶). In order to expand the structural information available for the motif B′ methyltransferases, we decided to undertake the structural determination of XMT and DXMT from C. canephora (robusta). A structural comparison between SAMT, XMT and DXMT should allow the identification of key structural features important for catalysis and substrate recognition of diverse compounds by this important family of plant methyltransferases. Here, we describe the successful cloning, purification and crystallization of XMT and DXMT, the key N-methyltransferases involved in caffeine biosynthesis in C. canephora (robusta).
2. Materials and methods
2.1. Cloning and expression
The cDNA clones pcccs30w12m12 and pcccp21sf19 encoding the sequences of C. canephora caffeine-pathway N-methyltransferases were obtained from the Cornell/Nestlé EST library (Lin et al., 2005 ▶). Resequencing confirmed that these cDNAs encoded the full-length N-methyltransferase sequences and these were renamed XMT and DXMT owing to their very close homologies with other coffee XMT and DXMT sequences in GenBank. The DNA fragments encoding both the XMT and DXMT proteins were obtained by PCR from the pcccs30w12m12 and pcccp21sf19 cDNA clones, respectively. The single set of primers 1 (5′-CAGCACTAGGGATCCATGGAGCTCCAAGAAGTCCTG-3′) and 2 (5′-TCGTCGATCACTCGAGTTACATGTCTGACTTCTCTGGCTT-3′) were used to amplify the relevant region and incorporate the desired restriction sites (BamHI/XhoI) for cloning into the expression vector pProEXHTb (Life Technologies). This plasmid was first transformed into the Escherichia coli cloning strain TOP10 using the ampicillin resistance of the plasmid for selection and the resulting colonies were screened by colony PCR. Those incorporating the XMT and DXMT inserts were sequenced in order to ensure that the correct sequence was cloned and no mutations were introduced. The nucleotide sequences reported in this paper have been submitted to the DDBJ/GenBank/EBI Data Bank with accession Nos. DQ422954 (XMT) and DQ422955 (DXMT).
The plasmids were subsequently transformed into a BL21(DE3)-RIL expression strain for protein production. The cells were grown in Luria–Bertani broth containing 100 µg ml−1 ampicillin and 34 µg ml−1 choloramphenical. Small-scale tests that confirmed the presence of an expressed protein of the correct molecular weight were used to find the optimal soluble expression conditions. Large-scale expression (2–4 l) was carried out by first growing the cells at 310 K. The cultures were then moved to 293 K when the OD600 was 0.6–0.8. After 1 h, expression was induced by addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 0.5 mM and incubation was continued overnight at 293 K. The cells were harvested at 4000g for 15 min at 277 K.
2.2. Purification
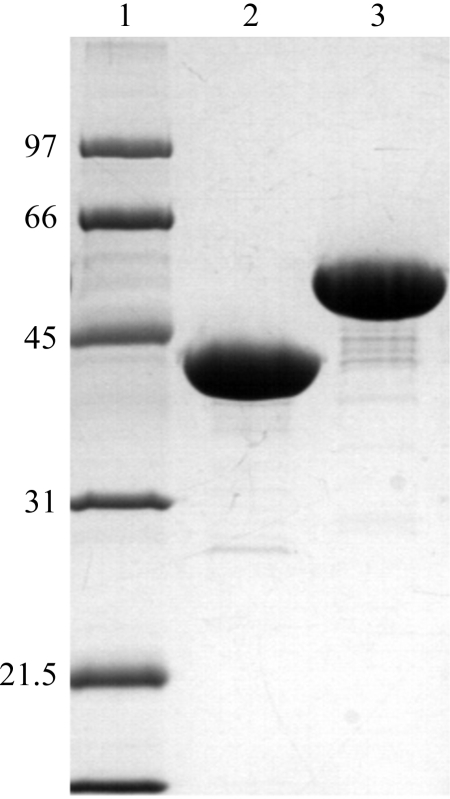
The XMT and DXMT proteins were both purified using the following protocol. The bacterial cells were resuspended in lysis buffer [50 mM Tris–HCl pH 8.0, 500 mM NaCl, 20 mM imidazole pH 8.0, 20 mM β-mercaptoethanol, 10%(v/v) glycerol and 1%(v/v) Tween 20] and stirred at 277 K for 1 h with lysozyme (0.5 mg ml−1). After sonication, the cell debris was removed by centrifugation at 20 000g for 20 min at 277 K. The protein was extracted from the clarified supernatant by mixing with pre-equilibrated Ni–NTA resin (Qiagen), loading onto a gravity-flow column and washing with ten volumes of lysis buffer and ten volumes of wash buffer [50 mM Tris–HCl pH 8.0, 500 mM NaCl, 20 mM imidazole pH 8.0, 20 mM β-mercaptoethanol, 10%(v/v) glycerol]. The His-tagged protein was eluted with elution buffer [50 mM Tris–HCl pH 8.0, 500 mM NaCl, 250 mM imidazole pH 8.0, 20 mM β-mercaptoethanol and 10%(v/v) glycerol]. Protein-containing fractions were pooled and dialysed by concentration into 50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 5 mM β-mercaptanol. The polyhistidine tag was removed by digestion with tobacco etch virus (TEV) protease for 2 h, followed by passage of the digestion mixture through Ni–NTA resin (Qiagen) to remove uncleaved protein and the TEV protease. The protein was then concentrated and loaded onto a Superdex-200 HR10/30 gel-filtration column (GE Healthcare). The protein eluted as a single peak and the protein-containing fractions were pooled and incubated overnight with 1 mM SAH containing either 1 mM xanthosine (XMT) or 1 mM theobromine (DXMT) in 10 mM HEPES buffer pH 7.5 and 5 mM DTT. Samples were then concentrated to an approximate concentration of 6–10 mg ml−1 by ultrafiltration and analysed by SDS–PAGE gel electrophoresis (Fig. 1 ▶).
Figure 1.
SDS–PAGE (12.5%) analysis of XMT and DXMT; approximately 20 µg of each protein was loaded. Lane 1 contains molecular-weight markers (kDa), lane 2 contains XMT and lane 3 contains DXMT.
2.3. Crystallization and preliminary X-ray data
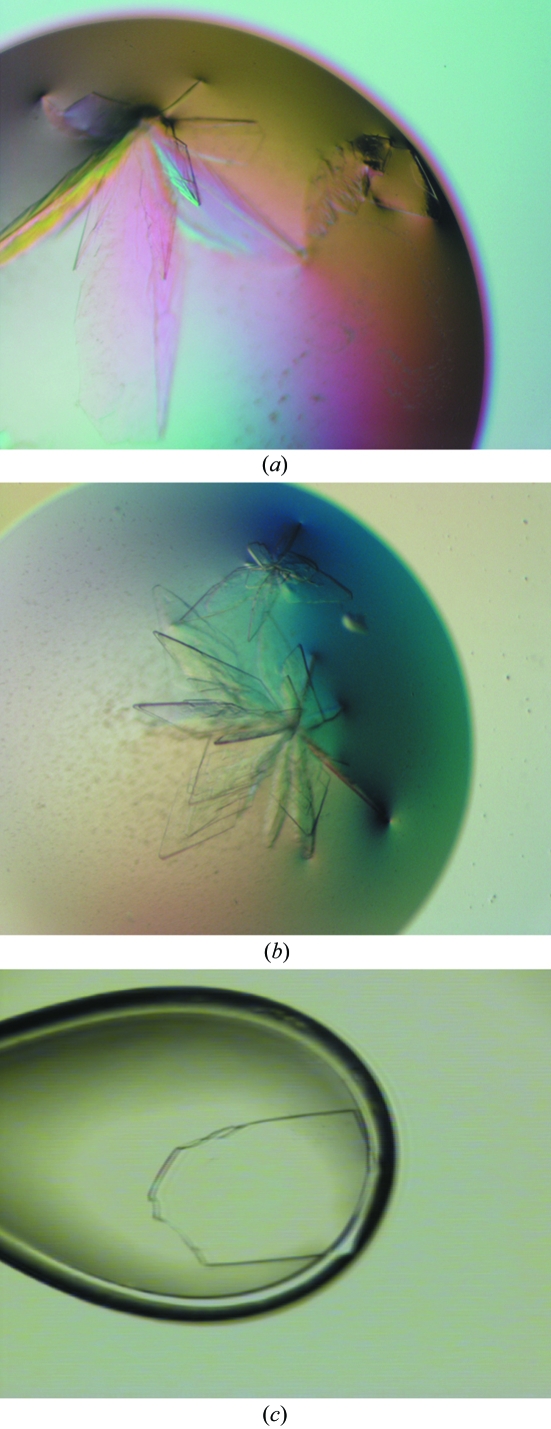
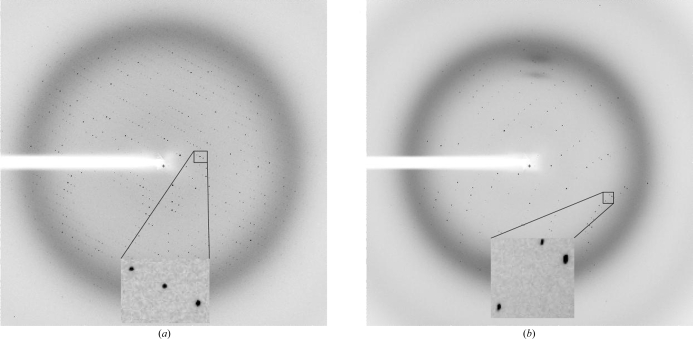
Initial crystallization screens were carried out using a Cartesian crystallization robot in the EMBL Grenoble high-throughput crystallization facility. The initial crystallization conditions were refined manually using Linbro plates (Hampton Research) and the conventional hanging-drop technique (McPherson, 1999 ▶). XMT and DXMT crystallized under very similar conditions, with the crystals forming flower-shaped crystal assemblies after 1–3 d at 293 K (Figs. 2 ▶ a and 2 ▶ b). These large assemblies could be broken into single plate-like crystals, some of which were suitable for X-ray diffraction experiments (Fig. 2 ▶ c). XMT crystallized from precipitant containing 17–22% PEG 3350, 100–200 mM Li2SO4 and 100 mM Tris–HCl pH 8.5 containing 2 mM DTT, 1 mM SAH and 1 mM xanthosine. DXMT crystallized from precipitant containing 23–28% PEG 3350, 200 mM Li2SO4, 100 mM Tris–HCl pH 8.5–8.7 containing 2 mM DTT, 1 mM SAH and 1 mM theobromine. The crystals were then transferred to cryogenic conditions containing 38% PEG 3350 before flash-freezing them at 100 K. All X-ray data were collected on beamline ID23-1 at the European Synchrotron Radiation Facility, ESRF, Grenoble, France (Nurizzo et al., 2006 ▶). XMT in complex with SAH and xanthosine crystallized in space group P212121 with two molecules per asymmetric unit and a 2.8 Å data set was collected (Fig. 3 ▶ a). DXMT in complex with SAH and theobromine crystallized in space group C2221 with one molecule in the asymmetric unit and a 2.5 Å data set was collected (Fig. 3 ▶ b). All data were integrated and scaled using the XDS suite (Kabsch, 1993 ▶) and a summary of the data statistics is given in Table 1 ▶.
Figure 2.
Crystals of (a) XMT and (b) DXMT as described in §2.3. (c) Photograph of a single cryoprotected crystal of DXMT (200 × 150 × 20 µm) as seen through the on-axis visualization unit of the mini-diffractometer on ID23-1. The crystal was manipulated as described in §2.3.
Figure 3.
X-ray diffraction pattern obtained from a crystals of (a) XMT and (b) DXMT on beamline ID23-1 at the ESRF using a 1 s exposure and a MAR CCD 225 mm detector.
Table 1. Crystal data and data-collection statistics.
Values in parentheses are for the outermost shell.
| Protein | XMT | DXMT |
|---|---|---|
| X-ray source | ID23-1 | ID23-1 |
| Wavelength (Å) | 0.9762 | 0.9762 |
| Unit-cell parameters (Å) | ||
| a | 57.29 | 50.47 |
| b | 115.74 | 105.59 |
| c | 119.26 | 140.95 |
| Space group | P212121 | C2221 |
| No. of molecules in ASU | 2 | 1 |
| Matthews coefficient (Å3 Da−1) | 2.4 | 2.2 |
| Solvent content (%) | 48.3 | 43.5 |
| Resolution range (Å) | 30–2.8 (3.0–2.8) | 30–2.5 (2.6–2.5) |
| Unique reflections | 78288 | 52943 |
| Observed reflections [I/σ(I) > 1] | 19533 | 13120 |
| Completeness (%) | 96.4 (88.6) | 97.4 (95.0) |
| Rmerge† | 9.0 (39.1) | 5.6 (29.3) |
| Mean I/σ(I) | 13.7 (4.3) | 17.2 (5.0) |
R
merge = 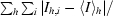
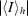 calculated for the whole data set.
calculated for the whole data set.
3. Results
Unigene sequences potentially encoding enzymes involved in caffeine synthesis were identified in the Cornell/Nestlé coffee EST database using a BLASTN search with a cDNA from C. arabica encoding a 7-methylxanthine methyltransferase (MXMT; accession No. AB048794). This search uncovered five different unigenes with very high levels of identity. Two apparently full-length cDNA clones (pcccs30w12m12 and pcccp21sf19) from one of these unigenes (unigene 123341) were completely sequenced. Although these sequences were highly related (83.5% identity at the DNA level and 81.8% identity at the protein level using ClustalW), they encoded two distinct full-length open reading frame (ORF) sequences for N-methyltransferases. Based on their homologies with other coffee cDNA sequences coding for biochemically characterized proteins, we assigned these C. canephora clones as XMT and DXMT, respectively. We have noticed that both cDNA sequences contain very short ORF sequences in their 5′ untranslated regions (5′UTR regions) and that in both cases the stop codon associated with this small ORF is situated just prior to the N-terminal methionine codon of the main ORF. This arrangement may have some biological function because it appears to be perfectly conserved in other characterized coffee cDNAs of this caffeine-synthesis gene family (see, for example, accession Nos. AB048793 and AB086415). It will be interesting in the future to determine if this short 5′UTR ORF has some influence on the transcriptional or post-transcriptional regulation of caffeine-synthesis genes.
Both XMT and DXMT proved to be insoluble using a simple lysis buffer (50 mM Tris–HCl pH 8.0, 200 mM NaCl and 5 mM β-mercaptanol). However, we could solubilize both proteins with the addition of 10% glycerol and 1% Tween 20, which is identical to the lysis buffer used for SAMT (Zubieta et al., 2003 ▶). Otherwise, the purification protocol followed the standard procedures outlined in §2. Both XMT and DXMT elute from a Superdex-200 HR10/30 gel-filtration column as a single peak corresponding to an apparent molecular weight of ∼80 kDa or approximately twice the calculated molecular weight of monomeric XMT and DXMT (42 and 43.4 kDa, respectively). This indicates that XMT and DXMT are homodimeric in solution, consistent with observations for the structural homologue SAMT (Zubieta et al., 2003 ▶).
The X-ray data from XMT suggested P212121 or a related space group with two molecules per asymmetric unit, corresponding to a Matthews coefficient (Matthews, 1968 ▶) of 2.4 Å3 Da−1 and a solvent content of 48.3%. Initial molecular-replacement trials using Phaser (McCoy et al., 2005 ▶) as implemented in CCP4 (Collaborative Computational Project, Number 4, 1994 ▶) with the SAMT structure (Zubieta et al., 2003 ▶) as a search model verified that the space group is indeed P212121 and that two molecules form a dimer in the asymmetric unit. The X-ray data from DXMT suggested space group C222 or C2221 with one molecule per asymmetric unit, corresponding to a Matthews coefficient (Matthews, 1968 ▶) of 2.2 Å3 Da−1 and a solvent content of 43.5%. Initial molecular-replacement trials using Phaser (McCoy et al., 2005 ▶) as implemented in CCP4 (Collaborative Computational Project, Number 4, 1994 ▶) with the SAMT structure (Zubieta et al., 2003 ▶) as a search model verified C2221 as the correct space group. Here, the biological dimer is preserved by a crystallographic twofold axis along b. The XMT and DXMT structures are currently under refinement and we are searching for better diffracting crystals in order to aid in their refinement.
Acknowledgments
We gratefully acknowledge the use of the EMBL Grenoble high-throughput crystallization facility and thank the EMBL Grenoble/ESRF Joint Structural Biology Group for access to and support at ESRF beamline ID23-1. We would also like to thank D. Vilanova and M. Lepelley for their assistance in the early part of this work.
References
- Ashihara, H. & Suzuki, T. (2004). Front. Biosci.9, 1864–1876. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Frischknecht, P. M. (1985). Phytochemistry, 3, 613–616.
- Hollingsworth, R. G., Armstrong, J. W. & Campbell, E. (2002). Nature (London), 417, 915–916. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800. [Google Scholar]
- Kato, M. & Mizuno, K. (2004). Front. Biosci.9, 1833–1842. [Google Scholar]
- Lin, C., Mueller, L. A., McCarthy, J., Crouzillat, D., Petiard, V. & Tanksley, S. D. (2005). Theor. Appl. Genet.112, 114–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed] [Google Scholar]
- McPherson, A. (1999). Crystallization of Biological Macromolecules. Cold Spring Harbour, NY, USA: Cold Spring Harbour Laboratory Press.
- Martin, J. L. & McMillan, F. M. (2002). Curr. Opin. Struct. Biol.12, 783–793. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Nurizzo, D., Mairs, T., Guijarro, M., Rey, V., Meyer, J., Fajardo, P., Chavanne, J., Biasci, J.-C., McSweeney, S. & Mitchell, E. (2006). J. Synchrotron Rad.13, 227–238. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Herai, Y., Koizumi, N., Kusano, T. & Sano, H. (2001). J. Biol. Chem.276, 8213–8218. [DOI] [PubMed] [Google Scholar]
- Shulaev, V., Silverman, P. & Raskin, I. (1997). Nature (London), 385, 718–721.
- Uefuji, H., Shinjiro, O., Yamaguchi, Y., Koizumi, N. & Sano, H. (2003). Plant Physiol.132, 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uefuji, H., Tatsumi, Y., Morimoto, M., Kaothien-Nakayama, P., Ogita, S. & Sano, H. (2005). Plant Mol. Biol.59, 221–227. [DOI] [PubMed] [Google Scholar]
- Zubieta, C., Ross, J. R., Koscheski, P., Yang, Y., Pichersky, E. & Noel, J. P. (2003). Plant Cell, 15, 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]