Glutaredoxin 2 from E. coli was cocrystallized with glutathione and data were collected to 1.60 Å. A mutant with the active-site residues Cys9 and Cys12 changed to serine was crystallized in the absence of glutathione and data were collected to 2.4 Å.
Keywords: glutaredoxin 2
Abstract
Glutaredoxin 2 (Grx2) from Escherichia coli is larger in size than classical glutaredoxins. It is extremely efficient in the catalysis of reduced glutathione-dependent disulfide reduction. A complex of Grx2 with reduced glutathione (GSH) has been crystallized. Data were collected to 1.60 Å. The crystals belong to space group P3221, with one Grx2–GSH complex in the asymmetric unit. The unit-cell parameters are a = b = 50.10, c = 152.47 Å. A Grx2 mutant, C9S/C12S, which cannot form a disulfide bond with GSH was also crystallized. The crystals diffracted to 2.40 Å and belong to space group P212121, with one molecule in the asymmetric unit. The unit-cell parameters are a = 28.16, b = 78.65, c = 89.16 Å.
1. Introduction
The glutaredoxin protein family is ubiquitously present in nearly every living organism from bacteria to humans. These enzymes are general GSH-dependent thiol–disulfide oxidoreductases (Fernandes & Holmgren, 2004 ▶). They can operate in a dithiol mechanism to reduce protein disulfides or in a monothiol and/or dithiol mechanism to reduce mixed disulfides formed between reduced glutathione (GSH) and proteins or low-molecular-weight thiols (Bushweller et al., 1992 ▶). Glutaredoxins have been implicated in a variety of cellular functions, including transcription, signal transduction, cell-cycle control, protection against oxidative stress and the reduction of the enzymes ribonucleotide reductase and 3′-phosphoadenylylsulfate reductase (reviewed in Fernandes & Holmgren, 2004 ▶). They are also involved in the glutathionylation of proteins (Ghezzi, 2005 ▶) and arsenate reduction (Mukhopadhyay & Rosen, 2002 ▶).
To date, four glutaredoxins (Grx1, Grx2, Grx3 and Grx4) have been characterized in Escherichia coli (Åslund et al., 1994 ▶; Holmgren, 1976 ▶; Fladvad et al., 2005 ▶). Grx4 has recently been identified as the only monothiol glutaredoxin in E. coli, with a suggested CGFS active site (Fladvad et al., 2005 ▶). Grx1, Grx2 and Grx3 are all dithiol glutaredoxins, with CPYC as their active site. Grx2 is atypical, differing from other classical glutaredoxins in its larger size and lower amino-acid sequence similarity to other glutaredoxins. It is extremely efficient in the catalysis of GSH-dependent disulfide reduction (Vlamis-Gardikas et al., 1997 ▶; Lundstrom-Ljung et al., 1999 ▶). Grx2 has been shown to be the most effective hydrogen donor for the reduction of arsenate by plasmid R773 ArsC, an arsenate reductase (Shi et al., 1999 ▶). The NMR structure of reduced Grx2 has been obtained (Xia et al., 2001 ▶) and the structure shows an overall similarity to glutathione S-transferases (GSTs), with the additional C-terminal residues forming a second larger domain that is lacking in Grx1 or Grx3. The NMR structures of the GSH complexes of human (Yang et al., 1998 ▶) and E. coli Grx1 (Bushweller et al., 1994 ▶) and E. coli Grx3 (Nordstrand et al., 1999 ▶) have been reported. The structure of the complex of human Grx2 (which, despite its name, is not closely related to E. coli Grx2) with GSH has been deposited with PDB code 2fls (C. Johansson et al., unpublished results). However, information on the GSH-complexed or oxidized structures of E. coli Grx2 is still lacking. Here, we describe crystallization of the Grx2–GSH complex and of a derivative lacking cysteine that is unable to form a complex with GSH.
2. Materials and methods
2.1. Protein expression and purification
The grx2 gene was cloned into pET28a vector (Novagen) and expressed in E. coli strain BL21(DE3) (Shi et al., 1999 ▶). The protein with a six-histidine tag was purified as described previously (Shi et al., 1999 ▶), with the addition of a final step of gel-filtration chromatography using Superdex G-75. The six-histidine-tag fusion peptide was removed using a Thrombin Cleavage Capture Kit (Novagen) before crystallization, leaving three residues (Gly-Ser-His) from the vector at the N-terminus. A Grx2 C9S/C12S derivative (Shi et al., 1999 ▶) was purified similarly.
2.2. Crystallization
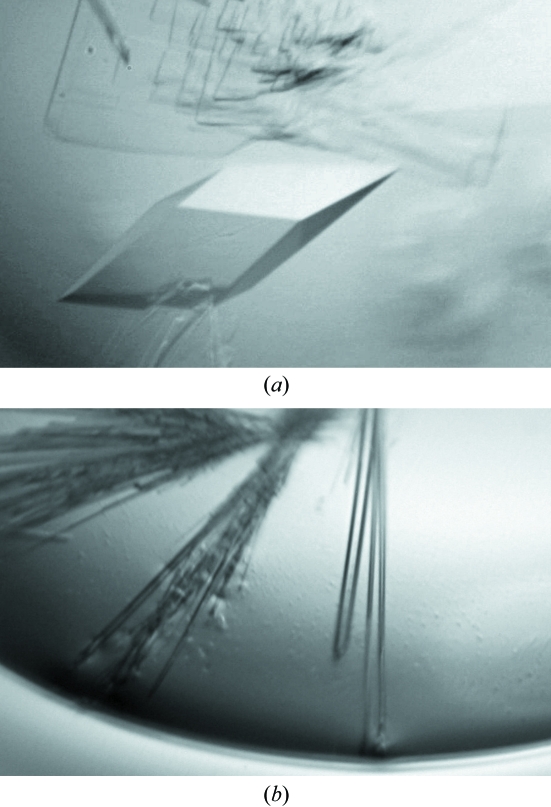
Purified Grx2 was buffer-exchanged into a buffer consisting of 10 mM Tris–HCl pH 7.5 containing 2 mM disodium ethylenediamine tetraacetate (EDTA), 5 mM dithiothreitol (DTT). Crystals were grown using hanging-drop vapor diffusion at 277 K with VDX plates (Hampton Research). About 200 solution conditions were tried before crystals were obtained. After incubation with 4 mM GSH and 20 mM sodium arsenate on ice for 10 min, 2 µl purified Grx2 (20 mg ml−1) was mixed with 2 µl well solution containing 0.1 M sodium citrate, 0.2 M ammonium acetate and 30%(w/v) PEG 4000 at a final pH of 5.9. The drop was then equilibrated against 0.5 ml well solution. The Grx2 C9S/C12S mutant was crystallized under similar conditions but in the absence of GSH and sodium arsenate. Crystals appeared in approximately two weeks (Fig. 1 ▶).
Figure 1.
Crystals of (a) Grx2–GSH and (b) Grx2 C9S/C12S. Approximate dimensions are 0.3 × 0.3 × 0.2 and 0.8 × 0.05 × 0.05 mm for the Grx2–GSH and Grx2 C9S/C12S crystals, respectively.
2.3. Data collection
Grx2–GSH or Grx2 mutant crystals were transferred to a cryoprotectant solution (well solution with an additional 5% PEG 4000) and flash-cooled in nitrogen gas and/or stored in liquid nitrogen. The Grx2–GSH data set was collected under cryogenic conditions at Argonne National Laboratory Advanced Photon Source (ANL-APS) beamline 32-ID-B. It was indexed and integrated using MOSFLM and scaled and merged using SCALA (Collaborative Computational Project, Number 4, 1994 ▶; Table 1 ▶). The Grx2 C9/C12S mutant data set was collected using an in-house Rigaku/MSC FR-D rotating-anode X-ray source equipped with an R-AXIS HTC image-plate detector at a crystal-to-detector distance of 150 mm. The X-rays were focused with the MSC Blue-2 Optics configuration with the focal point fixed at 120 mm beyond the crystal. A collimator with a 2 mm rear and 0.5 mm front aperture was used. The data were integrated, scaled and merged with the program CrystalClear 1.3.6 (Pflugrath, 1999 ▶).
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Grx2–GSH | Grx2 C9S/C12S | |
|---|---|---|
| Wavelength (Å) | 1.0000 | 1.5418 |
| Space group | P3221 | P212121 |
| Unit-cell parameters (Å) | a = b = 50.10, c = 152.47 | a = 28.16, b = 78.65, c = 89.16 |
| Resolution range (Å) | 22.47–1.60 (1.69–1.60) | 29.49–2.40 (2.49–2.40) |
| No. of observations | 185447 (4270) | 26609 (2816) |
| No. of unique reflections | 28685 (2853) | 8139 (785) |
| Completeness (%) | 94.8 (67.0) | 98.7 (99.1) |
| 〈I〉/〈σ(I)〉 | 20.6 (4.7) | 5.0 (1.8) |
| Rmerge† (%) | 6.2 (10.0) | 15.0 (40.7) |
R
merge = 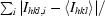
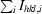 , where I
hkl,i is the intensity of the ith measurement of the reflection with Miller indices hkl and 〈I
hkl〉 is the mean intensity of that reflection.
, where I
hkl,i is the intensity of the ith measurement of the reflection with Miller indices hkl and 〈I
hkl〉 is the mean intensity of that reflection.
2.4. Initial phasing
The Grx2–GSH crystal structure was solved by molecular replacement using the program AMoRe (Navaza, 2001 ▶) implemented in the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). The first conformer of the Grx2 NMR structure (Xia et al., 2001 ▶) was used as an input probe. The best solution gave a correlation coefficient of 0.46. The second highest peak only had a correlation coefficient of 0.28. The Grx2 mutant structure was also solved by molecular replacement using the partially refined Grx2–GSH structure as the search model.
3. Results and discussion
We have crystallized the Grx2–GSH complex and the cysteine-free derivative C9S/C12S. The crystals of the Grx2–GSH complex diffracted to 1.6 Å. The Grx2–GSH crystals belong to space group P3221, with one complex in the asymmetric unit, giving a V M of 2.3 Å3 Da−1 (Matthews, 1968 ▶). The unit-cell parameters are a = b = 50.10, c = 152.47 Å. It is interesting to note that attempts to crystallize this complex in the absence of arsenate were unsuccessful, even though arsenate is not visible in the final structure. One possibility is that arsenate serves as an oxidant for disulfide-bond formation between Grx2 and GSH. Of relevance is the observation that the ectromelia virus glutaredoxin reduces dimethylarsenate to dimethylarsenite and a cysteine thiolate complex with dimethylarsenite was observed in the glutaredoxin structure (Bacik & Hazes, 2007 ▶).
Although the NMR structure of Grx2 in the reduced form has been determined (Xia et al., 2001 ▶), the region between residues 124 and 134 in the C-terminal domain was not well defined and the structure of the GSH-bound form was not determined. Our goal is to identify the conformational changes that occur in Grx2 as a result of interaction with GSH (and eventually with the ArsC arsenate reductase). This requires comparison of the structures of the reduced and oxidized forms in which all of the residues are well defined. However, we were only able to produce crystals of Grx2 in complex with GSH and not in either the reduced or the oxidized form. As an alternative, a C9S/12S derivative that is unable to support GSH-dependent ArsC-catalyzed arsenate reduction was utilized (Shi et al., 1999 ▶). Presumably, this protein is unable to form a complex with GSH and its structure may mimic that of the reduced form. Crystals of Grx2 C9S/C12S were obtained that diffracted to 2.4 Å. The C9S/C12S crystals belong to space group P212121 with one molecule in the asymmetric unit, giving a V M of 2.0 Å3 Da−1. The unit-cell parameters are a = 28.16, b = 78.65, c = 89.16 Å. In contrast to wild-type Grx2, neither arsenate nor GSH were required for crystallization of C9S/C12S.
Of the four E. coli glutaredoxins, only Grx1 and Grx3 have been extensively characterized structurally and functionally. The three-dimensional structures of Grx1 and Grx3 have been obtained by NMR in both the reduced, oxidized and mixed-disulfide forms (Bushweller et al., 1994 ▶; Sodano et al., 1991 ▶; Xia et al., 1992 ▶; Foloppe et al., 2001 ▶; Nordstrand et al., 1999 ▶, 2000 ▶). Although the NMR structure of Grx2 in the reduced form is available, considering the substantial differences in size and number of domains between Grx2 and Grx1 or Grx3, comparison of the crystal structures of Grx2 with and without GSH will be of considerable value in elucidating the mechanism of this atypical glutaredoxin. The refined structures of Grx2–GSH and the Grx2 C9S/C12S mutant and the implications for Grx2 function will be presented elsewhere.
References
- Åslund, F., Ehn, B., Miranda-Vizuete, A., Pueyo, C. & Holmgren, A. (1994). Proc. Natl Acad. Sci. USA, 91, 9813–9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacik, J. P. & Hazes, B. (2007). J. Mol. Biol.365, 1545–1558. [DOI] [PubMed] [Google Scholar]
- Bushweller, J. H., Aslund, F., Wuthrich, K. & Holmgren, A. (1992). Biochemistry, 31, 9288–9293. [DOI] [PubMed] [Google Scholar]
- Bushweller, J. H., Billeter, M., Holmgren, A. & Wuthrich, K. (1994). J. Mol. Biol.235, 1585–1597. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Fernandes, A. P. & Holmgren, A. (2004). Antioxid. Redox Signal.6, 63–74. [DOI] [PubMed] [Google Scholar]
- Fladvad, M., Bellanda, M., Fernandes, A. P., Mammi, S., Vlamis-Gardikas, A., Holmgren, A. & Sunnerhagen, M. (2005). J. Biol. Chem.280, 24553–24561. [DOI] [PubMed] [Google Scholar]
- Foloppe, N., Sagemark, J., Nordstrand, K., Berndt, K. D. & Nilsson, L. (2001). J. Mol. Biol.310, 449–470. [DOI] [PubMed] [Google Scholar]
- Ghezzi, P. (2005). Biochem. Soc. Trans.33, 1378–1381. [DOI] [PubMed] [Google Scholar]
- Holmgren, A. (1976). Proc. Natl Acad. Sci. USA, 73, 2275–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom-Ljung, J., Vlamis-Gardikas, A., Åslund, F. & Holmgren, A. (1999). FEBS Lett.443, 85–88. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, R. & Rosen, B. P. (2002). Environ. Health Perspect.110, Suppl. 5, 745–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza, J. (2001). Acta Cryst. D57, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Nordstrand, K., Åslund, F., Holmgren, A., Otting, G. & Berndt, K. D. (1999). J. Mol. Biol.286, 541–552. [DOI] [PubMed] [Google Scholar]
- Nordstrand, K., Sandstrom, A., Åslund, F., Holmgren, A., Otting, G. & Berndt, K. D. (2000). J. Mol. Biol.303, 423–432. [DOI] [PubMed] [Google Scholar]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- Shi, J., Vlamis-Gardikas, A., Åslund, F., Holmgren, A. & Rosen, B. P. (1999). J. Biol. Chem.274, 36039–36042. [DOI] [PubMed] [Google Scholar]
- Sodano, P., Xia, T. H., Bushweller, J. H., Bjornberg, O., Holmgren, A., Billeter, M. & Wuthrich, K. (1991). J. Mol. Biol.221, 1311–1324. [DOI] [PubMed] [Google Scholar]
- Vlamis-Gardikas, A., Åslund, F., Spyrou, G., Bergman, T. & Holmgren, A. (1997). J. Biol. Chem.272, 11236–11243. [DOI] [PubMed] [Google Scholar]
- Xia, B., Vlamis-Gardikas, A., Holmgren, A., Wright, P. E. & Dyson, H. J. (2001). J. Mol. Biol.310, 907–918. [DOI] [PubMed] [Google Scholar]
- Xia, T. H., Bushweller, J. H., Sodano, P., Billeter, M., Bjornberg, O., Holmgren, A. & Wuthrich, K. (1992). Protein Sci.1, 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Jao, S., Nanduri, S., Starke, D. W., Mieyal, J. J. & Qin, J. (1998). Biochemistry, 37, 17145–17156. [DOI] [PubMed] [Google Scholar]