A dimeric inert form of the replication initiator RepE of the F plasmid was expressed, purified and crystallized in complex with the repE operator DNA. Diffraction data were collected to 3.14 Å resolution.
Keywords: RepE dimer, transcriptional repressor, RepE–operator complex, high-salt purification
Abstract
The replication initiator factor RepE of the F plasmid in Escherichia coli is an essential protein that stringently regulates the F-plasmid copy number. The RepE protein has a dual function: its monomer functions as a replication initiator, while its dimer acts as a transcriptional repressor of the repE gene. The wild-type dimeric RepE protein was expressed as an N-terminal histidine-tagged protein, purified under native conditions with a high salt concentration and crystallized in complex with the repE operator DNA using the sitting-drop vapour-diffusion technique. The crystals diffracted to a resolution of 3.14 Å after the application of dehydration and crystal annealing and belong to space group P21, with unit-cell parameters a = 60.73, b = 99.32, c = 95.00 Å, β = 108.55°.
1. Introduction
The mini-F plasmid, which is a derivative of the F factor in Escherichia coli, replicates under strict control, maintaining a copy number of 1–2 per host chromosome (Kline, 1985 ▶). The plasmid-encoded initiator protein RepE (251 amino acids, 29 kDa) plays an important role in the regulation of mini-F replication. Two distinct oligomeric states, monomeric and dimeric, of RepE have been found and their functions are known to be different (Ishiai et al., 1994 ▶). The RepE monomer is a replication initiator that recognizes a specific 19 bp sequence DNA (iteron; Masson & Ray, 1986 ▶, 1988 ▶; Tokino et al., 1986 ▶). Within the replication origin (ori2), four directly repeated iterons (DR) are present and are bound by RepE molecules, which could cause local DNA melting at the 13-mer region with the assistance of a histone-like protein HU and subsequent initiation of mini-F replication (Kawasaki et al., 1996 ▶; Komori, Matsunaga et al., 1999 ▶). In contrast, the RepE dimer is an autogenous repressor that binds to the repE promoter/operator region located just upstream of the repE gene (Rokeach et al., 1985 ▶; Masson & Ray, 1986 ▶, 1988 ▶; Tokino et al., 1986 ▶). The operator of repE includes an inverted-repeat (IR) sequence, which shares 8 bp with the iterons. The RepE dimer recognizes the common 8 bp of the IR and inhibits the transcription of repE itself. Since RepE exists primarily as a dimer, in order to activate RepE as an initiator, conversion from the dimer to the monomer is accomplished by the molecular chaperones DnaK, DnaJ and GrpE (Kawasaki et al., 1990 ▶, 1991 ▶; Wada et al., unpublished results). The crystal structure of the active initiator form of RepE has been reported previously (Komori, Matsunaga et al., 1999 ▶) and suggested that the C-terminal domain recognizes the specific DNA sequence and that the N-terminal domain interacts with the DNA backbone. Based on the RepE monomer structure, a marked conformational change is required for dimerization. Structural information on the repressor form of RepE, however, remains unknown. Because most initiator proteins generally aggregate easily, wild-type and full-length initiators have not been targeted for structural studies. We have successfully expressed and purified wild-type RepE and here we report the crystallization and preliminary X-ray diffraction analysis of RepE bound to DNA containing an IR sequence in order to elucidate the crystal structure of dimeric RepE in complex with repE operator DNA.
2. Materials and methods
2.1. Expression and purification
E. coli strain M15 harbouring pREP4 plasmid (Qiagen, Hilden, Germany) was transformed with pKV7202 plasmid carrying an N-terminal His-tagged repE gene (Matsunaga et al., 1995 ▶) and grown at 310 K in Luria broth medium containing 50 µg ml−1 ampicillin and 25 µg ml−1 kanamycin. When the culture reached mid-log growth phase (optical density at 600 nm = 0.6), the culture was rapidly cooled on ice and protein expression was induced by the addition of isopropyl β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. After further growth for 22 h at 293 K, the cells were harvested by centrifugation at 2110g for 30 min at 277 K. All of the following purification steps of N-terminal His-tagged RepE (His6-RepE) were performed under cooled conditions at 277 K. The bacterial pellets were resuspended in lysis buffer [50 mM NaH2PO4 pH 8.0, 1 M KCl, 20%(v/v) glycerol and 10 mM imidazole] supplemented with 1 mg ml−1 lysozyme and 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (p-ABSF) and disrupted by sonication on ice. The cell lysate was centrifuged at 27 000g for 60 min and the supernatant was then filtered using a membrane filter with 0.45 µm pore size. After Ni2+–nitrilotriacetic acid (Ni–NTA) Superflow resin (Qiagen) equilibrated with lysis buffer was added to the supernatant of the cell lysate, the mixture was stirred for 60 min and loaded onto an Ni–NTA Superflow column. The column was then washed with a stepwise gradient of a wash buffer [50 mM sodium phosphate pH 8.0, 1 M KCl and 20%(v/v) glycerol] containing 30, 50 and 100 mM imidazole. His6-RepE was eluted with a wash buffer containing 200 mM imidazole. Several cycles of purification by means of Ni–NTA immobilized metal-affinity chromatography (IMAC) were performed. For a stock solution, glycerol was added to a final concentration of 35%(v/v) and purified His6-RepE was stored in small aliquots at 193 K.
2.2. Preparation of RepE–DNA complex
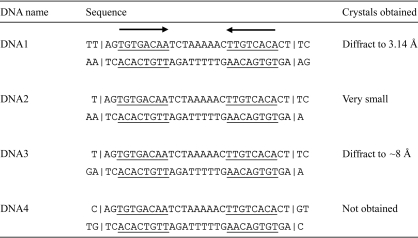
For RepE–DNA complex formation, oligonucleotides containing the inverted-repeat sequence in the repE operator recognized by the RepE dimer were prepared by Hokkaido System Science (Sapporo, Japan). Their sequences were designed based on the expectation that DNA duplexes form a pseudo-continuous helix in crystal packing (Joachimiak & Sigler, 1991 ▶), as shown in Fig. 1 ▶. DNA duplexes were prepared essentially as described by Komori, Sasai et al. (1999 ▶), except that the final concentrations were 2.4 mM. Each DNA duplex was added to His6-RepE solution in a 1.2-fold molar excess relative to the RepE dimer and the mixture was subsequently dialyzed against crystallization buffer [100 mM potassium citrate pH 6.2, 100 mM KCl, 10 mM MgCl2, 10%(v/v) glycerol and 2 mM dithiothreitol (DTT)] at 277 K and concentrated to approximately 5 mg ml−1 His6-RepE concentration. Before crystallization, the formation of the protein–DNA complex was confirmed by native polyacrylamide gel electrophoresis (PAGE), in which a band corresponding to the protein–DNA complex was stained with both Coomassie Brilliant Blue (CBB) and Mupid-Blue (Advance, Tokyo, Japan).
Figure 1.
DNA duplexes used in complex formation and crystallization trials. The common 8 bp sequences between iteron and operator are underlined and their directions are indicated by the arrows.
2.3. Crystallization
The crystallization conditions for the His6-RepE–operator complexes were screened by the sitting-drop vapour-diffusion method at 277 K. Initial conditions were found using a Hydra II Plus One (Matrix Technologies Corporation, Hudson, NH, USA) with commercially available screening kits (Hampton Research, Aliso Viejo, CA, USA; Sigma–Aldrich, St Louis, MO, USA). A drop was prepared by mixing equal volumes (0.3 µl for initial screening and 1.0 µl for optimization) of the His6-RepE–operator complex solution and the reservoir solution, allowing the mixture to equilibrate against the reservoir solution (80 µl for initial screening and 100 µl for optimization). The presence of the complex in the crystal component was identified by native PAGE using the washed and dissolved crystals.
2.4. X-ray diffraction analysis
A His6-RepE–DNA1 (Fig. 1 ▶) crystal mounted in a nylon loop was transferred and soaked for 36 h at 277 K in a dehydration solution containing a 1.2-fold higher concentration of each component of the crystallization condition. The dehydrated crystal was briefly immersed into the dehydration solution supplemented with 25–30%(v/v) glycerol as a cryoprotectant and then frozen by flash-cooling in a stream of nitrogen gas at 100 K. Prior to X-ray irradiation, we performed crystal annealing (Harp et al., 1998 ▶; Heras & Martin, 2005 ▶) by blocking the cryostream twice for 8 s each time. X-ray diffraction data were collected using an ADSC Quantum 315 CCD detector on SPring-8 beamline BL41XU at a wavelength of 1.0000 Å. A total of 180 frames were recorded with an oscillation angle of 1.0°, an exposure time of 3.0 s per frame and a crystal-to-detector distance of 350 mm. The diffraction intensities were integrated with the HKL-2000 (Otwinowski & Minor, 1997 ▶) program suite. The data-collection and processing statistics are summarized in Table 1 ▶.
Table 1. Data-collection and processing statistics of the His6-RepE–DNA1 complex.
Values in parentheses refer to the highest resolution shell.
| Wavelength (Å) | 1.0000 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 60.73, b = 99.32, c = 95.00, β = 108.55 |
| Resolution range (Å) | 50.0–3.14 (3.25–3.14) |
| Observed reflections | 42887 |
| Unique reflections | 16535 |
| Completeness (%) | 88.3 (73.6) |
| I/σ(I) | 11.0 (3.9) |
| Rsym† (%) | 6.7 (15.0) |
R
sym = 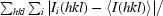
 , where I
i(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is the average value over multiple measurements.
, where I
i(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is the average value over multiple measurements.
3. Results and discussion
Wild-type RepE proteins tend to aggregate and more than 80% of the RepE expressed in E. coli was collected as inclusion bodies. RepE protein has therefore previously been purified under denaturing conditions using guanidine hydrochloride (Kawasaki et al., 1992 ▶; Kline et al., 1992 ▶; Ishiai et al., 1994 ▶; Matsunaga et al., 1997 ▶). However, in order to keep RepE proteins with their native fold for subsequent crystallization, we attempted to purify His6-RepE without denaturing. For bacterial culture conditions, the temperature was shifted from 310 to 293 K in order to decrease the protein-expression level during expression of the His6-RepE protein. Furthermore, during RepE purification a high salt concentration of 1 M was maintained in the buffers to prevent protein aggregation. Owing to these modifications, a significant improvement in the yield of purified RepE protein was achieved (from less than 1 mg to 25 mg of His6-RepE per litre of culture). After several cycles of purification by the IMAC method, His6-RepE attained over 90% purity as estimated by SDS–PAGE followed by staining with CBB. The result of gel-filtration chromatography for His6-RepE using a wash buffer containing 200 mM imidazole [eluted at 76 ml from Hi-Load 16/60 Superdex 200 (GE Healthcare Biosciences, Tokyo, Japan) at a flow rate of 0.3 ml min−1] indicates the presence of an ∼80 kDa protein but not of an ∼30 kDa protein, suggesting that His6-RepE forms dimers even under high-salt conditions (1 M).
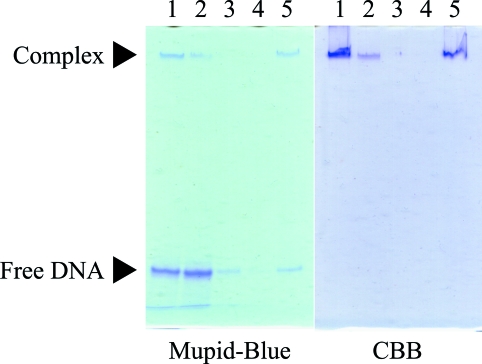
The complexes of His6-RepE with each of four DNA duplexes containing IR (31 or 33 bp; Fig. 1 ▶) were prepared and their complexation was confirmed by native PAGE as described above. Under normal conditions (pH 8.8), His6-RepE is unable to diffuse into a native polyacrylamide gel because the theoretical pI value is 9.5, whereas the complexes are able to move into the gel owing to the effect of the acidic oligonucleotides. As a result of staining the gels with CBB and Mupid-Blue independently to confirm the presence of His6-RepE and the DNA, respectively, specific sharp bands corresponding to the complexes were indeed detected at the same position in both gels (Fig. 2 ▶). No corresponding band was found in the lanes in which His6-RepE and DNA had been loaded independently onto the native gel (data not shown). The complex His6-RepE–DNA1 was estimated to have a molecular weight of ∼100 kDa by gel-filtration chromatography. In addition, it has been reported that the repE operator is preferentially bound by the dimeric form of RepE compared with the monomeric form (Ishiai et al., 1994 ▶). Therefore, we concluded that these complexes are constituted of the RepE dimer and the DNA duplex.
Figure 2.
Native PAGE of the crystals of the His6-RepE–DNA1 complex. Samples were loaded onto a 15% polyacrylamide gel (37.5:1 acrylamide:bisacrylamide) and electrophoresed. The gels were stained with either Mupid-Blue (left) or CBB (right) for the detection of DNA1 and His6-RepE, respectively. The arrows labelled Complex and Free DNA indicate the positions of the His6-RepE–DNA1 complex and free DNA1, respectively. Lane 1, solution used in crystallization; lane 2, crystallization mother liquor; lanes 3–4, washing solution; lane 5, dissolved crystals.
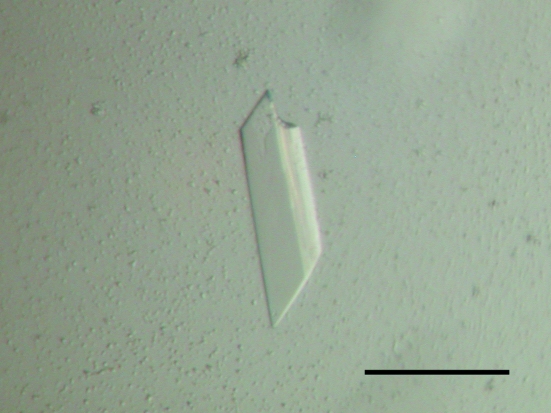
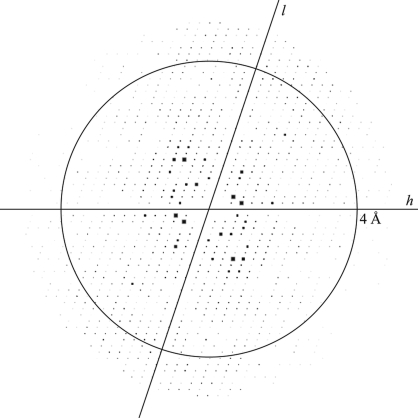
The crystallization conditions for the four complexes were screened by the sparse-matrix method. Although crystals of three complexes (complexes with DNA1, DNA2 and DNA3) were obtained, only one crystal (for the complex with DNA1) was suitable for X-ray analysis. After the presence of both His6-RepE and DNA1 in the obtained crystals had been confirmed (Fig. 2 ▶), the optimization of the crystallization conditions for His6-RepE–DNA1 afforded a final condition of 100 mM HEPES–NaOH pH 7.2, 200 mM NaCl, 10%(w/v) PEG 4000 and 5 mM SmCl3. Prismatic crystals grew in two weeks (Fig. 3 ▶; 0.12 × 0.04 × 0.02 mm). Prior to X-ray exposure, both a dehydration treatment (Schick & Jurnak, 1994 ▶; Heras & Martin, 2005 ▶) and a process of crystal annealing were indispensable to improve the resolution limit of X-ray diffraction from His6-RepE–DNA1 crystals (from approximately 8 to 3 Å resolution). Diffraction intensities were recorded to a maximum resolution of 3.14 Å using synchrotron radiation, but the diffraction patterns were anisotropic, showing diffraction to only 4.2 Å in the weakest direction (Fig. 4 ▶). The diffraction symmetry and the systematic absences of the diffraction data indicated that the His6-RepE–DNA1 crystals belong to the monoclinic space group P21, with unit-cell parameters a = 60.73, b = 99.32, c = 95.00 Å, β = 108.55°. Assuming the presence of one His6-RepE–DNA1 complex in the asymmetric unit, the V M value (Matthews, 1968 ▶) was calculated to be 3.3 Å3 Da−1, which corresponds to a solvent content of 63%. Although diffraction data were also collected at a wavelength of 1.6000 Å using one crystal, no significant peaks derived from Sm atoms were found either in the anomalous difference Patterson map or in the isomorphous difference Patterson map. Structure determination is currently in progress.
Figure 3.
Crystal of the His6-RepE–DNA1 complex. The scale bar is 100 µm in length.
Figure 4.
Pseudo-precession image of a His6-RepE–DNA1 crystal representing the h0l zone of reciprocal space. The circle indicates a resolution of 4 Å. The picture was generated with HKLVIEW (Collaborative Computational Project, Number 4, 1994 ▶).
Acknowledgments
The authors would like to thank Drs M. Kawamoto and N. Shimizu and the other staff at beamline BL41XU, SPring-8, Japan for their kind assistance with X-ray diffraction data collection. This work was supported by Grants-in-Aid for Scientific Research (to CW and KM) and by a grant from the National Project on Protein Structural and Function Analysis from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to KM).
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Harp, J. M., Timm, D. E. & Bunick, G. J. (1998). Acta Cryst. D54, 622–628. [DOI] [PubMed] [Google Scholar]
- Heras, B. & Martin, J. L. (2005). Acta Cryst. D61, 1173–1180. [DOI] [PubMed] [Google Scholar]
- Ishiai, M., Wada, C., Kawasaki, Y. & Yura, T. (1994). Proc. Natl Acad. Sci. USA, 91, 3839–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachimiak, A. & Sigler, P. B. (1991). Methods Enzymol.208, 82–99. [DOI] [PubMed] [Google Scholar]
- Kawasaki, Y., Matsunaga, F., Kano, Y., Yura, T. & Wada, C. (1996). Mol. Gen. Genet.253, 42–49. [DOI] [PubMed] [Google Scholar]
- Kawasaki, Y., Wada, C. & Yura, T. (1990). Mol. Gen. Genet.220, 277–282. [DOI] [PubMed] [Google Scholar]
- Kawasaki, Y., Wada, C. & Yura, T. (1991). J. Bacteriol.173, 1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, Y., Wada, C. & Yura, T. (1992). J. Biol. Chem.267, 11520–11524. [PubMed] [Google Scholar]
- Kline, B. C. (1985). Plasmid, 14, 1–16. [DOI] [PubMed] [Google Scholar]
- Kline, B. C., Sandhu, G. S., Eckloff, B. W. & Aleff, R. A. (1992). J. Bacteriol.174, 3004–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori, H., Matsunaga, F., Higuchi, Y., Ishiai, M., Wada, C. & Miki, K. (1999). EMBO J.18, 4597–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori, H., Sasai, N., Matsunaga, F., Wada, C. & Miki, K. (1999). J. Biochem. (Tokyo), 125, 24–26. [DOI] [PubMed] [Google Scholar]
- Masson, L. & Ray, D. S. (1986). Nucleic Acids Res.14, 5693–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson, L. & Ray, D. S. (1988). Nucleic Acids Res.16, 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga, F., Ishiai, M., Kobayashi, G., Uga, H., Yura, T. & Wada, C. (1997). J. Mol. Biol.274, 27–38. [DOI] [PubMed] [Google Scholar]
- Matsunaga, F., Kawasaki, Y., Ishiai, M., Nishikawa, K., Yura, T. & Wada, C. (1995). J. Bacteriol.177, 1994–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Rokeach, L. A., Sogaard-Andersen, L. & Molin, S. (1985). J. Bacteriol.164, 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick, B. & Jurnak, F. (1994). Acta Cryst. D50, 563–568. [DOI] [PubMed] [Google Scholar]
- Tokino, T., Murotsu, T. & Matsubara, K. (1986). Proc. Natl Acad. Sci. USA, 83, 4109–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]