A leucine-zipper with properties as apoptotic regulator in the ER has been crystallized. X-ray data to 2.5 Å resolution were collected, molecular replacement solutions were identified and refinement has been started.
Keywords: B-cell receptor-associated protein 31, leucine zipper, integral membrane proteins
Abstract
B-cell receptor-associated protein 31 (Bap31) is an integral membrane protein located in the endoplasmic reticulum (ER) that participates in the transport and quality control of membrane proteins and plays a role in determining cell sensitivity to ER stress and apoptosis. Its cytoplasmic region contains two target sites for caspase cleavage in certain apoptotic pathways. Here, the subcloning, expression, purification and crystallization of the Homo sapiens Bap31 leucine-zipper C-terminal fragment, which spans residues Gly160–Glu246, are reported. An N-terminally His-tagged protein was overexpressed in Escherichia coli and purified by chromatographic methods. X-ray diffraction data were collected in-house to 2.5 Å resolution. Crystals belong to space group P6122/P6522, with unit-cell parameters a = b = 70.7, c = 80.6 Å. Data analysis indicates the presence of one molecule per asymmetric unit.
1. Introduction
The endoplasmic reticulum (ER) is an essential intracellular organelle that plays a vital role in many cellular processes, including folding and post-translational modification of membrane-associated proteins, intracellular protein trafficking, sensing of stress and delivery of apoptotic signals (Groenendyk & Michalak, 2005 ▶). The ER contains resident proteins involved in the folding and quality control of newly synthesized proteins. When misfolded or unfolded proteins accumulate, a variety of ER stress-signaling pathways are turned on, including inhibition of protein synthesis as well as facilitation of protein degradation. If the cell is overwhelmed and unable to recover, the onset of the apoptotic pathway ensues. A disturbance in Ca2+ homeostasis appears to account for the start of the apoptosis process (Pinton & Rizzuto, 2006 ▶). Among other players, the cascade involves the Ca2+-dependent protease calpain cleaving and activating caspases, which interact and cleave several targets, including the ER protein transport regulator Bap31 (Breckenridge et al., 2003 ▶; Zuppini et al., 2002 ▶). Collectively, these proteins mediate crosstalk between the ER and the mitochondria via Ca2+ (Chandra et al., 2004 ▶).
Bap31 (B-cell receptor-associated protein 31) is widely spread through eukaryotes, showing homologs in several mammals, insects, worms, yeast and plants. Human Bap31 is a 246-amino-acid type III transmembrane protein located in the ER, consisting of three transmembrane domains and a cytoplasmic region running from residue 124 to the C-terminus (Ng et al., 1997 ▶). This region is presumed to be a target for caspase-8 cleavage at Asp164 and Asp238; it includes an overlapping putative pseudo death-effector domain (pDED) and leucine-zipper homology region running from positions 168 to 233 (Ng et al., 1997 ▶). The C-terminus contains a dilysine motif (KKEE) which has been implicated in protein retention or exit from the ER (Ng et al., 1997 ▶). Its transmembrane domain functions as an anterograde receptor for multiple cargo proteins such as class I MHC (Ladasky et al., 2006 ▶), the cytochrome P450 2C2 (Szczesna-Skorupa & Kemper, 2006 ▶), the tetraspanins CD9 and CD81 (Stojanovic et al., 2005 ▶), the β2 integrin CD11b/CD18 (Zen et al., 2004 ▶) or the IgD heavy chain (Schamel et al., 2003 ▶). In addition, Bap31 senses ER overload and signals apoptosis via its cytoplasmic region.
Several other proteins containing a pDED/leucine zipper have been identified, including BAR (bifunctional apoptosis regulator; Zhang et al., 2000 ▶), HIP-1 (huntingtin-interacting protein 1; Ybe et al., 2007 ▶) and HIPPI (HIP-1 protein interactor; Banerjee et al., 2006 ▶). A signaling model based on their heterotypic interaction has been advanced (Roth et al., 2003 ▶). The proposed partner of Bap31 is the ER transmembrane protein BAR, composed of a RING finger, a SAM domain and a pDED/leucine zipper (Zhang et al., 2000 ▶). The partner of HIP-1 is HIPPI. Recently, the crystallization of HIPPI pDED has been published (Banerjee et al., 2006 ▶). To shed light on the mechanism used by Bap31 and BAR to signal apoptosis upon ER stress, we here report the crystallization and preliminary X-ray diffraction analysis of the leucine-zipper homology region of human Bap31.
2. Materials and methods
2.1. Subcloning, overexpression and protein purification
Using bioinformatics software, the leucine-zipper region was predicted to run from Leu168 to Leu233 (Przybylski & Rost, 2002 ▶). Therefore, we subcloned the region spanning from Gly160 to the last residue, Glu246. The DNA coding fragment for the Bap31 leucine-zipper region was obtained by standard PCR methods using a full-length human Bap31 clone as template (GenBank accession No. NM_005745) and subcloned into the NdeI/BamHI sites of a modified pET15b bacterial expression vector (Novagen), thus adding an N-terminal 6×His tag to the protein (6×His-Met-Gly160). DNA sequencing corroborated the identity of the inserted fragment. The ligated plasmid was transformed into Escherichia coli strain BL21(DE3). Cells were grown at 310 K in LB medium to an optical density (OD) of 0.6 and overnight protein expression was carried out at 288 K upon addition of 0.5 mM IPTG. The Bap31 fragment was highly expressed and soluble as assessed by protein extraction with B-PER reagent (Pierce) followed by SDS–PAGE. Cell pellets from 1 l culture were resuspended in 25 ml lysis buffer (100 mM Tris pH 7.5) and lysed by sonication. Taking advantage of the hexahistidine tag and after centrifugation at 277 K, the filtered supernatant was loaded onto a nickel–NTA column and purified under native conditions using a gradient of imidazole by standard FPLC methods. Protein eluted at an imidazole concentration of 250 mM. Further purification was achieved by gel filtration on an S200 HR column (GE Healthcare) using 20 mM Tris pH 7.0 and 150 mM NaCl as elution buffer. The protein was concentrated to 20 mg ml−1 without visible precipitation. The protein purity was judged to be greater than 90% from SDS–PAGE analysis. MALDI–TOF mass spectrometry showed a molecular weight of 10 561 Da, compared with a calculated value of 10 559 Da. The yield was around 5 mg purified protein per litre of culture.
2.2. Crystallization and data collection
Crystals grew to maximum dimensions of 0.4 × 0.2 × 0.2 mm. within one week at room temperature (298 K) by sitting-drop vapor diffusion. Protein dissolved in gel-filtration buffer at 20 mg ml−1 was mixed with a solution containing 100 mM sodium acetate buffer pH 4.5, 200 mM ammonium acetate, 25%(w/v) PEG 4000 and 15%(v/v) glycerol and equilibrated against a reservoir of 500 µl of the same solution. Prior to data collection, the crystals were quickly dragged through a crystallization solution with glycerol at 25%(v/v) for cryoprotection and flash-frozen in liquid nitrogen. Crystals diffracted isotropically to 2.5 Å and a complete native data set was collected in-house at 100 K using a Cu rotating-anode X-ray source (Rigaku FR-E) equipped with Osmic optics and an R-AXIS IV image-plate detector. A total of 180 frames were collected with a crystal-to-detector distance of 200 mm. The exposure time was 2 min per frame with an oscillation of 1° per image. Data were indexed, integrated and scaled using the HKL package (Otwinowski & Minor, 1997 ▶). The crystal lattice is hexagonal, with unit-cell parameters a = b = 70.7, c = 80.6 Å. Merging statistics and systematic absences are consistent with space groups P6122 or P6522.
3. Results and discussion
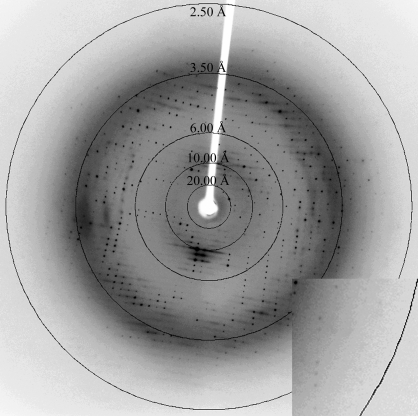
A bioinformatics analysis of human Bap31 confirmed the presence of three transmembrane helices in the N-terminal half of the protein followed by a 124-residue cytoplasmic domain. The latter is predicted to contain a leucine-zipper motif (Leu168–Leu233) flanked by two caspase-8 cleavage sites at Asp164 and Asp238. To gain further understanding of the apoptotic properties of this protein, a fragment of Bap31 comprising the entire leucine-zipper region (residues 160–246) was produced in E. coli as a soluble protein and purified to homogeneity. Crystals suitable for structural studies were grown by sitting-drop vapor diffusion (Fig. 1 ▶) and diffracted to 2.5 Å resolution (Fig. 2 ▶). Assuming the presence of one molecule in the asymmetric unit, the calculated solvent content was 55% (Matthews, 1968 ▶). Data-collection and processing statistics are summarized in Table 1 ▶.
Figure 1.
Image of a typical crystal of human Bap31 leucine-zipper homology region grown by the sitting-drop method.
Figure 2.
X-ray diffraction pattern of a human Bap31 leucine-zipper homology region crystal. Diffuse scattering in the a*b* plane is observable as horizontal streaks. The resolution shell shown on the inset at the bottom right corner corresponds to 2.5 Å.
Table 1. Summary of data-collection and processing statistics.
Values in parentheses are for the outer shell.
| Space group | P6122/P6522 |
| Unit-cell parameters (Å) | a = b = 70.7, c = 80.6 |
| Wavelength (Å) | 1.5418 |
| Resolution (Å) | 2.5 (2.59–2.50) |
| Measured reflections | 45292 |
| Unique reflections | 4411 |
| Redundancy | 10.3 |
| Completeness (%) | 99.2 (98.4) |
| Rmerge (%)† | 5.6 (73.5) |
| 〈I/σ(I)〉 | 34.7 (3.2) |
| Solvent content (%) | 55 |
| Mosaicity (°) | 0.9 |
R
merge = 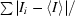
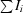 .
.
Similar results have recently been reported for the related pDED of HIPPI (Banerjee et al., 2006 ▶). With 15% sequence identity and 35% sequence similarity, both proteins show a conserved pattern of leucine residues reminiscent of a leucine zipper. Currently, our efforts are focused on identifying models suitable for molecular replacement as well as obtaining selenomethionine-labeled crystals for experimental phasing. The availability of the three-dimensional structure should be helpful in the design of experiments aimed at probing the basis of the apoptotic properties of this family of ER proteins.
Acknowledgments
We would like to thank Drs Jose M. de Pereda and Robert C. Liddington for helpful discussions. This work was supported by NIH grants AI65602 and AI55789 (to JP).
References
- Banerjee, M., Majumder, P., Bhattacharyya, N. P., Dattagupta, J. K. & Sen, U. (2006). Acta Cryst. F62, 1247–1250. [DOI] [PMC free article] [PubMed]
- Breckenridge, D. G., Stojanovic, M., Marcellus, R. C. & Shore, G. C. (2003). J. Cell Biol.160, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra, D., Choy, G., Deng, X., Bhatia, B., Daniel, P. & Tang, D. G. (2004). Mol. Cell. Biol.24, 6592–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenendyk, J. & Michalak, M. (2005). Acta Biochim. Pol.52, 381–395. [PubMed] [Google Scholar]
- Ladasky, J. J., Boyle, S., Seth, M., Li, H., Pentcheva, T., Abe, F., Steinberg, S. J. & Edidin, M. (2006). J. Immunol.177, 6172–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Ng, F. W., Nguyen, M., Kwan, T., Branton, P. E., Nicholson, D. W., Cromlish, J. A. & Shore, G. C. (1997). J. Cell Biol.139, 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pinton, P. & Rizzuto, R. (2006). Cell Death Differ.13, 1409–1418. [DOI] [PubMed] [Google Scholar]
- Przybylski, D. & Rost, B. (2002). Proteins, 46, 197–205. [DOI] [PubMed] [Google Scholar]
- Roth, W., Kermer, P., Krajewska, M., Welsh, K., Davis, S., Krajewski, S. & Reed, J. C. (2003). Cell Death Differ.10, 1178–1187. [DOI] [PubMed] [Google Scholar]
- Schamel, W. W., Kuppig, S., Becker, B., Gimborn, K., Hauri, H. P. & Reth, M. (2003). Proc. Natl Acad. Sci. USA, 100, 9861–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovic, M., Germain, M., Nguyen, M. & Shore, G. C. (2005). J. Biol. Chem.280, 30018–30024. [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa, E. & Kemper, B. (2006). J. Biol. Chem.281, 4142–4148. [DOI] [PubMed] [Google Scholar]
- Ybe, J. A., Mishra, S., Helms, S. & Nix, J. (2007). In the press. [DOI] [PMC free article] [PubMed]
- Zen, K., Utech, M., Liu, Y., Soto, I., Nusrat, A. & Parkos, C. A. (2004). J. Biol. Chem.279, 44924–44930. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Xu, Q., Krajewski, S., Krajewska, M., Xie, Z., Fuess, S., Kitada, S., Pawlowski, K., Godzik, A. & Reed, J. C. (2000). Proc. Natl Acad. Sci. USA, 97, 2597–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuppini, A., Groenendyk, J., Cormack, L. A., Shore, G., Opas, M., Bleackley, R. C. & Michalak, M. (2002). Biochemistry, 41, 2850–2858. [DOI] [PubMed] [Google Scholar]