Abstract
The yeast split-ubiquitin system has previously been shown to be suitable to detect protein interactions of membrane proteins and of transcription factors in vivo. Therefore, this technology complements the classical split-transcription factor based yeast two-hybrid system (Y2H). Success or failure of the Y2H depends primarily on the ability to avoid false-negative and false-positive hits that become a limiting factor for the value of the system, especially in large scale proteomic analyses. We provide here a systematic assessment of parameters to help improving the quality of split-ubiquitin cDNA-library screenings. We experimentally defined the optimal 5-fluoroorotic acid (5-FOA) concentration as a key parameter to increase the reproducibility of interactions and, at the same time, to keep non-specific background growth low. Furthermore, we show that the efficacy of the 5-FOA selection is modulated by the plating density of the yeast clones. Moreover, a reporter-specific class of false-positive hits was identified, and a simple phenotypic assay for efficient de-selection was developed. We demonstrate the application of this improved system to identify novel interacting proteins of the human Frizzled 1 receptor. We identified several novel interactors with components of the Wnt-Frizzled signalling pathways and discuss their potential roles as direct mediators of Frizzled receptor signalling. The present work is the first example of a split-ubiquitin interaction screen using an in-situ expressed receptor of the serpentine class, emphasizing the suitability of the described improvements in the screening protocol.
INTRODUCTION
The development of the yeast two-hybrid technology (Y2H) by Fields and Song (1) was a milestone in the analysis of protein interactions. It allowed for the first time the testing of protein interactions in living cells, and it is suitable for proteome-wide analyses of protein networks by using selectable reporter genes (2–5).
However, a major limitation of the Y2H system is that the protein–protein interaction under investigation has to be reproduced in the yeast nucleus, since it relies on the reconstitution of a transcription factor split into its DNA binding and activation domains. This limits the Y2H interaction studies e.g. of membrane proteins that have to be first truncated to soluble fractions, and frequently excludes screens with transcription factors that contain transactivation domains active in yeast.
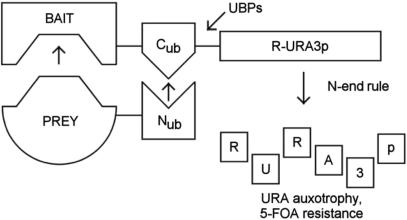
The yeast split-ubiquitin system has been developed to overcome the limitations of the classical Y2H system (6–9): The basic principle of this method is shown in Figure 1. The system represents a fragment complementation assay using the ubiquitin molecule split into C- and N-terminal halves (termed Cub and Nub, respectively) as interaction sensor. The bait protein of interest is fused as an N-terminal fusion to Cub, and the prey protein is expressed as either an N- or C-terminal fusion to Nub. The interaction of bait and prey protein forces the ubiquitin halves into close proximity. This results in the formation of a quasi-native ubiquitin, and triggers the proteolytic cleavage of the R-URA3p reporter molecule attached to the C-terminus of Cub by ubiquitin-specific proteases (UBPs), followed by proteolytic degradation of the now N-terminally destabilized URA3p (R-URA3p). As a consequence, the yeast cells loose their capability to grow in the absence of uracil, and thus permits negative selection in the presence of the otherwise toxic URA3p-specific antimetabolite 5-fluoroorotic acid (5-FOA).
Figure 1.
Principle of the yeast R-URA3p-based split-ubiquitin protein–protein interaction system. The interaction of bait and prey proteins leads to the reconstitution of ubiquitin, resulting in proteolytic degradation of the R-URA3p reporter molecule. This renders the yeast cells URA-auxotroph and resistant to 5-FOA. See text for details. UBPs, ubiquitin specific proteases.
The major advantage of this reporter system over the classical Y2H reporter system is that it does neither depend on nuclear translocation of the complex components nor on a transcriptional readout. Consequently, the R-URA3p based split-ubiquitin system can detect interactions at their natural site (in situ), e.g. at or in the membrane (9). It also facilitates the use of transcription factors as bait proteins (10). In contrast, a distinct variant of the split-ubiquitin system uses a transcription factor readout is restricted to the test of membrane-based protein interactions (and is therefore frequently termed ‘membrane-based Y2H’) (7). The high degree of sensitivity of the R-URA3p-based split-ubiquitin interaction system was recently documented through its use in a small-molecule—protein interaction system for drug discovery research (a split-ubiquitin—based yeast three-hybrid system) (11).
The success of the classical Y2H application in the past has relied strongly on methodological improvements particularly for large scale protein–protein interaction approaches. These improvements essentially aimed at increasing both the speed and the accuracy of the Y2H system: For instance, a significantly increased throughput could be achieved by the introduction of the interaction-mating approach instead of the various, less efficient and more laborious, transformation procedures to obtain bait- and prey-plasmid bearing yeast clones for the subsequent reporter assays (12,13). However, to obtain high fidelity in interpreting results from large scale interaction screens, the appearance of Y2H - intrinsic classes of false-positive and false-negative hits needs to be understood and subsequently reduced. Proteins with broad interaction capabilities represent one important class of false-positive hits, and can be specific for the version of the employed Y2H system (i.e. the split-transcription factor and the reporter version used). A fast and efficient way to sort out such false-positives is the use of the so-called dual-bait strategy (14). In this setup, a given prey protein is tested simultaneously against the actual and any unrelated bait within a single cell by employing different DNA-binding elements for the two baits combined with separate reporter genes.
Here we present a rationale to control the false-negative and false positive-rates in split-ubiquitin protein–protein interaction screens that use the R-URA3p reporter system. For this purpose, we used our past experience of more than 35 independent screenings employing a variety of different baits, and an established split-ubiquitin protein–protein interaction pair (p53–protein phosphatase 5) to fine-tune parameters of an optimized cDNA-library screening system. This way, we were able to identify the 5-FOA concentration as a critical parameter that allows controlling the rates of false-negative and false-positive interaction candidates. We experimentally define the optimal 5-FOA concentration, and demonstrate that it correlates to the plating density. Furthermore, we identified an R-URA3p reporter-specific class of false-positive hits, and present solutions to eliminate them by a simple and fast phenotypic assay.
We demonstrate the efficacy of this optimized split-ubiquitin cDNA-library screening protocol to find interactors of the human Frizzled 1 receptor. Frizzled (Fz) receptors share the structural organization of G protein-coupled receptors (GPCRs) and are comprised of seven transmembrane spanning domains, a complex N-glycosylated exofacial N-terminal region, and a cytoplasmic C-terminal tail. These receptors are activated through binding to ligands called Wnts, which are secreted lipid-modified signalling proteins. Wnt-Fz mediated signalling controls cell polarity, embryonic induction, specification of cell fate and moreover is thought to play a role in diseases such as cancer, osteoporosis and psychiatric disorders (15,16).
Wnt-Fz signaling has been classified into so called canonical (β-catenin dependent) and noncanonical (β-catenin independent) pathways (17–19). While the signalling components of canonical signalling are divergent from classical GPCR signal transduction, there are common features with non-canonical signalling. This includes e.g. sensitivity of Wnt-stimulated calcium flux and other signalling events to pertussis toxin (PTX), a specific inhibitor of G proteins of the Gi-class. However, direct intracellular mediators of both canonical and non-canonical Frizzled signalling have not yet been clearly defined. So far this precluded to classify β-catenin-independent signalling events into discrete pathways.
Using this optimized split-ubiquitin cDNA-library screening system, we describe the identification of novel interacting proteins of the human Fz1 receptor, which is a prototypical inducer of the canonical pathway (20,21). Several of these, or their respective homologs, were previously reported to interact physically and/or functionally with known components of Wnt-Fz signalling pathways, independently validating our screening strategy.
MATERIALS AND METHODS
E.coli and yeast strains
All plasmid constructions and amplifications were performed using the E. coli strain XL1-Blue (Stratagene). The yeast strain used for the split-ubiquitin assays was JD53 MATα his3-D200 leu2-3,112 lys2-801 trp1-D63 ura3-52 (22). For confocal microscopy and western blot analysis the yeast strain MH272-1da MATa his3 leu2-3,112 ura3-52 trp1 rme1 HMLa (23) was used.
Plasmid constructs
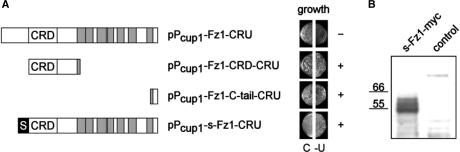
All bait constructs are based on the previously described vector pPcup1-ubc9-CRU (11), which is a low copy yeast–E. coli shuttle vector that carries a Cub-R-URA3 cassette (CRU), a copper-dependent promoter (PCUP1) for bait expression, and a HIS3 marker. All constructs were verified by sequencing. The human Fz1-derived bait constructs are depicted schematically in Fig. 5.
Figure 5.
Schematic overview of tested Frizzled 1 bait constructs and expression tests. (A) Full-length Frizzled (pPCUP1-Fz1-CRU) as well as respective deletion constructs (pPCUP1-Fz1-CRD-CRU and pPCUP1-Fz1-C-tail-CRU), and N-terminal fusion of the Ste2 N-terminal tail (termed ‘s’) to Fz1 (pPCUP1-s-Fz1-CRU) are represented. The position of the cysteine-rich domain (CRD) and predicted transmembrane regions (shaded boxes) are indicated. Expression of the bait constructs is shown by growth (+/−) on media lacking uracil (-U). Transformation control plates (C) contained uracil. (B) western blot analysis using s-Fz1-myc expressing yeast cells. Crude protein extracts were prepared and resolved on 4–12% gradient SDS-PAGE. Detection was performed using an anti-c-myc monoclonal antibody-peroxidase conjugate. Molecular weight standards in kDa are indicated. Aditional bands visible on the gel may represent glycosylated receptor, a frequent modification of serpentine receptors. The control extract was derived from a yeast strain transformed with the empty expression vector. See ‘Material and Methods’ for details.
pPCUP1-Fz1-CRU
This construct is based on the previously described vector pPCUP1-ubc9-CRU (11), which is a low copy yeast–E. coli shuttle vector that carries a CRU, a copper-dependent promoter (PCUP1) for bait expression, and a HIS3 marker. The human Fz1 coding sequence was amplified by PCR from a human whole brain cDNA preparation (Clontech) using primers Fz1-FW (5′-AATTGTCGACATGGCTGAGGAGGAGGCGCCTAAG-3′) and Fz1-RV (5′-AGCGGCCGCGACTGTAGTCTCCCCTTGTTTGC-3′), introducing SalI and NotI restriction sites, respectively. The Fz1 PCR product was eventually used to replace the Ubc9 insert in pPCUP1-ubc9-CRU using SalI-NotI restriction sites, resulting in pPCUP1-Fz1-CRU.
pPCUP1-Fz1-C-tail-CRU
Primers tail-FW (5′-AATTGTCGACATGTGGATCTGGTCCGGCAAG-3′), introducing SalI and a start codon, and Fz1-RV (see above) were used to amplify the 3′-terminal 87 bp of the Fz1 coding sequence from pPCUP1-Fz1-CRU by PCR. The Fz1-C-tail PCR product was eventually used to replace the ubc9 insert in pPCUP1-ubc9-CRU using SalI-NotI restriction sites, resulting in pPCUP1-Fz1-C-tail-CRU.
pPCUP1-Fz1-CRD-CRU
Primers CRD-FW (5′-AATTGTCGACATGGACCACGGCTATTGCCAG-3′), introducing SalI and a start codon, and CRD-RV (5′-AGCGGCCGCCCAAATGCCAATCCAGGTGC-3′), introducing NotI, were used to amplify a fragment of 645 bp covering the cysteine-rich-domain of Fz1 including the downstream portion seizing the predicted first helix from pPCUP1-Fz1-CRU by PCR. The resulting Fz1-CRD PCR product was eventually used to replace the ubc9 insert in pPCUP1-ubc9-CRU using SalI-NotI restriction sites, resulting in pPCUP1-Fz1-CRD-CRU.
pPCUP1-s-Fz1-CRU
The 5′ 138 bp of the Saccharomyces cerevisiae Ste2 coding sequence (termed ‘s’ for signal sequence), comprising the receptor's complete N-terminal extracellular tail, were amplified from a yeast genomic DNA preparation using primers Ste2-FW (5′-AATTGTCGACATGTCTGATGCGGCTCCTTC-3′), introducing SalI, and Ste2-RV (5′-GTACTGTTAACTAAACCTTG–3′), introducing a HpaI site. Human Fz1 cDNA lacking the 5′ 333 bp of the coding sequence, comprising the region upstream of the receptor's cysteine rich domain (CRD), was amplified from pPCUP1-Fz1-CRU using primers Fz1-HpaI (5′-ATATGTTAACGACCACGGCTATTGCCAGCC-3′), introducing HpaI, and Fz1-RV. In a triple ligation set up, SalI-HpaI restricted Ste2 fragment and HpaI-NotI restricted Fz1 fragment were used to replace the ubc9 insert in PCUP1-ubc9-CRU using SalI-NotI restriction sites, resulting in pPCUP1-s-Fz1-CRU.
pPCUP1-p53-CRU
This bait construct encoding the full length human p53 gene was cloned via SalI-NotI in an analogous way to the constructs described above, and was kindly provided by GPC Biotech AG (Munich).
pPADH-NubI-PP5
Full length human protein phosphatase 5 was identified as a p53 interactor in a previous split-ubiquitin screen performed at GPC Biotech AG (Munich) using above described pPCUP1-p53-CRU bait construct in a human fetal brain NubI-split-ubiquitin cDNA-library (see below).
pYES2-s-Fz1-myc
The s-Fz1 fusion gene was amplified by PCR from pPCUP1-s-Fz1-CRU using primers DD-11 (5′-ATAAGCTTATGGCTGAGGAGGAGGCGCC-3′) and DD-12 (5′-TAGGATCCCTACAGATCCTCTTCTGAGATGAGTTTTTGTTCGACTGTAGTCTCCCCTTGTTTG-3′), introducing a HindIII site, a C-terminal c-myc-tag followed by a STOP codon, and a BamHI site, respectively. The HindIII-BamHI digested s-Fz1-myc fragment was ligated to HindIII-BamHI restricted yeast expression vector pYES2 (Invitrogen), resulting in pYES2-s-Fz1-myc.
pYES2-s-Fz1-GFP
The s-Fz1 fusion gene was amplified by PCR from pPCUP1-s-Fz1-CRU using primers DD-13 (5′-ATAAGCTTATGTCTGATGCGGCTCCTTCATTG-3′) and DD-18 (5′-TAGAATTCGACTGTAGTCTCCCCTTGTTTG-3′) introducing HindIII and EcoRI sites, respectively. Enhanced green fluorescent protein (eGFP) was amplified by PCR from pEGFP-C1 (Clontech) using primers DD-19 (5′-ATGAATTCGTGAGCAAGGGCGAGGAGC-3′) and DD-20 (5′-TAGGATCCTTACTTGTACAGCTCGTCCATGC-3′), introducing EcoRI and BamHI sites, respectively. HindIII-EcoRI digested s-Fz1 and EcoRI-BamHI digested eGFP fragments were ligated to HindIII-BamHI restricted yeast expression vector pYES2 (Invitrogen), resulting in pYES2-s-Fz1-GFP.
pGEX-p53
The p53-cDNA was excised via Sal-NotI from pPCUP1-p53-CRU, and ligated to SalI-NotI-restricted pGEX-4T-3 (Amersham Biosciences), resulting in pGEX-p53.
pASK-PP5
The full length cDNA of human protein phosphatase 5 was amplified by PCR from pPADH-NubI-PP5 using primers PP5-FW(5′-ATGAATTCGCGATGGCGGAGGGCGAGAGGACT-3′) and PP5-RV(5′-ATCTCGAGCATTCCTAGCTGCAGCAGCGTGTTG-3′), introducing EcoRI and XhoI sites respectively, and ligated to EcoRI-XhoI restricted pASK-IBA5plus (IBA GmbH, Goettingen), resulting in pASK-PP5.
Western blot analysis of yeast extracts
Yeast strain MH272-1da (23), transformed with pYES2-s-Fz1-myc or with the empty vector pYES2, was cultivated in 50 ml SD-U medium to the exponential growth phase. To induce protein expression, the cultures were transferred to galactose-containing minimal medium for 5 h. Cells were pelleted and resuspended in 300 μl lysis buffer (100 mM Tris–HCl pH 8.0, 20% glycerol, 1% Triton X-100, 1 mM dithiothreitol, 1 mM phenylmethylsulphonylfluoride). Cells were disrupted by vortexing at 4°C for 10 min following addition of acid-washed glass beads (0.45 mm diameter). The crude protein extract was cleared by centrifugation at 3000 × g for 5 min. The protein concentration was determined using the Biorad protein assay (Biorad, Munich). The extract of 20 μg were electrophoresed using a 4–12% gradient NuPAGE Bis–Tris gel system (Invitrogen, Karlsruhe). The proteins were electroblotted to a nitrocellulose membrane. The membrane was blocked in Tris-buffered saline (TBS, 0.1% Tween 20) containing 5% non-fat dry milk, and subsequently incubated with a mouse anti-c-myc monoclonal antibody conjugated to peroxidase (Roche, Basel) at a 1:1000 dilution in TBS, 0.1% Tween 20, 1% bovine serum albumin for 1 h at room temperature. Detection was performed using the Immune-Star HRP substrate kit (Biorad, Munich) and Hyperfilm (Amersham Biosciences, Freiburg).
Glutathione S-transferase (GST) pull-down experiment
The preparation of the GST-p53 fusion-protein coupled to glutathione-agarose was performed according to the manufacturer's instructions for the GST gene fusion system (Amersham Biosciences): In brief, E. coli expression strain BL21 was transformed with pGEX-p53. Protein expression in an exponentially growing culture in 2YT medium (400 ml) including ampicillin was induced over 2 h by adding IPTG (0.1 mM). Eventually, the cells were harvested by centrifugation at 4°C, and resuspended in 20 ml phosphate-buffered saline (PBS) containing a protease inhibitor cocktail (Sigma). Cells were disintegrated by sonication under cooling, and the lysate was cleared by centrifugation at 10.000 × g at 4°C for 10 min. Fifty percent of 400 μl GSH-agarose (Sigma) were added to the sonicate, and incubated with gentle agitation at room temperature. Subsequently, the GSH-agarose was pelleted by centrifuging for 5 min at 500 × g, and washed three times with 10 ml PBS at 4°C.
The purification of the Strep-tagged PP5 protein was performed according to the manufacture's instructions for the Strep-Tactin affinity tag system (IBA GmbH, Goettingen): In brief, E. coli expression strain BL21 was transformed with pASK-PP5. Protein expression in an exponentially growing culture in 2YT medium (100 ml) including ampicillin was induced over 2 h by adding anhydrotetracycline (200 μg l). Eventually, the cells were harvested by centrifugation at 4°C, and resuspended in 5 ml ice-cold 100 mM Tris-HCl pH8.0, 1 mM EDTA containing a protease inhibitor cocktail (Sigma). Cells were disintegrated by sonication under cooling, and the lysate was cleared by centrifugation at 10.000 × g at 4°C for 10 min. The cleared lysate of 5 ml were applied to a streptactin-sepharose column (1 ml bed volume) equilibrated with 100 mM Tris–HCl pH8.0, 1 mM EDTA. The column was eventually washed five times with 500 μl−1 100 mM Tris-HCl pH 8.0, 1mM EDTA. Elution of the bound protein was performed using six times 500 μl 100 mM Tris-HCl pH 8.0, 1 mM EDTA containing 2.5 mM desthiobiotin, and the eluates were pooled.
GST-p53, or GST-loaded GSH-agarose of 100 μl (prepared from BL21 transformed with pGEX as control) were washed two times with 1 ml 100 mM Tris–HCl pH8.0, 1 mM EDTA, and eventually incubated each with 1 ml Strep-PP5 containing eluate, and incubated at 4°C for 1 h. At the end of the incubation, the beads were settled by gravity, and the supernatant was collected as the unbound fraction. The resin was extensively washed with 100 mM Tris–HCl pH8.0, 1 mM EDTA, and subsequently eluted with the addition of 1 ml SDS-containing sample loading buffer, representing the bound fraction. Corresponding aliquots of Strep-PP5 containing eluate (‘input’-fraction), ‘unbound’ and ‘bound’ fractions were electrophoresed using a 12% SDS-PAGE gel system. The proteins were electroblotted to a nitrocellulose membrane. The membrane was blocked in 3% non-fat dry milk in 1 × PBS, 0.1% Tween 20 (Sigma) for 1 h at room temperature. Eventually, the membrane was incubated with a Strep-Tactin-HRP conjugate (IBA GmbH, Goettingen) diluted 1:100 000 in PBS, 0.1% Tween 20 for 1 h. Detection was performed using the Immune-Star HRP substrate kit (Biorad, Munich) and Hyperfilm (Amersham Biosciences, Freiburg). Eventually, the membrane was blocked again, and redetected using anti-GST-POD conjugate (Sigma) diluted 1 : 20 000 in 1 × PBS, 0.1% Tween. Purity and integrity of the GST-p53 and Strep-PP5 fusion proteins were monitored using silver-stained SDS-PAGE gels (data not shown).
Confocal microscopy
Microscopic inspection of agarose-embedded s-Fz1-GFP expressing yeast strain MH272-1da (23) following galactose-induction was performed using a Zeiss LSM 510 META confocal microscope.
Survival assays using the p53-PP5 model protein interaction
The survival assays were carried out by a procedure analogous to the actual cDNA-library screens described in the appendix section of this article. In brief, yeast strain JD53 was sequentially co-transformed with plasmids as indicated in the ‘Results’ section using a high-efficiency transformation procedure (24). Plating at a density of ∼600 co-transformants/cm2 medium area (standard screening plating density) was performed onto large screening dishes (Nunc 22.5 × 22.5 cm BioAssay Dish), containing either SD-LEU-HIS medium, or SD-LEU-HIS, 100 mM Cu2SO4, with 5-FOA concentrations as indicated in the ‘Results’ section. The evaluation was done as described in the ‘Results’ section.
Human fetal brain NubI split-ubiquitin cDNA-library
The employed human fetal brain NubI—split ubiquitin cDNA library was kindly provided by GPC Biotech AG (Munich, Germany) and was constructed as follows: Split-ubiquitin prey expression vectors are based on plasmid PADNS (25), and are derivatives of PADNX-Nub-BC plasmids as previously described (10). These plasmids carry 2µ origins, ampicillin resistance genes and the wildtype N-terminal part of ubiquitin (NubI) (6) driven by an ADH promoter. pPADH-NubI contains unique SalI and NotI restriction sites in the multiple cloning site (MCS) followed by stop codons in all three reading frames.
Total human fetal brain poly(A)+ RNA was purchased from Clontech (St-Germain-en-Laye), and cDNA synthesis was performed using poly (T)-primers introducing a NotI restriction site. After second strand synthesis, a SalI adapter ligation was performed, followed by NotI digestion and cDNA size-fractionation chromatography. Individual ligation set ups of resulting different cDNA fractions to each of the three different frame versions of SalI-NotI restricted pPADH-NubI prey vector were performed. Following small scale E. coli (DH10B, Gibco) electroporation with aliquots of individual ligation reactions, size distribution of cDNA inserts was monitored using PCR. Eventually, fractions were pooled to cover a size distribution ranging from ∼500 to 3000 bp, and finally a total of ∼1 × 10E6 independent clones was generated, from which plasmid maxi—preparations were performed (Qiagen, Hilden).
Optimized split-ubiquitin cDNA-library screening procedure
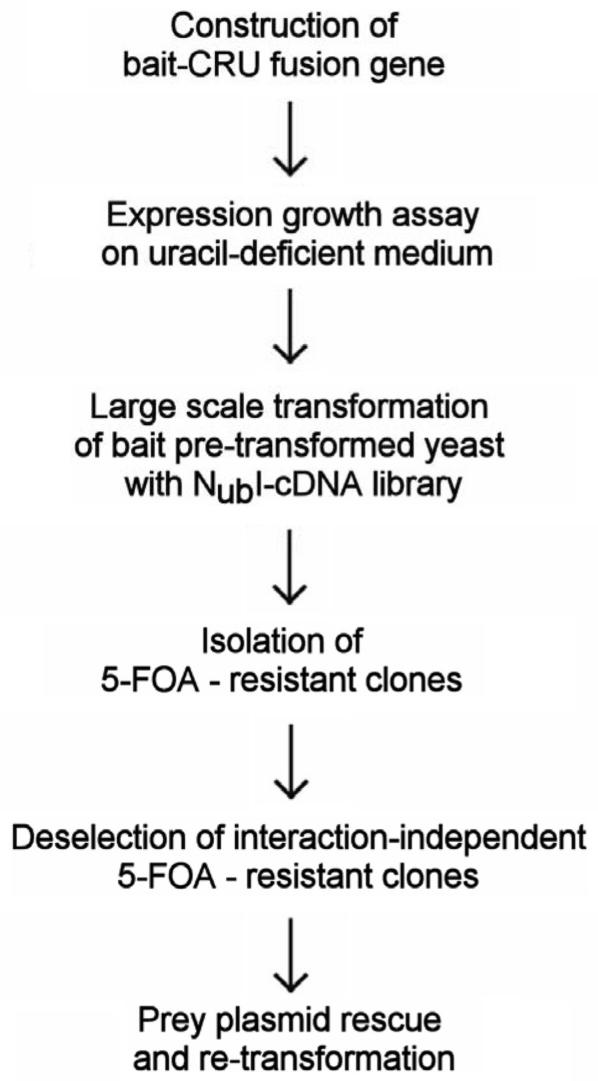
A detailed experimental protocol is provided in the Supplementary section of this article, and is schematically represented in Figure 2.
Figure 2.
Optimized flowchart for split-ubiquitin protein-protein interaction cDNA-library screens. See text for details.
RESULTS
We set out to define generally applicable conditions to minimize both false-negative and false-positive hits in the split-ubiquitin cDNA-library screening system based on the R-URA3p reporter (9) (Figure 1). We found that variations of two parameters that are interdependent strongly affect the quality of the screening results: (i) The optimal 5-FOA concentration used for selecting interaction pairs, (ii) the plating density of the transformations that, in turn, affects the 5-FOA selection pressure. We determined optimal conditions for both parameters and tested these in an interaction screening assay using the human Fz1 receptor that in the past has been shown to be not suitable for Y2H interactor analyses. We find that the resulting workflow of our improved screening protocol (Figure 2) is generally applicable and has, in the meantime, been successfully used in >35 interactor screens. Furthermore, we proved experimental details in the ‘Material and Methods’ and the Supplementary sections accompanying this article to facilitate the general application of the protocol.
Definition of the optimal 5-FOA concentration for split-ubiquitin cDNA-library screens as a key parameter to control the hit rate
In the many screens we performed so far with distinct bait constructs, the 5-FOA concentration used in the selection strategy strongly affected the ratio of 5-FOA resistant clones compared to the total number of screened yeast clones (We will term this ratio ‘survival rate’ in the following text). It also affected the rate of false-positive hits, which are defined as bait-prey interactions that cannot be reproduced in an independent retesting using the split-ubiquitin system. In general, increasing the 5-FOA concentration from 0.5 to 1.0 g per liter of medium led to survival rates that were reduced by at least one order of magnitude (typically from 1 : 30 000 down to 1 : 500 000).
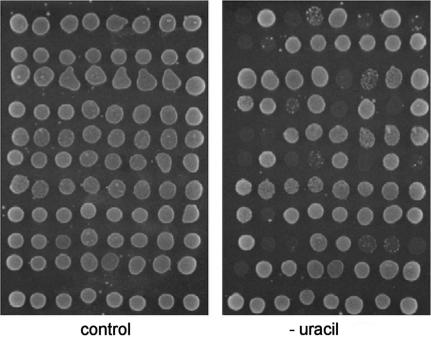
Surprisingly, the false-positive rate increased generally up to >95% at 5-FOA concentrations above 0.8 gl−1, whereas one would intuitively suggest that the reduced survival rate would result in a higher specificity and a reduced number of false-positives. Analyses of these 5-FOA resistant clones revealed that they contained mutated bait plasmids with an apparently defective R-URA3 reporter cassette. This was indicated by the absence of any growth on medium lacking uracil, paralleled by 5-FOA resistance that is not conferred by any prey plasmid in the re-tests (for an example see Figure 7).
Figure 7.
De-selection of false-positives resulting from mutations in R-URA3p. To identify prey-plasmid independent URA3-auxotrophic clones, passive prey-plasmid loss was induced. Clones were eventually spotted onto medium lacking uracil (-uracil). After 48 h incubation, clones that failed to grow were excluded from further analyses. The control plate contained uracil.
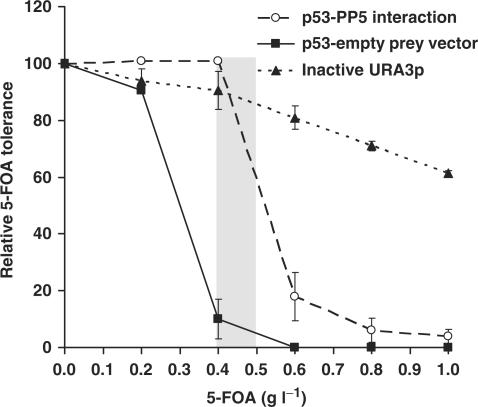
We reasoned that this enrichment of URA3-negative clones at higher 5-FOA concentrations may be the result of a higher 5-FOA tolerance as those clones that express interacting bait and prey combinations. To test this systematically we compared the survival rates of clones expressing an inactive URA3p with clones expressing interacting bait-prey combinations at different 5-FOA-concentrations. For this experiment we chose the human tumor suppressor gene p53 as bait protein (p53-CRU) and human protein phosphatase 5 (PP5) as prey (NubI-PP5). This pair of interacting proteins was identified and characterized in a previous screen (Figure 3). Indeed, clones bearing only the inactive URA3-cassette, but no bait/prey combinations, showed a considerably higher 5-FOA tolerance (IC50 5-FOA: >1.0 g−1l; inhibitory 5-FOA concentration resulting in a survival rate of 50%) than clones expressing the strongly interacting p53-PP5 combination, (IC50 5-FOA: ∼0.5 gl−1) (Figure 4). Consistently, URA3-negative clones grew better to become significantly larger colonies than the p53-PP5 interaction clones on screening agar plates. An equivalent observation was also made in the patch growth assay.
Figure 3.
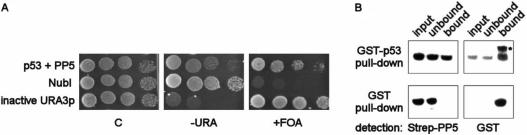
Interaction of human p53 and protein phosphatase 5 (PP5). (A) Split-ubiquitin interaction assay. Clones expressing p53 (p53-CRU bait construct) and PP5 (Nub-PP5 prey) were spotted in serial dilutions onto medium lacking uracil (-URA), medium containing 5FOA (+FOA), and control plates selective for the presence of plasmids (C). Interaction is indicated by lack of growth on -URA, paralleled by 5-FOA resistance. A clone co-transformed with p53 and empty prey vector (NubI) served as negative control. A previously identified bait construct with an inactive URA3p phenocopying a true interaction event was also included. (B) GST-pull down experiment. Purified Strep-tagged PP5 (Strep-PP5) expressed from E. coli was incubated with GSH-agarose beads loaded with E. coli expressed GST-p53 fusion protein, or GST alone. Corresponding aliquots of Strep-PP5 ‘input’, ‘unbound’ and ‘bound’ fractions were electrophoresed using SDS-PAGE, and electroblotted to nitrocellulose. Detection was performed first by using a Strep-tactin-HRP conjugate, followed by redetection using an α-GST-HRP conjugate. The asterisk indicates the specific signal.
Figure 4.
Differential 5-FOA tolerance for yeasts expressing interactors and after mutational loss of R-URA3p. Clones expressing interacting p53 (bait)-PP5 (prey), p53 and empty Nub-prey-vector (no interaction), or an inactive URA3p (false-positive) were plated at the standard plating density (∼600 co-transformants/cm2 medium) on SD-HIS-URA (transformation control plate) and on 5-FOA (as indicated). Relative 5-FOA tolerance levels were calculated as the ratio of emerging 5-FOA resistant clones with respect to the control plate (survival rate in %). Data represent the mean of two independent experiments +/− range. The grey area indicates the range of 5-FOA concentrations found optimal for the screens.
We conclude that there is a residual URA3p-activity that generates a background even in those clones in which an interaction occurs. This, in turn, results in a considerable increase in 5-FOA sensitivity that is even higher than that seen in clones that, due to mutations or contaminations, lack a functional URA-3 gene. From this experiment we strongly suggest to not exceed an optimal 5-FOA concentration of 0.5 gl−1. Above this threshold concentration, the chances to isolate valid interactions decrease strongly. As a cause we identified an enrichment of false-positive clones resulting from URA3 inactivation that is independent of the N-end rule degradation of URA3 as a consequence of ubiquitin reconstitution via bait-prey interactions. In the Fz1 interaction screen shown below we describe a simple and rapid method to phenotypically sort out this class of false-positives.
In order to experimentally define the minimal 5-FOA concentration required to suppress non-specific background growth, we tested the combination of a plasmid bearing p53-CRU and the empty NubI-prey plasmid in the 5-FOA tolerance assay to mimick the absence of p53 interaction. As expected, tolerance of these clones for 5-FOA was lower than the other combinations tested (IC50 5-FOA ∼0.3 gl−1). This suggests a minimal 5-FOA concentration of ∼0.4 gl−1 at which most background growth is suppressed (Figure 4). We found equivalent minimal 5-FOA concentrations to be useful in a variety of bait constructs not related to p53, such as the yeast proteins Mrs6p, Sec16p and Sec63p (data not shown). In conclusion, we have defined experimentally an optimal 5-FOA concentration of 0.5 gl−1 at which a balance between the maximal growth of interaction clones and a tolerable non-specific background growth coexists.
Yeast plating density affects the selection pressure by 5-FOA
We found another critical property of the R-URA3p reporter that is modulated by the yeast transformation protocol of the libraries. We observed in various screens that the 5-FOA survival rates of clones harboring interacting bait/prey combinations strongly depend on their plating density (defined as number of co-transformants per cm2 medium area), which is a variable of the transformation efficiency. In order to demonstrate this relationship systematically, we devised additional 5-FOA tolerance experiments: For this purpose, we again used the p53-PP5 model interaction and kept the 5-FOA concentration constant at the optimal rate of 0.5 gl−1 as described above. Instead, we reduced the plating density from 600 co-transformants per cm2 medium area (corresponding to our standard transformation protocol, as used above) down to 60 by a 10-fold dilution of the transformation mixture. Surprisingly, we observed a dramatically reduced survival rate from approximately 60% down to 10% (data not shown). We suggest that a decreased number of transformants expressing URA3p may result in an increased ratio of 5-FOA molecules per cell. This exerts a stronger selective pressure, and consequently results in lowered survival rates. Based on these results, we suggest a 5-FOA concentration of 0.5 gl−1 as optimal for a plating density of 600–900 transformants/cm2. Our experiments also showed that lowering the plating density through low-efficiency transformation methods (e.g. by double transformations of bait and library instead of a sequential transformation as used here; see ‘Supplementary Data’) results in too stringent selection conditions, probably as a result of a local excess of 5-FOA. As a consequence, URA3-negative clones will have a growth advantage and will be enriched (data not shown). On the other hand, we found that considerably higher plating densities (>2000 co-transformants per cm2 medium area, which can be achieved using yeast mating protocols) yield unwanted interaction—independent background growth due to a lowered ratio of 5-FOA molecules per cell (data not shown).
In conclusion, we suggest to first define transformation efficiencies for any new bait/library screenings. Based on the resulting plating density, the optimal 5-FOA concentration that allows maximal survival of interaction events and, at the same time, minimizes non-specific background growth, should be determined individually.
Split-ubiquitin NubI-human fetal brain cDNA-library screen using the human Frizzled 1 receptor as bait
Using the parameters defined above, we performed a split-ubiquitin interactor screen using a NubI-human fetal brain cDNA-library and the human Fz1 receptor as bait. We reasoned that a screen using this bait that in the past proved to be non-accessible for standard Y2H screenings may serve as a stringent control for the quality of the protocol. The chart depicting the screening workflow that is generally applicable is shown in Figure 2.
Cloning of baits. A simple growth assay to determine bait expression
We used the full length cDNA of human Fz1 to construct the pPCUP1-Fz1-CRU bait expression vector (Figure 5A) in which the Fz1 cDNA is expressed as a translational fusion to the URA3 gene. We reasoned that the generation of URA3p could be used as an indicator for Fz1 expression. In order to verify solid expression of this construct, we therefore tested URA3p levels in a growth assay. Surprisingly, the transformants with pPCUP1-Fz1-CRU did not grow on minimal media lacking uracil (100 μM Cu2+ for induction, used as standard concentration). This immediately prevented the use of this construct in our split-ubiquitin interaction assays (Figure 5A). Whereas most bait constructs in our hands required a standard copper ion concentration of 100 μM for induction, some baits were fully expressed only upon addition of up to 200 μM Cu2+. In the case of pPCUP1-Fz1-CRU, however, copper concentrations >200 μM were not effective (data not shown). We have observed similar difficulties with only six other bait constructs not related to Fz1, representing <20% of all tested proteins. These included CHIP (a cytosolic E3/E4 ligase), PINK1 (a Parkinson's Disease associated kinase with predominantly mitochondrial localization), the Ret protooncogene (a transmembrane receptor) and inversin (a cytosolic component of the Wnt signalling pathway). A correlation between the particular cellular localization of these proteins and expression problems could not be detected. In all cases but one the expression problem could be solved by truncating the bait constructs and by choosing distinct 5′ ends. One suggestion is that the 5′ ends of some bait cDNAs, providing the translation initiation sites of the bait-URA3 constructs, may prevent sufficient expression levels. Another possibility is that some proteins, in particular membrane proteins, may be improperly targeted to the yeast secretory pathway.
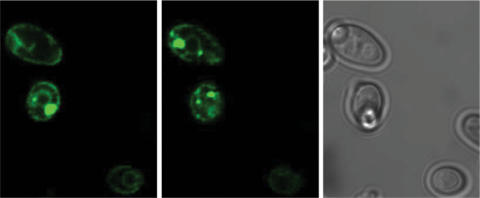
To distinguish between both possibilities, we selectively expressed Fz1 receptor truncations that contain domains which are known, or are predicted to, mediate protein interactions. Firstly, we expressed the N-terminal cysteine-rich-domain (CRD), which is characteristic for Frizzled receptors and has been implicated in Wnt-ligand binding (26). Secondly, we tested expression of the C-terminus of the receptor, since this may be critical for contacting downstream effectors on the cytoplasmic face of the plasma membrane. For this purpose, pPCUP1-Fz1-CRD-CRU and PCUP1-Fz1-C-tail-CRU plasmids were constructed (Figure 5A). Both constructs resulted in sufficient growth on uracil selective media indicating that this time sufficient amounts of bait proteins were expressed to use them in subsequent library screenings (Figure 5A). Although we did not analyze expression problems more systematically, in the case of F1 the data are consistent with improper targeting of the full-length clone. In order to force targeting of Fz1 to the correct subcellular localization, we designed a chimeric receptor bait, in which the amino acid sequence upstream of the CRD domain was replaced with the extracellularly localized, N-terminal tail of the yeast α-factor receptor Ste2p (termed ‘s’-sequence in the following). Expression of this s-Fz1 chimeric receptor complemented uracil auxotrophy, suggesting that the fusion protein was made in sufficient amounts and that Ura3p obviously interacts with its cytoplasmically localized substrate orotidine-5'-phosphate (Figure 5A). This was confirmed in a western blot using a myc-tagged version of the s-Fz1 receptor where we obtained a signal of expected size (60 kDa) (Figure 5B). In order to prove the correct subcellular localization of the s-Fz1 receptor (in the cytoplasmic membrane), we expressed the bait in a fusion construct with enhanced green fluorescent protein (eGFP) and monitored the GFP signal. The protein was localized both at the plasma membrane and in vesicle-like structures in the cell, comparable to the localization described previously for GFP-tagged Ste2p (27) (Figure 6).
Figure 6.
Confocal micrographs of yeast cells expressing s-Fz1-GFP. Cells were immobilized by agarose-embedding, and the fluorescence signal was recorded from two different planes. s-Fz1-GFP can be detected at the membrane and in intracellular spots that most likely are vesicle-like structures, similarly to the localization of Ste2p (27). The right panel shows the brightfield view.
We conclude from these studies that it is necessary, before performing the library screenings, to verify for each bait that sufficient levels of protein are expressed. We suggest using the complementation of uracil auxotrophy of the bait constructs as a simple test for bait expression and trying out alternative protein subfragments, in case the expression of full-length clones fails.
Screening for protein interactors of s-Fz1
We screened for protein interactors of s-Fz1 using a poly(T)-primed human fetal brain cDNA-library fused C-terminally to NubI. We generated approximately 3 × 10E6 HIS3-LEU2 positive co-transformants, corresponding to 3-fold library coverage. Using the conditions described above, we isolated a total of 88 5-FOA resistant clones which corresponds to a survival rate of 1 : 34 090. This was comparable to previous split-ubiquitin screens with other baits performed in our lab.
Elimination of false-positive interaction candidates by URA3 inactivation
As stated above, one class of false-positive signals in our split-ubiquitin screens could be derived from URA3-negative plasmids. We devised a fast phenotypic test to sort out these candidates prior to more time-consuming analyses. For this purpose, we first tested whether there exists a correlation of URA-auxotrophy to the presence of the respective prey plasmid: Clones that were auxotroph for URA should contain a prey plasmid, otherwise they are regarded as false-positives. Random loss of this prey plasmid should result in URA-prototrophy. Instead of relying on a counter-selectable marker typically used in classical yeast two-hybrid applications, our negative selection scheme allowed us to test URA-auxotrophy following the passive loss of the prey plasmid.
For this purpose, the primary hit clones were sub-cultured at non-selective conditions to induce passive prey-plasmid loss, and phenotypic analyses were performed (Figure 7). Accordingly, 63 of 88 clones showed URA-prototrophy, corresponding to a very low false positive rate of 28.4%, which was similar to previous screens performed in our lab. One explanation for this remaining population may be a constant random mutation rate of the URA3 gene in the various bait strains. A higher rate of false-positives would have been indicative of too stringent selection conditions (such as too high 5-FOA concentration, see above). Moreover, contamination with foreign yeast plasmids without URA3 could also account for significantly higher false-positive rates, and can easily be identified using the presented procedure.
Phenotypic re-testings of identified prey plasmids
In our next selection step we retransformed the remaining 63 clones to test their ability to confer URA-auxotrophy and 5-FOA resistance. Surprisingly, only 16 clones showed the expected phenotype (Figure 8), although all 63 clones had behaved correctly in the URA-prototrophy tests (see above). Based on our previous experience the relatively large proportion of 75% false-positives is not unusual. These typically arise from clones picked from background colony growth. This is the result of the relatively small experimental window for the optimal 5-FOA concentration to control background growth while, at the same time, maximizing the survival of interactors (compare Figure 4).
Figure 8.
Positive phenotypic re-tests of isolated prey-plasmids. Following prey-plasmid rescue, individual re-transformations of s-Fz1-CRU bait and prey plasmids were performed. Duplicate clones were spotted each in serial dilutions onto medium lacking uracil (-URA), medium containing 5-FOA (+FOA), and control plates selective for the presence of plasmids (C). Interaction is indicated by lack-of-growth on –URA, paralleled by 5-FOA resistance. As negative control served a clone co-transformed with s-Fz1-CRU and empty prey vector (NubI). A previously identified bait construct with an inactive URA3p phenocopying a true interaction event was also included.
Analyses of the s-Fz1 prey-data set
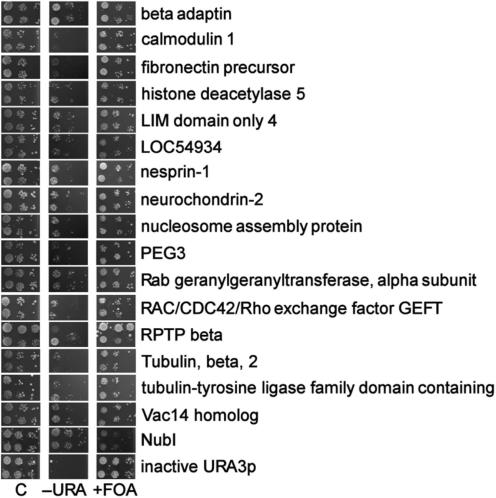
The 16 re-transformation positive clones were sequenced, and peptide sequence database searches were performed using BLASTx (National Center for Biotechnology Information—NCBI). The extracted results are shown in Table 1.
Table 1.
Proteins identified as Frizzled1 interactors in the split-ubiquitin screen
| Prey protein definition | Accession | Prey full length | Nub-fusion point | Pathway association with |
|---|---|---|---|---|
| Beta adaptin | AAA40797 | 937 | 155 | ? |
| Calmodulin 1 | AAP36156 | 150 | 1 | Wnt/Ca2+ signaling |
| Fibronectin precursor | CAA26536 | 2328 | 1218 | Planar cell polarity + Wnt/cAMP signaling |
| Histone deacetylase 5 | AAD29047 | 1122 | 669 | Canonical pathway |
| Hypothetical protein LOC54934 | AAH13900 | 390 | 289 | ? |
| LIM domain only 4 | AAH03600 | 165 | 41 | ? |
| Nesprin-1 | AAN60442 | 8797 | 450 | ? |
| Neurochondrin-2 | BAA77831 | 712 | 546 | Wnt/Ca2+ signaling |
| Novel protein containing a tubulin-tyrosine ligase family domain | CAI22724 | 553 | 1 | ? |
| Nucleosome assembly protein | BAA08904 | 506 | 77 | ? |
| PEG3 | AAC83176 | 615 | 414 | ? |
| Rab geranylgeranyltransferase, alpha subunit | NP_004572 | 567 | 354 | ? |
| RAC/CDC42/Rho exchange factor GEFT | AAO49463 | 580 | 206 | Wnt/Rho signaling |
| Receptor-type protein tyrosine phosphatase beta, RPTP beta, PTP zeta | AAB26530 | 2307 | 1447 | Canonical pathway |
| Tubulin, beta, 2 | AAH29529 | 445 | 1 | Canonical pathway |
| Vac14 homolog | NP_060522 | 782 | 183 | ? |
? refers to poorly characterized protein. Further experiments are needed to link it to Wnt pathway.
A general observation was that, in all retransformation-positive clones, cDNA inserts were fused in-frame with the Nub moiety. In contrast, cDNA inserts derived from the false-positive fraction contained almost exclusively 5′-untranslated regions plus the respective downstream coding sequence. This demonstrates that the method provided allows an efficient de-selection of false-positive clones.
Taken together, we provide a combination of improvements of the original split-ubiquitin protocol, together with several selection steps to reduce potential false-positive interactors. This easy-to-reproduce protocol was tested to find interactors of human Fz1, a protein that was previously notoriously problematic to be used in Y2H screens. After the systematic elemination of false-positive interactors, 16 candidates were obtained.
DISCUSSION
Here we describe a systematic analysis of R-URA3p reporter properties critical for the efficiency and robustness of split-ubiquitin cDNA-library screening. One unexpected finding testing a p53–PP5 model interaction was that URA-deficient cells are less sensitive for toxic concentrations of 5-FOA if they expressed prey proteins (compared to clones in which URA probably was mutated). Therefore, in order to enhance the efficiency of screening procedures, the optimal 5-FOA concentration needs to be thoroughly titrated for each individual bait plasmid. On the one hand, this 5-FOA concentration needs to be high enough to minimize non-specific background. On the other hand, we found that choosing conditions that are too stringent strongly enriches for false-positive hits derived from URA3-inactivation, probably as a consequence of spontaneous mutation events. For a rapid identification of this reporter-specific class of false-positive preys, we have provided the protocol for a simple phenotypic test that functions independently of counter-selection for the prey plasmid. Both phenotypic testing and counter-selection significantly enhanced the reliability of split-ubiquitin interactions screens, as could be demonstrated recently in a number of cases [reviewed in (28)].
We also provide evidence that the 5-FOA selection conditions can be improved by lowering the density of yeast cells plated after transformation. This, on the other hand, requires the determination of transformation efficiencies individually for each bait and library prior to screening. A flow chart describing individual steps of improvement to enhance speed, reliability and fidelity of split-ubiquitin screens sums up our results (Figure 2).
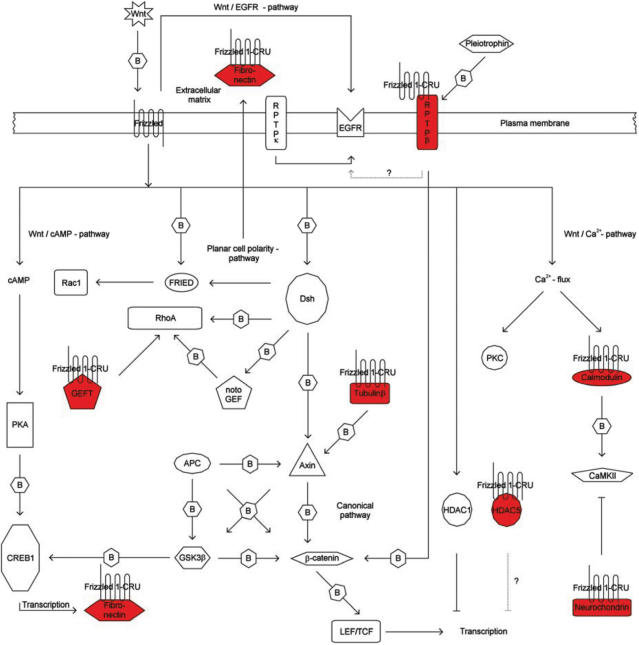
In the second part of our experiments we have shown the practicability of our improved protocol by identifying proteins that are capable of interacting with Fz1. Although the experimental placement of these proteins in the signalling pathway of Fz1 is not the goal of this manuscript, we would like to note that, with few exceptions, these have previously been described to act in Fz1-associated pathways and therefore are good candidates which could represent novel missing links of the pathway (Figure 9; see also ‘Supplementary Information;). For example, a novel interaction between Fz1 and the guanine nucleotide exchange factor GEFT was found (29). Rho family GTPases were reported previously to play a role in Drosophila PCP signalling, and Rho, Rac and Cdc42 take also part in vertebrate non-canonical Wnt signalling [reviewed in (17)]. However, the precise mechanism of activation of Rho family GTPases in Wnt signalling has not yet been elucidated. It is well known that activation of Rho family proteins requires the exchange of bound GDP for GTP mediated by the Dbl family of guanine nucleotide exchange factors (GEFs). GEFT has been described as RhoA-specific GEF, which is however in conflict with an earlier report suggesting Rac1 and Cdc42 specificity (30).
Figure 9.
Schematic representation of candidate interactors and their putative postion in the Frizzled signaling pathway. Frizzled-CRU bait and prey proteins (shown in red) are indicated. Arrows with solid lines represent associations based on literature knowledge, dotted lines represent speculative associations. Arrows marked with ‘B’ represents ‘physical interaction (binding)’. Abbreviations: APC, adenomatous polyposis of the colon; CaMKII, calcium/calmodulindependent protein kinase II; cAMP, cyclic adenosine monophosphate; CREB, cAMP responsive element binding protein; Dsh, Dishevelled; EGFR, epidermal growth factor receptor; FRIED, frizzled interaction and ectoderm development; GEFT, RAC/CDC42/Rho exchange factor guanyl-nucleotide exchange factor; GSK3β, glycogen synthase kinase-3; HDAC, histone deacetylase; LEF/TCF, lymphoid enhancer factor/T-cell transcription factor; notoGEF, Xenopus notochord expressed Rho-specific guanine nucleotide exchange factor; PKA, protein kinase A; PKC, protein kinase C; Rac, ras-related C3 botulinum toxin substrate; Rho, Rho (Ras homology) subfamily of Ras-like small GTPases; RPTP, receptor-type protein tyrosine phosphatase; Wnt, wingless-type MMTV integration site family member.
Our finding of a physical association of Frizzled1 and GEFT may give a new hint on the mechanism of direct signal transduction of Wnt to Rho family GTPases. The proposed role of GEFT in the regulation of cell morphology, cell proliferation, and transformation further supports its potential role in Wnt-Fz signalling (30).
Additionally, in Xenopus laevis the related Rho guanine nucleotide exchange factor xNET1 was found to physically interact with the canonical Wnt Fz signalling component Dishevelled (which itself is a Frizzled interactor), and has been implicated in gastrulation movements (31).
Recently, a notochord expressed Rho-specific guanine nucleotide exchange factor (notoGEF) was shown to have a direct role in PCP-controlled gastrulation cell movements (Kosuke Tanegashima and Igor Dawid, NIH; personal communication).
With the exception of the Rab geranylgeranyltransferase α-subunit and β-adaptin other interactors found are only poorly characterized and cannot be linked to the Wnt/Fz1 pathway without additional experiments. There are many questions remaining from the Fz1 screen that are obviously not the main focus of this manuscript and need to be addressed systematically elsewhere. However, we will briefly introduce the most prevailing issues since similar questions will arise in any split-ubiquitin or related screen: Why did we not isolate some of the published interactors, whereas we identified a few interacting proteins that cannot be easily explained based on the known functions of Fz1. It needs to be noted that we did not identify in our screen the following previously reported interactors of Fz1: Wnt, Norrin, Dishevelled, PSD95, Kermit, GOPC, MAGI3 (see http://www.stanford.edu/∼rnusse/pathways/binding.html). Different reasons may account for this finding. First, interactor binding to the ectodomain is unlikely to work using the split-ubiquitin system, since the ubiquitin domains would meet extracellularly, where the protein degradation machinery is absent. Second, PDZ domain proteins (including, e.g. Dishevelled, Kermit) typically recognize the free C-terminal residues of their targets that are not accessible due to the fusion with the CRU domain. Third, both Wnt and Norrin are secreted proteins, localizing the Nub moiety extracellularly, whereas Cub was fused to the C-terminal, intracellular end of s-Fz1. Fourth, a low abundance of specific mRNA in the non-normalized human fetal brain library that was used may be a trivial explanation. Alternatively, misfolding of the expressed interaction domains in the yeast, or sterical interference with the ubiquitin or URA domains, may force the interaction domain into an incorrect structure. This would also result in problems in any other screening system using tagged proteins or protein fusions. Some of these issues could be overcome by increasing the number of screened yeast clones, and/or by using an alternatively constructed prey-plasmid library using N-terminal fusions of cDNA inserts to Nub.
Surprisingly, we identified as a candidate interactor an extracellular factor, the matrix protein fibronectin. A simple explanation would be that both proteins interact during their transport through the ER and Golgi compartment. In another example, the TM proteins presenilin, Pen-2, nicastrin and Aph-1 of the γ-secretase complex involved in APP cleavage also interact first in the ER to become later localized to the cytoplasmic membrane (32). Alternatively, the fibronectin clone that was selected did not represent a full length protein, suggesting that this protein fragment could have been mislocated to the interior of the cell, where it interacts with the predicted extracellular part of Fz1. It is not known at present, but cannot be excluded, that a subfraction of s-Fz1-CRU, either as a consequence of its synthetic secretion signal, the CRU domain, or as part of its biological role, may also be localized to the cytoplasm. Futher experiments will be required to solve these issues.
Clearly, the potential novel Fz1 interactors need further functional validation which can be achieved by a combination of different experimental approaches. Physical interaction may first be confirmed by a complementary biochemical approach such as co-immunoprecipitation. In vitro functional validation methods may include cell culture-based reporter gene assays specific for signalling phenomena, such as the TOP flash assay for canonical signalling (20,21). Typically, in such assays functional interactions can be monitored by measuring reporter gene expression following the activation/inactivation of signalling components. Furthermore, interactions can be validated using suitable model organisms (such as Drosophila, Xenopus or C. elegans) that encode homologs of the Wnt-Fz signal transduction cascade.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank GPC Biotech AG (Munich, Germany) for providing us with strain JD53, the bait vector, a human fetal brain cDNA-library and control plasmids. We further would like to thank N. Penski and U. Topf for technical assistance, and E. Schmidt and the anonymous reviewers for helpful suggestions. Funding to pay the Open Acess publication charges for this article was provided by the European Union 6th Framework Network of Excellence LIFESPAN (LSHG-CT-2007-036894), the BMBF (FRISYS), DFG SFB592, SFB746, and GRK1104, and the Fonds der Chemischen Industrie.
Conflict of interest statement. None declared.
REFERERENCES
- 1.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 2.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 3.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stagljar I, Korostensky C, Johnsson N, te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunnwald M, Varshavsky A, Johnsson N. Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum. Mol. Biol. Cell. 1999;10:329–344. doi: 10.1091/mbc.10.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wittke S, Lewke N, Muller S, Johnsson N. Probing the molecular environment of membrane proteins in vivo. Mol. Biol. Cell. 1999;10:2519–2530. doi: 10.1091/mbc.10.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laser H, Bongards C, Schuller J, Heck S, Johnsson N, Lehming N. A new screen for protein interactions reveals that the Saccharomyces cerevisiae high mobility group proteins Nhp6A/B are involved in the regulation of the GAL1 promoter. Proc. Natl. Acad. Sci. USA. 2000;97:13732–13737. doi: 10.1073/pnas.250400997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirnberger D, Unsin G, Schlenker S, Reichel C. A small-molecule-protein interaction system with split-ubiquitin as sensor. Chembiochem. 2006;7:936–942. doi: 10.1002/cbic.200500544. [DOI] [PubMed] [Google Scholar]
- 12.Bendixen C, Gangloff S, Rothstein R. A yeast mating-selection scheme for detection of protein-protein interactions. Nucleic Acids Res. 1994;22:1778–1779. doi: 10.1093/nar/22.9.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cagney G, Uetz P, Fields S. High-throughput screening for protein-protein interactions using two-hybrid assay. Methods Enzymol. 2000;328:3–14. doi: 10.1016/s0076-6879(00)28386-9. [DOI] [PubMed] [Google Scholar]
- 14.Serebriiskii I, Khazak V, Golemis EA. A two-hybrid dual bait system to discriminate specificity of protein interactions. J. Biol. Chem. 1999;274:17080–17087. doi: 10.1074/jbc.274.24.17080. [DOI] [PubMed] [Google Scholar]
- 15.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 16.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev. Cell. 2003;5:367–377. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 18.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HY, Malbon CC. Wnt-frizzled signaling to G-protein-coupled effectors. Cell. Mol. Life. Sci. 2004;61:69–75. doi: 10.1007/s00018-003-3165-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazit A, Yaniv A, Bafico A, Pramila T, Igarashi M, Kitajewski J, Aaronson SA. Human frizzled 1 interacts with transforming Wnts to transduce a TCF dependent transcriptional response. Oncogene. 1999;18:5959–5966. doi: 10.1038/sj.onc.1202985. [DOI] [PubMed] [Google Scholar]
- 21.Zilberberg A, Yaniv A, Gazit A. The low-density lipoprotein receptor-1, LRP1, interacts with the human frizzled-1 (HFz1) and downregulates the canonical Wnt signaling pathway. J. Biol. Chem. 2004;279:17535–17542. doi: 10.1074/jbc.M311292200. [DOI] [PubMed] [Google Scholar]
- 22.Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An Essential Yeast Gene Encoding a Homolog of Ubiquitin activating enzyme. J. Biol. Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- 23.Bickle M, Delley PA, Schmidt A, Hall MN. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 1998;17:2235–2245. doi: 10.1093/emboj/17.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gietz RD, Woods RA. Transformation of yeast by the Liac/ss carrier DNA/PEG method. Methods Enzym. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 25.Colicelli J, Birchmeier C, Michaeli T, O'Neill K, Riggs M, Wigler M. Isolation and characterization of a mammalian gene encoding a high-affinity cAMP phosphodiesterase. Proc. Natl. Acad. Sci. USA. 1989;86:3599–3603. doi: 10.1073/pnas.86.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 27.Stefan CJ, Blumer KJ. A syntaxin homolog encoded by VAM3 mediates down-regulation of a yeast G protein-coupled receptor. J. Biol. Chem. 1999;274:1835–1841. doi: 10.1074/jbc.274.3.1835. [DOI] [PubMed] [Google Scholar]
- 28.Reichel C, Johnsson N. The split-ubiquitin sensor: measuring interactions and conformational alterations of proteins in vivo. Methods Enzymol. 2005;399:757–776. doi: 10.1016/S0076-6879(05)99050-2. [DOI] [PubMed] [Google Scholar]
- 29.Lutz S, Freichel-Blomquist A, Rumenapp U, Schmidt M, Jakobs KH, Wieland T. p63RhoGEF and GEFT are Rho-specific guanine nucleotide exchange factors encoded by the same gene. Naunyn. Schmiedebergs. Arch. Pharmacol. 0000;369:540–546. doi: 10.1007/s00210-004-0926-5. [DOI] [PubMed] [Google Scholar]
- 30.Guo X, Stafford LJ, Bryan B, Xia C, Ma W, Wu X, Liu D, Songyang Z, Liu M. A Rac/Cdc42-specific exchange factor, GEFT, induces cell proliferation, transformation, and migration. J. Biol. Chem. 0000;278:13207–13215. doi: 10.1074/jbc.M208896200. [DOI] [PubMed] [Google Scholar]
- 31.Miyakoshi A, Ueno N, Kinoshita N. Rho guanine nucleotide exchange factor xNET1 implicated in gastrulation movements during Xenopus development. Differentiation. 2004;72:48–55. doi: 10.1111/j.1432-0436.2004.07201004.x. [DOI] [PubMed] [Google Scholar]
- 32.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]