Abstract
Hox genes determine anterior–posterior specificity of an animal body. In mammals, these genes map onto four chromosomal loci in a clustered manner, and their expression is regulated in a coordinated manner according to their chromosomal structure. In the present study, we analysed the Hoxb9 promoter and found that promoter activity in cultured cells is linked to secondary structure formation of promoter DNA. In nuclear extracts, we also detected binding activity specific for secondary-structured DNA. We successfully isolated a candidate gene encoding this specific DNA-binding protein, FBXL10, and demonstrated the effects of the gene product on Hoxb9 promoter activity. Our results suggest that DNA can regulate gene expression by other, non-sequence-specific modes of genetic coding.
INTRODUCTION
Many genes related to developmental processes have temporally and spatially restricted expression profiles that correspond to their roles during the development of multicellular organisms. Failure to regulate a gene often has deleterious consequences that may affect the life of an organism. Although much information exists on transcription regulation, details of regulation mechanisms taking place at the genome level remain elusive. Indeed, it remains a mystery how countless enhancers and promoters interact in an orderly fashion on a single macromolecule of DNA such as a chromosome.
Hox genes encode a group of transcription factors at work during development. These factors are required for anterior–posterior specification of body segments. In mammals, 39 Hox genes have been identified, and about 10 of these genes form a gene cluster on the same strand of DNA. These gene clusters map onto four genomic loci called HoxA, B, C and D complexes (1,2). These genes show a characteristic genomic organisation in which the order of their chromosomal transcription units mirrors the spatial order of their expression domains along the anterior–posterior axis of the developing embryo. Hox genes located at the 3′ extremity of the complex are activated in anterior embryonic domains, whereas genes located at progressively more 5′ positions are transcribed in more posterior areas. This phenomenon, called ‘spatial colinearity’, was originally described in Drosophila and further extended to all bilaterians exhibiting an anterior–posterior axial polarity (3–6). A similar type of colinearity can be observed over time in vertebrates such that Hox genes located at the 3′ extremity are activated earliest and genes located at progressively more 5′ positions are transcribed later, according to their order on the genome (7–9). Thus, ‘temporal colinearity’ is a characteristic of Hox gene regulation in vertebrates.
The spatial and temporal regulation of the Hox complex has been proposed to be a multi-step process, with each step being initiated by progressive release from heterochromatic silencing (8–10). Although the mechanistic basis of the progressive activation of genes is largely unknown, we have demonstrated that interaction between the 5′-upstream region of the Hox complex and promoters of resident Hox transcription units are important for early silencing (8,9). To elucidate the mechanisms underlying this higher-order regulatory system, we performed a functional analysis of one key DNA element—the Hox promoter. All mammalian Hox complexes are known to contain 4th, 9th and 13th paralogous genes. Therefore, we selected the Hoxb9 promoter as our study system based on an assumption that DNA structures surrounding important regions are better conserved than DNA structures in functionally neutral regions. Here, we report that the functional promoter fragment forms a secondary structure. We subsequently isolated factors that bind this promoter.
MATERIALS AND METHODS
Cell cultures and luciferase activity assays
P19 cells were cultured as described previously (11). HeLa cells and NIH3T3 cells were cultured in Dulbecco's Modified Eagle's Medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (JRH).
Luciferase assays were carried out with the Dual-Luciferase Reporter Assay system (Promega). Hoxb9 upstream fragments were ligated into a pGL4.12 vector (Promega). Transfection efficiency was monitored using a Renilla reniformis luciferase reporter driven by a TK promoter (pGL4.74, Promega). Cells were transfected with constructs and siRNAs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. Chemiluminescence was measured with a luminometer LB410 (Berthold Technologies, Inc.). Values are presented as means and standard deviations of at least four independent experiments.
RNAi
For RNAi, the siRNAs used were Stealth™ RNAi (Invitrogen) designed according to the manufacturer's protocol. The siRNA sequences against human FBXL were as follows:
iRhF10, AAGUAAGUGAGACUGGAUCUCCACC;
iRhF11, UUGGCCACUGUACUAUAGGAACUCC.
To examine the effects of these siRNAs on Hoxb9 promoter activity, we extracted total RNA from HeLa cells with TRIzol (Invitrogen).
The siRNA sequences against mouse Fbxl were as follows:
iRmF10-a, AAGUAAGUGAGACUGGAUCUCCACC;
iRmF10-b, UUCACACUCACUUCUCCGCUUGGCA;
iRmF11, UAUAUUUGUUGGUUUGGAGCUUCUC.
Twenty-four hours after transfecting siRNA into P19 cells, we started retinoic acid (RA) treatment. We isolated RNA 72 h after siRNA transfection (48 h after RA treatment). First-strand cDNA synthesis was carried out using 5 μg of total RNA and SuperScript III First-Strand Synthesis SuperMix (Invitrogen) according to the manufacturer's instructions. We assessed Hox and Fbxl genes expression in HeLa and P19 cells by real-time PCR (RT–PCR) using a Corbett RG-3000 and Invitrogen Platinum SYBRGreen PCR kit. We calibrated the real-time PCR results by Rotor-Gene 6 Software (Corbett Research) with standard cDNAs whose concentrations are known. We also carried out real-time PCR with β-actin as a control in HeLa and P19 cells.
We used the following primers in which the first letter of each primer—h or m—represent human or mouse, respectively; hm primers are used both for human and mouse.
hβ-actin-F, TGGACATCCGCAAAGACCTG;
hβ-actin-F, ACATCTGCTGGAAGGTGGAC;
mβ-actin-F, CATGTTTGAGACCTTCAACAC;
mβ-actin-R, GTGATGACCTGGCCGTCAGG;
mHoxb9-F, TTTGCGAAGGAAGCGAGGAC;
mHoxb9-R, AAGAGTGAGCTGGGGAAGGG;
hmFbxl10-F, CAAGGAGCAGAAGATGAACCG;
hmFbxl10-R, TGGGGCTTCTCGTATTTCCG;
hmFbxl11-F, ACTGCTGTCGGGTCTTCCACT;
hFBXL11-R, CGGTTTCCTGTTGAATCTGGG;
mFbxl11-R, GATTTTCCTGTTCAGTCTGGGC.
Gel mobility shift assay
We performed electrophoretic mobility shift assay (EMSA) as described previously (11). Nuclear extracts were isolated as described previously (12). For competition studies, 12.5-, 25-, 50-, and 100-fold molar ratios of cold DNA against the probe were added.
South-western screening
South-western screening was carried out according to the procedure established by Vinson and colleagues (13), with slight modifications. Instead of using 106 c.p.m./ml of probe, we used 104 c.p.m./ml of probe in order to facilitate the detection of specific binding to the probe. We isolated 55 clones after screening a P19 cDNA library and selected one clone that displayed fragment A-specific binding, but not fragment B-specific binding, upon further South–western screening.
Antibodies
We generated a polyclonal antibody against FBXL10 by immunizing rabbits with a peptide corresponding to amino acids 923–938 of mouse Fbxl10. Mouse monoclonal antibody (mAb) to ubiquityl-histone H2A and mouse mAb against FLAG were purchased from Upstate and Sigma, respectively.
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed based on a protocol described previously (9). After P19 cells were cultured with or without 0.3 μM of RA for 48 h in Petri dishes, we collected the cells for ChIP assay. Immunoprecipitation was performed with either anti-Fbxl10 or anti-ubiquityl-histone H2A antibody. To isolate bound chromatin, we used Dynabeads conjugated with Protein A and G for anti-Fbxl10 antibody and Dynabeads rat anti-Mouse IgM for ubiquitylated histone (Dynal Biotech). Samples were washed, reverse crosslinked and digested with proteinase K. Purified DNA samples were analysed by RT–PCR.
RESULTS
Correlation between promoter activity and secondary structure formation of the Hoxb9 promoter
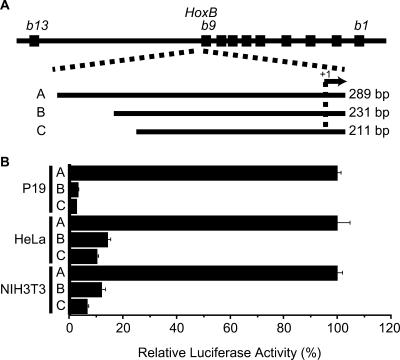
Transfection assays using P19, HeLa and NIH3T3 cell lines demonstrated that Hoxb9 promoter activity required a 289-bp fragment that contains the 274-bp fragment upstream of the transcription initiation site (−274 to +15 bp from transcription initiation site; fragment A in Figure 1A and B). Shorter fragments (fragment B, −216 to +15 bp; fragment C, −196 to +15 bp) significantly decreased promoter activity (Figure 1A and B). These results generally agree with those previously reported, except that the differences in promoter activity between fragments A and B were much greater than those we observed previously (11). This is probably because, in the present study, we used a different vector backbone in the luciferase constructs and we used high-turnover luciferase assays to assess promoter activity.
Figure 1.
Luciferase assays of Hoxb9 promoter activity. (A) Diagram of DNA fragments used for analysis. The top-most line represents a map of the HoxB complex. We assessed the effects of three different fragments (A, B and C) on promoter activity. (B) Relative luciferase activities of promoter assay constructs in P19, HeLa and NIH3T3 cells.
Although the promoter activities of fragments A and B clearly differed, EMSA and footprinting failed to show that nuclear extracts contain binding activity on the DNA spanning from positions −210 to −274 of promoter fragment A (11). Instead, we found that the 296-bp fragment A separated into multiple bands when electrophoresed on native polyacrylamide gels in the absence of protein, while fragments B and C did not separate into distinct multiple bands, suggesting that fragment A becomes highly structured (Figure 2A), perhaps attaining a secondary structure. We confirmed by sequencing that the multiple bands from fragment A share the same sequence. This structural change may be responsible for altering its mobility on EMSA gels. Similarly sized DNA fragments, including fragments from λ-phage and mammalian DNA such as those from the human c-myc promoter, did not show similar heterogeneous mobility patterns in native gels. Among the multiple bands derived from fragment A, the mobility of the fastest-moving band corresponded to the mobility of a same-sized linear DNA fragment (289 bp). To get insight into the secondary structure of the slower-moving band, we conducted digestion experiments with S1 nuclease and mung bean nuclease, which enabled us to identify single-stranded residues within the DNA fragment. The slower-moving bands were much more sensitive to these nucleases than the fastest-moving band (linear DNA fragment). These results indicate that the slower-moving band contains single-stranded residues. We could not detect, however, signs of digestion at specific residues. The fragment represented by the slow-moving band may contain multiple sites that are sensitive to S1 nuclease digestion. Alternatively, the strands close to the ends of this fragment may have dissociated.
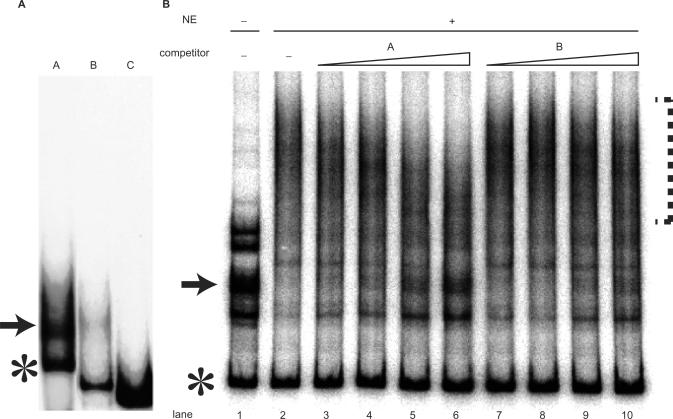
Figure 2.
Hoxb9 promoter fragment forms a secondary structure. (A) Native gel electrophoresis of fragments A, B and C revealed that only the fragment having promoter activity, fragment A, separated into multiple, but discrete, bands. Bands consisting of slow-moving secondary-structured DNA are indicated by arrows, and bands consisting of fast-moving linear DNA are indicated by asterisks. (B) EMSA performed in the absence (−) or presence (+) of nuclear extracts (NE) from P19 cells and competition assay for protein–DNA binding. Because a protein in the NE specifically binds the highly structured DNA fragment (indicated by arrow), the slow-moving band disappeared and formed a smear (indicated by dotted line) in the presence of NE, as shown in lanes 1 and 2. Lanes 3–10 show results from competition assays. The concentrations of competitor DNA used were 12.5-, 25-, 50- and 100-fold molar ratio relative to the probe (lanes 3 and 7; 4 and8; 5 and 9; 6 and 10, respectively). Adding fragment A as a competitor caused the band (arrow) to reappear (lanes 3–6). Fragment B as a competitor, however, was much less effective in causing the band to reappear, indicating that fragment B failed to compete successfully.
Although the detailed secondary structure of the Hoxb9 promoter fragment (fragment A) is yet to be determined, we do know that this fragment contains a GC-rich region, which is a prime candidate for triplex DNA formation. To take on a triplex structure, however, this short DNA fragment must be circular, not linear, in order to attain the torsion needed for triplex formation; thus, the secondary structure of this fragment is unlikely to be triplex (14). Nevertheless, the link we observed between secondary structure formation and promoter activity suggests that secondary structure formation confers promoter activity to the Hoxb9 fragment.
To examine secondary structure formation of the Hoxb9 promoter in another species and to confirm the importance of secondary structure formation, we also isolated the corresponding fragment from the human genome. The human counterpart of this fragment derived from the HOXB9 promoter showed about 92% sequence identity to the fragment derived from the MmHoxb9 promoter (Supplementary Figure S1A) and exhibited more promoter activity than the mouse promoter (data not shown). Moreover, we also observed that the HsHOXB9 promoter fragment underwent secondary structure formation similar to that as the MmHoxb9 promoter fragment (Supplementary Figure S1B). These results indicate that the characteristic ability to undergo secondary structure formation is conserved over species.
Specific binding activity against secondary structured Hoxb9 promoter in nuclear extracts
EMSA performed in the presence of nuclear extracts from P19 and HeLa cells revealed that these extracts had specific binding activity for the slow-moving band of fragment A (Figure 2B). Although the slow-moving band (arrows in Figure 2) disappeared when binding activity was assessed in the presence of nuclear extracts (Figure 2B), the fast-moving band (asterisks in Figure 2) was unaffected (Figure 2B). These findings indicate that binding activity was structure specific rather than sequence specific, as the DNA represented by the fast-moving and slow-moving bands shared identical sequences. Although the addition of nuclear extracts completely abolished the slow-moving band, no distinct new bands were formed, suggesting that the DNA–protein complex contained multiple proteins. To confirm whether the binding activity was specific, we performed competition assays using as competitors fragment A (which forms the secondary structure) and fragment B (which showed much less secondary structure formation) (Figure 2B). Adding fragment A to the assay mixture caused the slow-moving band (indicated by arrow in the Figure 2B) to reappear, suggesting that DNA–protein complex is dissociated by competition. On the other hand, adding fragment B did not cause the slow-moving band to reappear (Figure 2B). Taken together, these findings indicate that (i) fragment A, which functions as a promoter, is capable of taking on a secondary structure; and (ii) protein(s) in the nuclear extracts specifically bind the secondary-structured fragment A.
Cloning of secondary-structured DNA-specific binding protein
To determine the significance of these findings, we attempted to clone the secondary structured DNA-specific binding protein(s). We isolated a clone after screening a P19 cDNA library using the South–western method (Figure 3A) (13) and verified its binding activity further using EMSA (Figure 3B). Database searches revealed that the clone is identical to Fbxl10, human PCCX2, DKFZp434I0535 and JEMMA. Fbxl10 encodes an F-box, a motif for E3 ubiquitin ligase (15). It also encodes a bipartite nuclear localization signal (NLS) as well as leucine-rich domains (Figure 3A). The presence of cxxc and PHD-finger domains suggests that FBXL10 may be related to chromatin modification factors such as those encoded by Polycomb-group (PcG) and trithorax-group (trxG) genes. Recently, FBXL10 was identified as the histone demethylase, JHDM1b, and several following works reported that the jmjC domain is responsible for histone demethylation activity (16–19). Thus, the presence of a jmjC domain in Fbxl10 further supports the idea that Fbxl10 participates in chromatin modification.
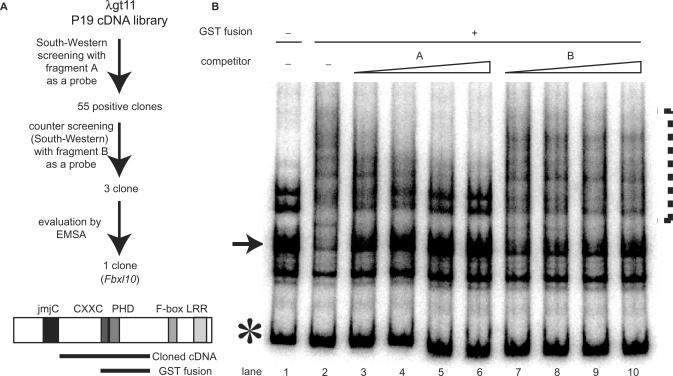
Figure 3.
Cloning of Fbxl10, which encodes a protein that binds the highly structured Hoxb9 promoter fragment. (A) Scheme of cloning strategy and schematic diagram of the obtained Fbxl10 gene. Cloning was done by using South–western methods and fragment A as a probe. The 55 clones we isolated were counter-selected by South–western methods and fragment B as a probe. Further confirmation was carried out with EMSA. We obtained the partial sequence of Fbxl10 through South–western cloning. Sequence analysis of full-length Fbxl10 cDNA revealed several motifs: jmjC, cxxc, Zn-finger, PHD Zn-finger and F-box domains and leucine-rich repeats (LRR). (B) EMSA with GST-fusion protein derived from the cloned fragment. The fragment used to produce the GST-fusion protein is indicated as a bold black line located below Fbxl10 (panel A). Competition assays were carried out as described in the legend of Figure 2. As observed with our EMSA analyses performed with nuclear extracts (NE) (see Figure 2), fragment A competed successfully for binding, while fragment B did not. Arrow points to the slow-moving band; asterisk marks the band representing linear DNA.
Another Fbxl10 homologue, Fbxl11 (known as JHDM1a), shares high homology with all the functional domains of Fbxl10.
We fused in-frame the part of Fbxl10 containing the cxxc and PHD finger domains with GST (heavy line in Figure 3A), and subjected the resulting fusion protein to EMSA. As with EMSA performed in the presence of nuclear extracts, EMSA with the GST-fusion protein resulted in a band shift similar to that of the slow-moving band of fragment A (Figure 3B), however, without yielding new discrete bands, only a smear. These findings suggest that a homo–multimer complex formed around the GST–Fbxl10 fusion protein. Moreover, fragment A not B, successfully out-competed binding to the GST-fusion protein as observed in the EMSA experiments performed with nuclear extracts (Figure 3B). From these experiments, we concluded that Fbxl10 is likely to be the DNA-binding protein within the nuclear extracts that specifically bound the secondary-structured fragment A.
Effect of secondary-structured DNA-specific binding protein on Hoxb9 promoter activity
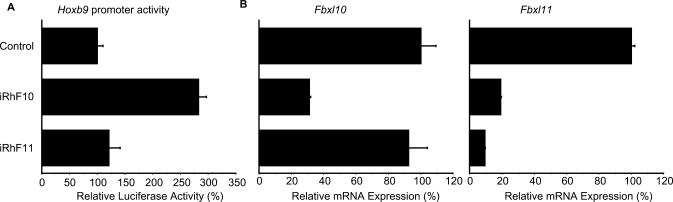
We performed a functional analysis of Fbxl10 and Fbxl11 using the RNAi gene-knockdown technique. We designed siRNAs against human FBXL10 (iRhF10 siRNAs) and FBXL11 (iRhF11 siRNAs) and examined the influence of these siRNAs on Hoxb9 promoter activity by co-transfecting them into HeLa cells with luciferase constructs containing fragment A. Transfection of FBXL10 and FBXL11 siRNAs reduced intrinsic FBXL10 and FBXL11 expression (Figure 4). We observed cross-repression of FBXL10 and FBXL11 by these siRNAs. The siRNA iRhF10 effectively reduced FBXL10 and FBXL11 mRNA expression (Figure 4). Although iRhF11 effectively down-regulated FBXL11, it had a relatively weak effect on FBXL10. Luciferase activity of the reporter construct containing fragment A showed 2.8-fold up-regulation compared to that of the control when HeLa cells were co-transfected with the reporter construct and iRhF10 (Figure 4). In contrast, luciferase activity did not change significantly when HeLa cells were co-transfected with the construct and iRhF11 (Figure 4). These results suggest that Fbxl10 is the primary factor that influences Hoxb9 promoter activity. We could not exclude, however, the influence of Fbxl11 on Hoxb9 promoter activity.
Figure 4.
Effects of FBXL genes on Hoxb9 promoter activity in HeLa cells. (A) Effects of siRNAs on FBXL expression. We designed siRNAs—iRhF10 and iRhF11—and used RT–PCR to examine the effects of these siRNAs on Hoxb9 promoter activity. Specific siRNAs against FBXL10 increased promoter activity, whereas siRNAs specifically against FBXL11 either did not increase or weakly increased promoter activity. (B) Treatment with siRNAs influences intrinsic FBXL expression. Each of these siRNAs down-regulated FBXL expression, as evident by the definitive reduction of RT–PCR signals. RT–PCR samples were normalized relative to β-actin expression.
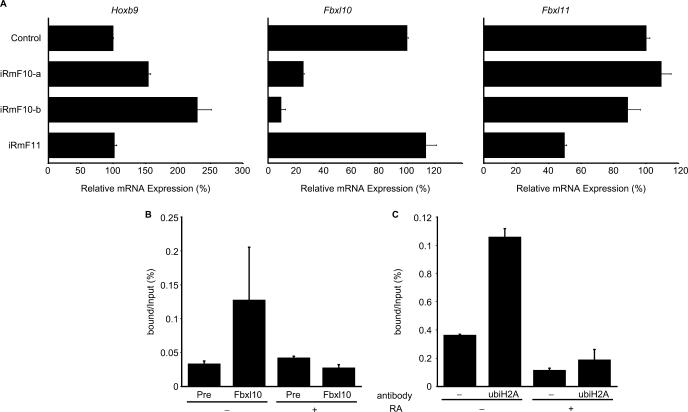
We further examined the effects of Fbxl10 and Fbxl11 genes on the native Hox locus using P19 embryonal carcinoma (EC) cell cultures. Although undifferentiated P19 cells showed little or no expression of Hoxb9 gene, P19 EC cells express Hoxb9 gene when differentiation is induced by RA treatment (11). To determine how Fbxl10 and Fbxl11 genes affect the regulation of native Hoxb9 gene, we knocked down either Fbxl10 or Fbxl11 expression in RA-treated P19 cells with siRNA and measured Hoxb9 gene expression by RT–PCR (Figure 5). Forty-eight hours after RA induction, a time at which Hoxb9 gene is expressed (Figure 5) (11), Hoxb9 exhibited elevated expression when Fbxl10 expression was suppressed by treatment with Fbxl10-specific siRNA (iRmF10-a or iRmF10-b) (Figure 5). Despite having a high degree of sequence homology with Fbxl10, Fbxl11 in general did not influence Hoxb9 expression (Figure 5; iRmF11). These results support our hypothesis that Fbxl10 interacts with the Hoxb9 promoter whereas Fbxl11 does not, even though the structures of their protein products resemble each other.
Figure 5.
Influence of Fbxl genes on the native Hoxb9 induction system. (A) The influences of Fbxl genes were analysed by introducing siRNAs. We designed three siRNAs—iRmF10-a, iRmF10-b and iRmF11—and used RT–PCR to examine the effects of these siRNAs on intrinsic Hoxb9 and Fbxl expression. The siRNAs were introduced into P19 cells during the 24-h pre-incubation period and during the 48-h RA-induction period (total exposure time: 72 h). RT–PCR samples were normalized relative to β-actin expression. Transfection of Fbxl10-specific siRNAs elevated Hoxb9 expression (left graph), whereas transfection of Fbxl11-specific siRNAs did not significantly influence Hoxb9 expression. The specificity of these siRNAs on target genes is shown in the remaining graphs as indicated. (B) Binding of FBXL10 protein to native Hoxb9 promoter in P19 cells. ChIP analysis revealed that FBXL10 binds Hoxb9 promoter in undifferentiated P19 cells but not in differentiated P19 cells, which express Hoxb9. (C) Histone H2A ubiquitylation status of the Hoxb9 promoter differs as P19 cells differentiate. ChIP was carried out with anti-ubiquitylated histone H2A antibody. Similar to the profile observed for FBXL10, ubiquitylated histone H2A seems to accumulate at the Hoxb9 promoter region when P19 cells are in an undifferentiated state.
To examine the direct interaction between FBXL10 protein and the Hoxb9 promoter, we performed ChIP assays using anti-FBXL10 antibody (Figure 5B). However, we observed FBXL10 binding onto the Hoxb9 promoter in undifferentiated P19 cells. FBXL10 binding to the Hoxb9 promoter disappeared in differentiated P19 cells, which express Hoxb9. These results correspond well to our siRNA findings showing Fbxl10 repression activity on the Hoxb9 promoter and strongly suggest that Fbxl10 is indeed a Hox regulator. To further investigate mechanisms underlying the regulation of Hoxb9 gene expression during differentiation, we conducted ChIP assays using various anti-histone antibodies. While we did not detect differences in ChIP assays performed with anti-K9 acetylated histone H3, anti-K36 trimethylated histone H3, anti-K36 dimethylated histone H3 and anti-K36 monomethylated histone H3 antibodies, we did detect reduced binding to the Hoxb9 promoter when ChIP was performed with anti-ubiquitylated histone H2A antibody (Figure 5C). This reduced binding of ubi-H2A was similar to the reduced binding of FBXL10 we observed in differentiated P19 cells (Figure 5B). These observations suggest the possibility that Fbxl10 functions as a histone ubiquitylase not a histone demethylase during Hox regulation.
Analysis of Fbxl10 domains required for regulation of Hoxb9 gene
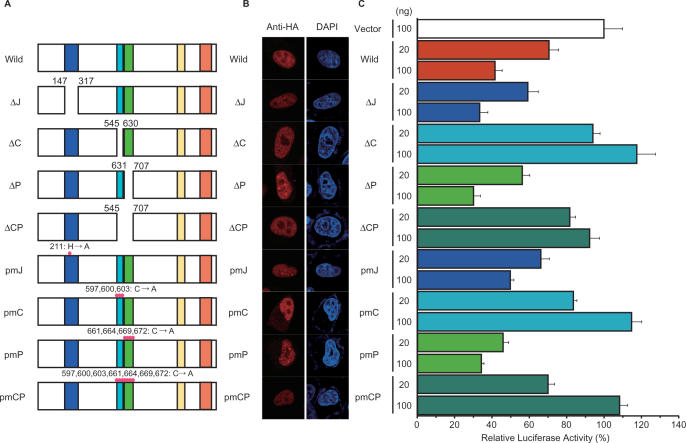
To determine the importance of the cxxc and PHD domains, which we used to make fusion proteins for DNA-binding assays (Figure 3), we created several mutants of Fbxl10 having either deletion or point mutations in the jmjC domain, cxxc domain and/or the PHD domain (Figure 6A). All of these mutant proteins localized to the nucleus, as did wild-type Fbxl10 (Figure 6B). Next, to observe how these mutants influence promoter activity, we introduced the constructs expressing these mutants into HeLa cells along with a luciferase reporter construct driven by Hoxb9 promoter fragment A.
Figure 6.
Assessment of domains involved in the specific regulation of Hoxb9. (A) Schematic diagrams of Fbxl10 mutants. ΔJ, C and ΔP represent deletion mutants of the jmjC domain, cxxc domain, and the PHD domain, respectively. ΔCP represents a deletion mutant of both cxxc and PHD domains. Three other mutants, pmJ, pmC, pmP and pmCP, have point mutations in jmjC, cxxc, PHD and both the cxxc and PHD domains, respectively. (B) Subcellular localization of jcp1 mutants. Each mutant protein was fused with an HA-tag at C-terminal and then expressed in HeLa cells. Fusion proteins were localized by immunocytochemistry with anti-HA antibody. All mutants localized to the nucleus. (C) Effect of jcp1 mutants on Hoxb9 promoter activity. We co-transfected HeLa cells with mutant-expressing plasmids and Hoxb9 luciferase reporter plasmids and examined the influence of each mutation by measuring luciferase activity.
Elevated expression of wild-type Fbxl10 down-regulated luciferase activity (Figure 6C). Luciferase activity remained unaffected in cells transfected with the vector construct (negative control) (Figure 6C). Both deletion and point mutants of the cxxc domain (ΔC, ΔCP, pmC and pmCP in Figure 6) failed to affect luciferase activity, whereas the mutants of the jmjC or PHD domains influenced luciferase activity in a manner similar to wild-type Fbxl10. Thus, the repressive activity of Fbxl10 was completely abolished by mutations within the cxxc domain, but mutations in the jmjC and PHD domains did not influence the activity of Fbxl10 in this assay system. We observed a similar effect of Fbxl10 on human HOXB9 promoter and similar loss of Fbxl10 repressive activity resulting from point mutations in the cxxc domain (Supplementary Figure 1C). Although we could not demonstrate the importance of the jmjC and PHD domains, these results indicated that the cxxc domain plays a critical role in Fbxl10-mediated regulation of Hoxb9.
The secondary structure formation of other Hox promoters
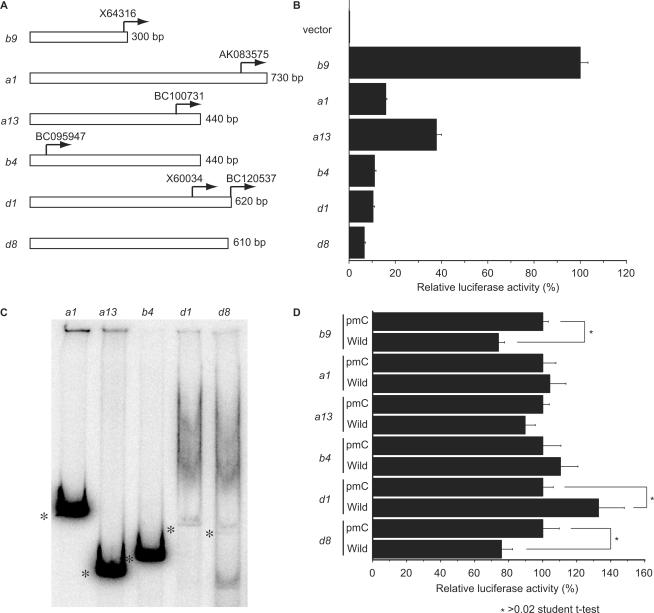
We expected that secondary structure formation was not restricted to the Hoxb9 promoter; thus, we extensively searched for other promoters within the Hox complex capable of taking on secondary structures. We isolated several DNA fragments containing the sequences surrounding predicted transcription initiation sites of several Hox genes and briefly analysed the promoter activity of these fragments by measuring luciferase activity (Figure 7A and B). We identified several potential Hox promoter fragments having lengths of about 300–700 bp (Figure 7B). These fragments clearly displayed promoter activity when compared to that of the control luciferase vector lacking a promoter, although most of these promoters showed weaker activity than the Hoxb9 promoter (Figure 7B). Some of these fragments with promoter activity, such as Hoxd1 and Hoxd8, also showed heterogeneous mobility in native gels, as observed with Hoxb9 promoter fragment A (Figure 7C), although some fragments from Hoxa1, Hoxa13 and Hoxb4 did not. At the very least, these results suggest that promoter regions of some Hox genes also have the potential to take on secondary structures, as with the case of Hoxb9. As we observed with Hoxb9, overexpression of Fbxl10 also influenced the promoter activities of Hoxd1 and Hoxd8 (Figure 7D). In contrast, promoter fragments that did not take on secondary structures, such as Hoxa1, Hoxa13 and Hoxb4, did not influence promoter activity (Figure 7D). Overexpression of Fbxl10 activated the Hoxd1 promoter but repressed Hoxb9 and Hoxd8 promoters (Figure 7D). Although we clearly observed a correlation between secondary structure formation of Hox promoters and Fbxl10, these results suggest that the Hox promoter response to Fbxl10 varies.
Figure 7.
Higher structure formation of Hox promoters. (A) Examples of isolated Hox promoter fragments. Arrows indicate the 5′ end of cDNA sequences annotated in the NCBI sequence library. The transcription initiation site of Hoxb9 was determined previously (11). (B) Relative luciferase activity of isolated promoter fragments. The promoter intensities of these fragments are shown relative to the luciferase activity measured in cells transfected with Hoxb9 constructs, which was set to 100. The negative control (luciferase vector lacking a promoter fragment) did not show any traces of activity. (C) Native gel electrophoresis of Hox promoter fragments. The fragments from Hoxa1, Hoxa13 and Hoxb4 did not exhibit multiple bands, while the fragments from Hoxd1 and Hoxd8 separated into multiple bands, as observed with Hoxb9 (Figure 2). Asterisks mark linear portions of the DNA fragments. (D) Effects of Fbxl10 on Hox promoters. Each luciferase construct was co-transfected with an Fbxl10 expression construct. Co-transfection experiments with the pmC construct represented control experiments (Figure 5). Overexpression of wild-type Fbxl10 influenced luciferase activity in cells co-transfected with either Hoxd1 or Hoxd8 promoter fragments, fragments that take on a higher structure like Hoxb9. By contrast, wild-type Fbxl10 and pmC Fbxl10 failed to significantly influence luciferase activity in cells co-transfected with Hoxa1, Hoxa13 or Hoxb4 promoter fragments. Asterisks indicate significant differences compared to cells expressing the control vector according to the Student's t-test (P < 0.03).
DISCUSSION
Secondary structure formation of Hox promoters
We have previously proposed that multiple DNA–site interactions are important for the coordinated expression of Hox genes (8–10,20). This hypothesis also posited a crucial role of regulated positional movement of chromosomal loci in this Hox gene system. Recent reports have emphasized the significance of chromosomal position within the nucleus during various biological processes (21,22). Chambeyron and colleagues (23,24) demonstrated the importance of chromosomal positioning of Hox genes during development, correlating chromosomal position with transcription activity. In this scheme, DNA–DNA interactions (direct or indirect via protein–protein interactions) can crucially determine the relative position of DNA loci.
The mechanisms underlying chromosomal movement remain completely unknown. However, it is clear that the interaction between the repressive region (8) and promoters of resident Hox genes are decisive factors in this regulatory process. Here, we focused on the Hoxb9 promoter and found a novel characteristic of Hox promoter DNA—the Hox promoter forms secondary structures. Although, the relationship between this phenomenon, chromosomal movement and the formation of secondary DNA structures remains to be determined, our results suggest that the secondary structure formation of promoter DNA is important for Hox gene regulation. Indeed, we observed that several DNA fragments from Hox complexes displayed a similar heterogeneity in mobility when assessed in native gels, but DNA fragments from non-Hox regions or other organisms, such as bacteria, did not display this mobility shift (data not shown). The nature of the higher structures formed by various Hox promoter fragments remains elusive. However, it is doubtful that these structures represent DNA triplets, since the secondary structure of the short DNA fragments we analysed was stable and detectable by native gel electrophoresis. Even the linear form of this fragment lacked the torsion derived from circular forms of DNA, which is required for triplet formation (14). We believe that the formation of secondary structures may be a novel type of genetic coding, although secondary structure formation to some degree is dependent on DNA sequence.
Secondary-structured DNA-specific binding protein and gene transcription
Isolation of a clone (Fbxl10) encoding a protein that specifically bound to a secondary-structured Hox promoter fragment and determination of its influence on Hox promoter activity shed light onto the significant role of secondary structures of promoter DNA in transcription regulation. Furthermore, promoter analysis using other Hox promoters indicated that Fbxl10 regulates multiple Hox genes not just Hoxb9 gene. The molecular structure of this protein suggests that it is a chromatin factor, having shared homology with the cxxc and PHD Zn-finger domains of PcG and trxG genes. The Fbxl10 and Fbxl11 genes also contain sequences encoding an F-box domain, a protein motif found in a component of E3 ubiquitin ligase, strongly suggesting that Fbxl10 and Fbxl11 proteins participate in protein ubiquitylation processes. Recently, histone ubiquitylation has been shown to be important in histone modifications involved in transcription regulation (25). Indeed, Gearhart and colleagues (26) suggested that FBXL10/JHDM1b is a component of the BCOR (BCL6 corepressor) complex and is responsible for histone H2A monoubiquitylation. In addition to these motifs, Fbxl10 contains a leucine-rich repeat, indicating that it may also interact with other proteins.
The presence of a jmjC domain in Fbxl10 further supports the idea that it is involved in chromatin modification. Recently, Tsukada and colleagues (16) reported that JHDM1a (Fbxl11 in the present work) functions as a histone demethylase and that the jmjC domain is responsible for histone demethylation activity. It is probable, therefore, that Fbxl10 (also known as JHDM1b) also has histone demethylase activity, since Fbxl10 and Fbxl11 (JHDM1a) share high homology throughout their molecular structures, including in their jmjC histone demethylase domains (16). It is also probable that Fbxl10 is involved in transcription activation processes (16). The coexistence of demethylation activity and ubiquitylation activity in one molecule, Fbxl10, is somewhat controversial. Indeed, our analysis of the Hoxd1 and Hoxb9 promoters suggests that Fbxl10 can function differentially to activate or repress promoter activity depending on the promoter or context. Multiple domains of Fbxl10 and Fbxl11 proteins may be involved in facilitating the protein–DNA or protein–protein interactions required for complex transcription regulation.
High homology in the molecular structures of Fbxl10 and Fbxl11 suggests that the products of these genes may have functional similarities. However, Fbxl11 did not significantly influence Hoxb9 regulation in our transient promoter assays or in our assessment of the transcription profile of the native Hoxb9 gene in P19 EC cells. These results suggest that, despite their high homology, Fbxl10 and Fbxl11 differ in their abilities to regulate downstream genes.
The structural composition of Fbxl10 and Fbxl11 proteins (e.g. numerous domains for interacting with other proteins) and the complexity of their activity on transcription suggest that Fbxl10 and Fbxl11 proteins may function as a structural ‘hub’ for a multi-protein–DNA complex, and that they may also serve as a functional ‘hub’, coordinating different functional protein complexes for regulating gene transcription.
With regard to Hoxb9 transcription, in the present study Fbxl10 likely functioned as a histone ubiquitylase rather than a demethylase, because ChIP analysis showed that ubiquitylated histone H2A differentially bound the Hoxb9 promoter, a situation that reflects Hoxb9 expression during P19 EC cell differentiation. However, we also observed that trichostatin A and butyrate abolished Fbxl10-mediated repression of Hoxb9 promoter, suggesting that HDAC (histone deacetylase) is also involved in Hox regulation, as shown in the recently reported case of c-jun (27). Recently, other jmjC proteins (Utx and JMJD3) were identified as demethylase Hox regulators of K27 trimethylated histone H3 (28,29). According to these schemes, at least three different histone modification activities may intersect through Fbxl10. To fully understand the regulatory role of Fbxl10, we need to continue to analyse the regulatory mechanisms of Fbxl10 on Hox genes.
Based on the present results, we propose an alternative type of information coding in chromosomal DNA, one that is based on secondary structure rather than on sequence. Although the structure of DNA depends to some degree on nucleotide sequence, we do not yet have sufficient information to correlate sequence with structure. However, the presence of proteins that specifically bind secondary DNA structures and the potential role of these proteins in gene transcription provide further support for the importance of secondary structure formation in DNA.
ACKNOWLEDGEMENTS
We thank T. Kojima and T. Tabata for sharing information and helpful discussions and members of the Kondo laboratory for sharing resources. We also thank M. Ota and Y. Zhang for reading the manuscript. T.K. also thanks D. Duboule and M. Muramatsu for continuous encouragement. This work is supported by a grant from the Human Frontier Scientific Program Organization grant RGY0316/2001-M to T.K. Funding to pay the Open Access publication charges for this article was provided by institutional budget from RIKEN BSI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Duboule D. Guidebook to the Homeobox Genes. Oxford: Oxford University Press; 1994. [Google Scholar]
- 2.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–200. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 3.Lewis E. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 4.Akam ME. Hox and HOM: Homologous gene clusters in insects and vertebrates. Cell. 1989;57:347–349. doi: 10.1016/0092-8674(89)90909-4. [DOI] [PubMed] [Google Scholar]
- 5.Duboule D, Dollé P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989;8:1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 7.Izpisúa-Belmonte J-C, Falkenstein H, Dollé P, Renucci A, Duboule D. Murine genes related to the Drosophila AbdB homeotic gene are sequentially expressed during development of the posterior part of the body. EMBO J. 1991;10:2279–2289. doi: 10.1002/j.1460-2075.1991.tb07764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo T, Duboule D. Breaking colinearity in the mouse HoxD complex. Cell. 1999;97:407–417. doi: 10.1016/s0092-8674(00)80749-7. [DOI] [PubMed] [Google Scholar]
- 9.Mishra RK, Yamagishi T, Vasanthi D, Ohtsuka C, Kondo T. Involvement of Polycomb-group genes in establishing HoxD temporal colinearity. Genesis. 2007;45:570–576. doi: 10.1002/dvg.20326. [DOI] [PubMed] [Google Scholar]
- 10.Kondo T, Zákány J, Duboule D. Control of colinearity in AbdB genes of the mouse HoxD complex. Mol. Cell. 1998;1:289–300. doi: 10.1016/s1097-2765(00)80029-5. [DOI] [PubMed] [Google Scholar]
- 11.Kondo T, Takahashi N, Muramatsu M. The regulation of the murine Hox-2.5 gene expression during cell differentiation. Nucleic Acids Res. 1992;20:5729–5735. doi: 10.1093/nar/20.21.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinson CR, LaMarco KL, Johnson PF, Landschulz WH, McKnight SL. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 1988;2:801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- 14.Kohwi-Shigematsu T, Kohwi Y. Detection of triple-helix related structures adopted by poly(dG)-poly(dC) sequences in supercoiled plasmid DNA. Nucleic Acids Res. 1991;19:4267–4271. doi: 10.1093/nar/19.15.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 17.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 18.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 19.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 20.Yamagishi T, Ozawa M, Ohtsuka C, Ohyama-Goto R, Kondo T. Evx2-Hoxd13 intergenic region restricts enhancer association to Hoxd13 promoter. PLoS ONE. 2007;2:e175. doi: 10.1371/journal.pone.0000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 22.Tanemura K, Ogura A, Cheong C, Gotoh H, Matsumoto K, Sato E, Hayashi Y, Lee HW, Kondo T. Dynamic rearrangement of telomeres during spermatogenesis in mice. Dev. Biol. 2005;281:196–207. doi: 10.1016/j.ydbio.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 23.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chambeyron S, DaSilva NR, Lawson KA, Bickmore WA. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 26.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koyama-Nasu R, David G, Tanese N. The F-box protein Fbl10 is a novel transcriptional repressor of c-Jun. Nat. Cell Biol. 2007;9:1074–1080. doi: 10.1038/ncb1628. [DOI] [PubMed] [Google Scholar]
- 28.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 29.Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]