Abstract
Platelet-derived growth factors are a family of mitogens and chemoattractants comprising of four ligand genes (A-, B-, C-, D-chains) implicated in many physiologic and pathophysiologic processes, including atherosclerosis, fibrosis and tumorigenesis. Our understanding of the molecular mechanisms, which regulate PDGF-C transcription remains incomplete. Transient transfection analysis, conventional and quantitative real-time PCR revealed the induction of PDGF-C transcription and mRNA expression in smooth muscle cells (SMCs) exposed to the peptide hormone angiotensin (ATII), which induces Egr-1. Occupancy of a G + C-rich element in the proximal region of the PDGF-C promoter was unaffected by ATII. Instead we discovered, using both nuclear extracts and recombinant proteins with EMSA and ChIP analyses, the existence of a second Egr-1-binding element located 500 bp upstream. ATII induction of PDGF-C transcription is mediated by the angiotensin type 1 receptor (AT1R) and Egr-1 activation through this upstream element. DNAzyme ED5 targeting Egr-1 blocked ATII-inducible PDGF-C expression. Moreover, increased PDGF-C expression after exposure to ATII depends upon the differentiation state of the SMCs. This study demonstrates the existence of this novel ATII-AT1R-Egr-1-PDGF-C axis in SMCs of neonatal origin, but not in adult SMCs, where ATII induces Egr-1 but not PDGF-C.
INTRODUCTION
Platelet-derived growth factor-C (PDGF-C) (1), like PDGF-D are the two most recently identified members of the PDGF family of growth factors, which includes the well-characterized PDGF-A and PDGF-B. PDGFs are important regulators of cell proliferation and survival in many types of mesenchymal cells including smooth muscle cells (SMCs), connective tissue cells and fibroblasts. Studies over the last two decades have implicated PDGF-A and -B in pathophysiologic processes such as atherosclerosis, restenosis, fibrosis and tumorigenesis (2,3). Since its discovery in the same year as PDGF-D, PDGF-C has been found to participate in fibrotic disease (4,5), angiogenesis (6,7), embryogenesis (8–10), palate formation (11) and platelet activation (12). The human PDGFC gene is located on chromosome 4q32, which encodes a 345 amino acid protein with a two-domain structure: an N-terminal CUB-domain and a C-terminal growth factor domain (GFD). PDGF-CC is produced as a latent factor, requiring thereby activation by proteolysis to release the GFD from the CUB domain (1). PDGF-C mRNA is expressed in most human adult tissues, with highest levels in heart, kidney and pancreas and smaller amount are found in placenta, skeletal muscle and prostate (1,5,7).
Angiotensin II (ATII), the effector peptide of the renin-angiotensin system, is involved in blood pressure control, vascular tone and growth factor induction. Additionally, ATII is a pro-atherogenic factor as it is capable of stimulating vascular SMC proliferation through the generation of complex signaling events (13) that affect the expression of pathophysiologically relevant genes such as PDGF-A (14), PDGF-B (15) and PDGF-D (16). Vascular SMCs respond to ATII multiphasic manner: within seconds, ATII can activate PLC and Ca2+ mobilization; within minutes, protein kinase C (PKC) and phospholipase D (PLD) are activated; and within hours, NADH/NADPH oxidase activity is stimulated (17). The SMC response to ATII is influenced by its differentiation state (17). For example, in both cultured newborn rat arterial medial SMCs and rat arterial neointimal SMCs, PDGF-B mRNA expression is induced by ATII, but no change in B-chain expression is observed in rat adult SMCs (15). SMC heterogeneity is a well-known feature of this cell type (18). The ‘contractile’ state, which is typical of the differentiated artery (19,20), whereas the ‘synthetic’ state is characteristic of developing or pathologic arteries and the SMCs exhibit an epithelioid shape with enhanced proliferative and migratory activity.
ATII has previously been shown to regulate PDGF-A (14), PDGF-B (15) and PDGF-D (16) transcription, however the ATII-inducible expression of each isoform may be mediated by distinct mechanisms. In the case of PDGF-D, ATII acts through reactive oxygen species (ROS), specifically H2O2 and Ets-1, whereas ATII-inducible PDGF-B expression, although not yet fully elucidated, has been shown to be dependent on Ras, ERK and c-Jun-terminal kinase (JNK) signaling. Furthermore, ATII activates the extracellular signal-related kinase (ERK) pathway to stimulate Egr-1 and PDGF-A expression via a G+C-rich region (located −76 to −47 bp) in the proximal PDGF-A promoter bearing Egr-1-binding elements (14,21). This element is strikingly similar to that in the proximal PDGF-C promoter (22). Egr-1 mediates inducible PDGF-A and PDGF-C transcription in cells exposed to FGF-2 through this proximal element (22,23). Since this element controls inducible PDGF-A expression in cells exposed to a variety of other agonists and conditions [such as ATII (14), PMA (21) and shear stress (24)], we hypothesized that this element in the PDGF-C promoter regulates altered expression in response to other stimulants. It is presently unknown whether ATII regulates the PDGF-C gene. This demonstration is important, as it would place ATII as an agonist governing the expression of every known PDGF ligand.
This study reveals the existence of a second functional Egr-1-binding element in the PDGF-C promoter located ∼500 bp upstream of the proximal G + C-rich element previously shown by us to mediate FGF-2-inducible PDGF-C transcription (22). We report that ATII induction of PDGF-C transcription is mediated by the angiotensin type 1 receptor (AT1) and Egr-1 activation through this upstream element. Moreover, we demonstrate that ATII-inducible PDGF-C expression is SMC differentiation state-dependent and that the two Egr-1-binding elements operate independently.
MATERIALS AND METHODS
Chemicals
ATII (Sigma) was dissolved in water (Baxter), aliquoted and stored at –80°C. The ATII receptor inhibitor PD123319 was purchased from Sigma and Losartan was a generous gift from Merck, Sharp & Dohme (Sydney, Australia).
Cell culture
WKY12-22 rat neonatal (pup) aortic SMCs and adult rat aortic SMCs were cultured in Waymouth's MB752/1 medium (Life Technologies, Inc.), pH 7.4, supplemented with 10 U/ml penicillin, 10 µg/ml streptomycin and 10% fetal calf serum at 37°C and 5% CO2 in an Air Jacket CO2 incubator (model 60A0100a, Thermoline). At 90% confluence, the cells were rinsed twice with phosphate-buffered saline (PBS) solution and passaged by incubating with 0.05% trypsin and 0.02% ethylenediaminetetraacetic acid (EDTA) in Hank's balanced salt solution (BioWhittaker) for 1–3 min at 37°C and then tapping firmly. The cells were resuspended in growth medium and placed in 75 cm2 flasks. The SMCs were growth-arrested in serum-free media for 24 h prior to stimulation with ATII (10−7 M). In ATII receptor inhibitor studies, WKY12-22 cells were incubated with Losartan (1 µM) or PD123319 (1 µM) for 1 h and then treated with ATII for 2 h before mRNA extraction.
Total RNA preparation and reverse transcriptase reaction
Cells were washed twice with cold PBS and total RNA was extracted with TRIzol® (Life Technologies, Inc.). cDNA was synthesized from 5 µg of RNA using the Super Script III First Strand Synthesis Kit (Invitrogen, Carlsbad, USA) as per manufacturer's instructions. cDNA was stored at −20°C.
Semi quantitative polymerase chain reaction (PCR)
PCR was performed utilizing the Platinum Tag polymerase kit. cDNA was amplified for PDGF-C, Egr-1 products. The set of primers used for rat PDGF-C was: forward, 5′ -GGC ATG AGA GAG TTG TCA CTA TAT CTG GTA-3′, and reverse, 5′ -GTC CAA GTC TAT CTG CCA TCG ATC T-3′ with denaturing temperature of 98°C for 1 min, followed by 22 cycles of 96°C for 30 s, 60°C for 30 s, 72°C for 30 s and extending at 72°C for 5 min, producing a 481 bp fragment. For rat Egr-1 amplification, primers used were: forward, 5′-GCC TTT TGC CTG TGA CAT TT-3′, and reverse, 5′-AGC CCG GAG AGG AGT AAG AG-3′, with amplification conditions of 98°C for 1 min; 94°C for 30 s, 60°C for 30 s, 72°C for 30 s repeated 30 times; 72°C for 1 min, producing a 230 bp amplicon. Rat beta-actin was used as a housekeeping gene. Primers used were, forward, 5′-AGC CAT GTA CGT AGC CAT CC-3′, and reverse, 5′-CTC TCA GCT GTG GTG GTG AA-3′. The cycling conditions were 98°C for 1 min, followed by 20 cycles at 94°C for 30 s, 58°C for 30 s, 72°C for 30 s, finishing with 72°C for 2 min, amplifying a 228 bp product. PCR products were electrophoresed on 1.5% (w/v) agarose gel in 1 × TBA-EDTA at 90 V for 55 min in TBE buffer. Products were visualized by transillumination using the Gel Doc 2000 photographic system and Quantity One version 4.1.1 software.
Quantitative real-time PCR
Real-time quantitative PCR was performed using ABI PRISM7700 Sequence Detection System in a final volume of 25 µl containing 1 µl of cDNA, 12.5 µl of SYBR Green Master Mix (Applied Biosystems), 0.5 µM of forward and reverse primers (Sigma) in DNAse-free water at the following PCR conditions: 50°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Primer sequences for rat GAPDH were (forward) 5′-ACA AGA TGG TGA AGG TCG GTG -3′; (reverse) 5′-AGA AGG CAG CCC TGG TAA CC-3′; and for rat PDGF-C (forward) 5′-CAG CAA GT GCA GCT CTC CA-3′; (reverse) 5′-GAC AAC TCT CTC ATG CCG GG -3′. Primer product size was verified on a 2% agarose/TBE gel. As an internal control and for normalization purposes, the level of mRNA expression for GAPDH, a housekeeping gene, was measured simultaneous to PDGF-C expression for each sample. The results are presented as relative amounts of PDGF-C mRNA versus the housekeeping mRNA value for the same sample. Statistical analysis was performed by paired t-test (*denotes P < 0.05) and compared with untreated controls.
Western blot analysis
WKY 12-22 SMCs were grown in 100 mm petri dishes, rendered growth quiescent by incubating in serum-free medium for 24 h and agonist was added for the times indicated or transfection was performed (pCB6-Egr-1 and CMV-Sp1). SMCs were washed twice with cold PBS and lyzed with RIPA buffer containing protease inhibitors (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 1% deoxycholate, 0.1% Triton X-100, 5 mg/ml leupeptin, 100 mM PMSF, 10% Trasylol, 0.5 M EDTA). The suspension was centrifuged for 10 min and the supernatant was collected. Protein estimation was carried out by BCA protein assay kit (Pierce, Rockford, IL, USA). Six microliters of 4 × SDS protein loading sample buffer along with 2 µl of 0.5 M DTT and 20 or 10 µg of lysates (dependent upon the experiment) were boiled for 5 min. Proteins were resolved in 10% SDS–PAGE and transferred onto Immobilon-P transfer membranes (Millipore). Membranes were blocked overnight with 5% skimmed milk in 1% PBS, 0.05% Tween-20 at 4°C. Membranes were then incubated with antibodies to Egr-1 (1:1000), Sp1 (1:1000) and beta-actin (1:30000) for 1 h at 22°C followed by incubation with their respective secondary antibody [anti-rabbit, anti-goat and anti-mouse IgG conjugated with horseradish peroxidase (HRP)] for 1 h. Membranes were incubated with chemiluminescence (Perkin Elmer Life Sciences, Shelton, CT) for 1 min and then exposed the film for 1–5 min.
PDGF plasmid constructs
The ‘Firefly’ luciferase-based reporter construct pPDGF-C-797 was described previously (22). A mutant form of this construct (pPDGF-C-797.mutEgr-1) was prepared using Quick Change site-directed mutagenesis kit (Stratagene) with the following primers (forward: 5′- CGG GAG AGC CGC TGA GCA TAA AAA TAT CGC CAG GCG CGC GCT C-3′ and reverse: 5′-GAG CGC GCG CCT GGC GAT ATT TTT ATG CTC AGC GGC TCT CCC G-3′) targeting the −560 to −518 bp region of the PDGF-C promoter (relative to ATG site). The integrity of the reporter construct was confirmed by nucleotide sequencing. CMV-Sp1 and pCB6-Egr-1 were generous gifts of Robert Tjian and Vikas Sukhatme, respectively.
Transient transfections
SMCs were seeded in 100 mm dishes and grown to 60% confluence and serum-arrested for 24 h before transfection with FuGENE6 (Roche Applied Science) at a ratio of 3 µl of FuGENE6 per microgram of DNA, with 5 µg of reporter construct pPDGF-C-797 and 0.5 µg of pRL-null, an internal control vector. In overexpression studies, SMCs were co-transfected with 1, 2 or 3 µg of either pCB6, pCB6-Egr-1, CMV-gutless, CMV-Sp1 or both pCB6 and CMV-gutless or pCB6-Egr-1 and CMV-Sp1. After incubation at 37°C for 24 h, luciferase activity was quantified using the Dual-Luciferase Reporter Assay (Promega). ‘Firefly’ luciferase activity was normalized to ‘Renilla’ data generated from pRL-null. When the effect of ED5 (0.4 µM), or its scrambled counterpart, ED5SCR was assessed, a ‘double hit’ transfection was used to maximize DNAzyme suppression of Egr-1 expression (first transfection 18 h prior to agonist stimulation of Egr-1, and the second, at the time of the stimulus). This pre-emptive delivery approach enables cellular uptake of the DNAzyme before Egr-1 is acutely induced by ATII. This routinely results in >75% uptake in our laboratory as determined by fluorescence microscopy with transfected FITC-labeled DNAzyme (25).
Preparation of nuclear extracts and electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared and EMSA was performed as previously described (22).
Statistical methods
Data was analyzed for statistical significance using Student's t-test, and considered significant when P < 0.05. Error bars represent the mean ± SE.
Chromatin immunoprecipitation analysis (ChIP)
ChIP assays were performed with human SMCs at 80% confluency using a modification of a protocol previously described (26). Briefly, cross-linking was performed by the addition of 37% (w/v) formaldehyde for 15 min, followed by glycine incorporation to a final concentration 125 mM. The cells were washed with PBS, followed by 1 ml of immunoprecipitation (IP) buffer with protease inhibitors (26). Lysates were sonicated on ice for six rounds of 15 × 1 s pulses (output level 7) and then centrifuged at 14 000 r.p.m. for 10 min. Supernatants were fractioned evenly into tubes and incubated with anti-Sp1 and anti-Egr-1 antibodies (Santa Cruz Biotechnology, Inc.) for 30 min at 22°C, then at 4°C for 15 min, followed by centrifugation for 10 min at full speed. Prewashed Protein-A and -G beads were added and the slurry was rotated for 45 min at 4°C. The beads were washed five times with IP buffer without inhibitors prior the addition of 100 µl of 10% Chelex-100 and boiled for 10 min. Samples were digested with proteinase K (100 µg/ml) for 30 min at 55°C, then boiled for 10 min centrifuged and supernatant collected. DNA was purified by phenol–chloroform extraction and ethanol precipitation. DNA was dissolved in water and used for PCR reaction in 1 mM MgCl2, 0.1 mM dNTPs, 0.1 µM of primers and 1 U Platinum Taq Polymerase (Invitrogen). Cycling conditions were: 98°C for 1 min, 40 cycles of 98°C for 30 s, 58°C for 30 s and 72°C for 30 s followed by 72°C for 2 min. PDGF-C promoter containing the putative Egr-1-binding site was amplified (330 bp) with the following primers: forward 5′-TAG AGG TGT TCC GTG GAA GG-3′ and reverse 5′-TTG TCC CCT CCC CTT CTC TA-3′.
RESULTS AND DISCUSSION
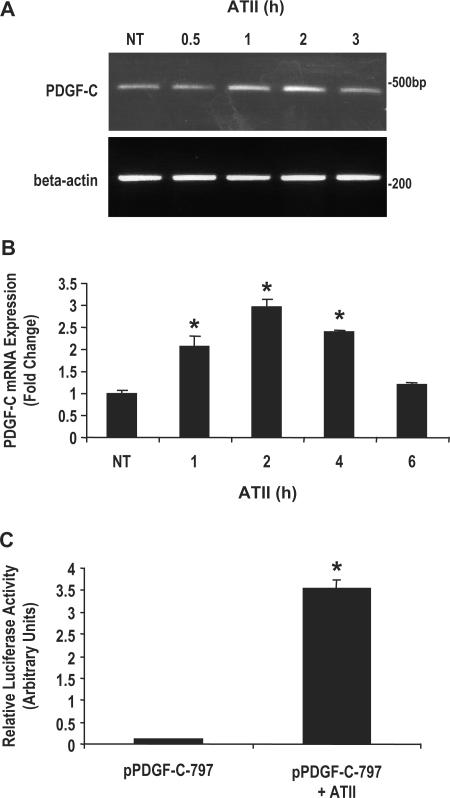
ATII has previously been shown to induce PDGF-A (14), PDGF-B (15) and PDGF-D (16) transcription in SMCs. To begin to investigate whether ATII can influence the expression of PDGF-C, we isolated mRNA from rat neonatal aortic SMCs (WKY12-22 cells) exposed to ATII (10−7 M) for various times. Conventional RT-PCR revealed that PDGF-C was basally expressed and was transiently induced by ATII, maximally at 2 h (Figure 1A). Quantitative real-time PCR demonstrated a 2-fold increase in PDGF-C mRNA levels at 1 h and maximal 3-fold induction by 2 h (Figure 1B). To establish that PDGF-C is under the transcriptional control of ATII, transient transfection was performed in WKY12-22 SMCs with pPDGF-C-797, a ‘Firefly’ luciferase-based reporter construct bearing 797 bp of the PDGF-C promoter, prior to exposure to ATII (10−7 M). Treatment with ATII induced PDGF-C promoter activity (Figure 1C).
Figure 1.
ATII induces PDGF-C expression and promoter activation in neonatal SMCs. Total RNA was isolated from neonatal (WKY12-22) SMCs cells treated with or without ATII (10−7 M) for various times. (A) Conventional RT-PCR or (B) real-time PCR was performed for PDGF-C as described in the Materials and Methods section. The graph represents PDGF-C expression relative to the expression of GAPDH to correct for cDNA concentration between samples. (C) Transient transfection analysis was performed in WKY12-22 SMCs transfected with 5 µg of pPDGF-C-797 and treated with or without ATII (10−7 M) for 24 h. The data are representative of three or more independent determinations. Error bars represent the mean ± SE. Asterisk (*) denotes P < 0.05 compared to control.
ATII induction of PDGF-A expression is mediated via ERK-dependent Egr-1 binding to a G + C-rich region of the PDGF-A promoter at overlapping Egr-1/Sp1-binding sites (−76 to −47 bp) (14). To determine whether ATII induces Egr-1 binding to a similar region in the proximal PDGF-C promoter, we performed EMSA using nuclear extracts from WKY12-22 SMCs treated with or without ATII (10−7 M) for 1 h and a 32P-labeled double-stranded oligonucleotide probe [32P-Oligo C(−35/−1)] previously shown to bind FGF-2-inducible Egr-1 (22). Four nucleoprotein complexes formed, three major (Na, Nc, Nd) and one minor (Nb) (Figure 2A). Supershift analysis using antibodies targeting Sp1 revealed protein complex Na contains Sp1 (Figure 2A). However, to our surprise, antibodies to Egr-1 (or ATF4) did not alter the binding profile, despite the induction of Egr-1 mRNA and protein expression by ATII (Figure 2B and C). As a control for the antibody, we performed EMSA using nuclear extracts from WKY12-22 SMCs treated with FGF-2 (25 ng/ml) together with 32P-Oligo C(−35/−1). As previously obtained using extracts from FGF-2-treated rat SMCs (22), six nucleoprotein complexes (N1–N6) formed using 32P-Oligo C(−35/−1), including one complex containing supershiftable Egr-1 protein (N3) (Figure 2D). These results indicate that although ATII induces Egr-1 in WKY12-22 SMCs, Egr-1 does not interact with the proximal G + C-rich region of the PDGF-C promoter.
Figure 2.
ATII does not induce binding to the Egr-1/Sp1-binding site (−35/−1 bp) in the PDGF-C promoter. (A) EMSA was performed using nuclear extracts from WKY12-22 SMCs treated with or without ATII (10−7 M) for 1 h and 32P-Oligo C(−35/−1). Supershift analysis using antibodies targeting Sp1 demonstrate that nucleoprotein complex Na contains Sp1. Antibodies directed towards Egr-1 reveal that none of the complexes contain Egr-1 protein. (B) RT-PCR demonstrates that Egr-1 is inducibly expressed in WKY12-22 SMCs exposed to ATII (10−7 M) for the indicated times. (C) Western blot analysis demonstrates that ATII increases Egr-1 protein expression in WKY12-22 SMCs. (D) EMSA using 32P-Oligo C(−35/−1) and nuclear extracts from WKY12-22 SMCs treated with FGF-2 (25 µg/ml) for 1 h. Nucleoprotein complexes N1 and N2 contain Sp1, and N3 contains Egr-1 as demonstrated by antibody-based band elimination.
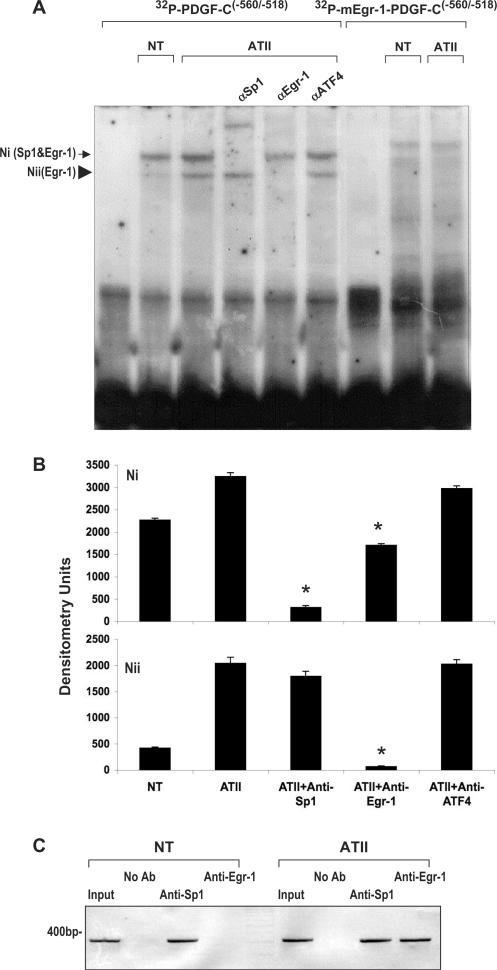
After further examination of the PDGF-C promoter, we identified a second Egr-1-binding element (5′-CGCCCC CGC-3′) upstream (at position −543/−535) (Figure 3). This putative binding motif resides within the PDGF-C luciferase-based reporter construct used in Figure 1C, which was activated by ATII. To establish whether ATII induces binding to this putative element, we performed EMSA using nuclear extracts from WKY12-22 SMCs treated with or without ATII (10−7 M) for 1 h, and a 32P-labeled oligonucleotide probe [32P-PDGF-C(−560/−518)] whose sequence spans −543/−535 (Figure 4A). Two complexes, one more intense (Ni) than the other (Nii), formed when extracts from untreated SMCs were incubated with 32P-PDGF-C(−560/−518) (Figure 4A, lane 2 and Figure 4B). Complex Nii was induced upon ATII treatment (Figure 4A, lane 3 and Figure 4B), and antibody-based complex elimination analysis revealed it to contain Egr-1 protein (Figure 4A, lane 5 and Figure 4B). Complex Ni, whose intensity was also increased by ATII, contains both Sp1 and Egr-1, with Sp1 the predominant occupant. Sp1 antibodies supershifted the complex, whereas Egr-1 antibodies reduced its intensity (Figure 4A, lanes 4 and 5 and Figure 4B). In support of these data, Sp1 and Egr-1 are known to physically interact (27). In contrast, antibodies to ATF4 had no significant effect on the binding profile of either complex (Figure 4A, lane 6 and Figure 4B). These results indicate that Sp1 interacts basally, and Egr-1 inducibly, with site −560/−518 in the PDGF-C promoter. Moreover, consistent with EMSA, ChIP analysis revealed that under basal conditions Sp1 is bound to the authentic PDGF-C promoter (Figure 4C), and that both Egr-1 and Sp1 are bound to the promoter upon exposure to ATII (Figure 4C). The identity of the amplicon as a fragment of the PDGF-C promoter was confirmed by nucleotide sequencing (data not shown).
Figure 3.
Proximal region of the PDGF-C promoter. A consensus Egr-1-binding element was found −543 to −535 bp in the PDGF-C promoter upstream from the G + C-rich element (located −35 to −1 bp) previously found to contain an overlapping Egr-1/Sp1 site (22). The Egr-1 consensus element is denoted by bold type and underline (contained with EMSA probe, PDGF-C(−560/−518) denoted by italics), and the G + C-rich element is defined by bold type and underline. The relative base pairs are represented by numbers in bold type and the curved arrow represents the putative transcriptional start site (as defined by UCSC Genome Database).
Figure 4.
ATII induces Egr-1 binding to the PDGF-C promoter. (A) EMSA was performed using nuclear extracts from neonatal SMCs treated with or without ATII (10−7 M) for 1 h and 32P-PDGF-C(−560/−518) or 32P-mEgr-1-PDGF-C(−560/−518). Two nucleoprotein complexes (Ni and Nii) formed after incubation of 32P-PDGF-C(−560/−518) with nuclear extracts from cells untreated or treated with ATII. Supershift/band elimination analysis using antibodies to Sp1 and Egr-1 reveal complex Ni contains Sp1 and Egr-1, and complex Nii to contain only Egr-1. Antibodies to ATF4 did not promote shift in any complex. In reactions containing nuclear extracts from untreated or ATII-treated SMCs and 32P-mEgr-1-PDGF-C(−560/−518) (mutant form of the oligonucleotide in which consensus Egr-1 site mutated), neither complex formed. (B) Assessment of band intensities in the 32P-PDGF-C/ATII groups by scanning densitometry. Complex densities for Ni and Nii are indicated. NT denotes non-ATII-treated. (C) ChIP studies show that under basal conditions Sp1 is bound to the authentic PDGF-C promoter, and that both Egr-1 and Sp1 are bound to the promoter upon exposure to ATII. Asterisk (*) denotes P < 0.01 relative to ATII.
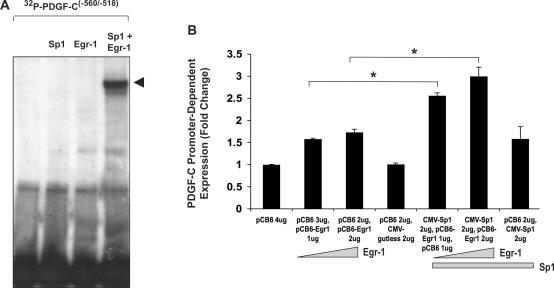
In order to confirm the interaction of Sp1 and Egr-1 with this region of the PDGF-C promoter (−560/−518 bp) and to determine whether Sp1 and Egr-1 interact competitively or cooperatively for binding to the overlapping site, EMSA was performed using human recombinant Sp1 and Egr-1 and 32P-PDGF-C(−560/−518) (Figure 5A). In binding reactions containing either recombinant Sp1 or Egr-1, a nucleoprotein complex was not observed. However, a single complex was detected when both Sp1 and Egr-1 were incubated with the oligonucleotide 32P-PDGF-C(−560/−518) (Figure 5A). This data suggests that, individually, Sp1 and Egr-1 are unable to bind this novel element and that presence of the two transcription factors is required for the formation of a single nucleoprotein complex. Interestingly, only one major complex formed when 32P-PDGF-C(−560/−518) was incubated with recombinant Egr-1 and Sp1 (Figure 5A, arrow), which compares with complex Ni (but not Nii) containing both nuclear Sp1 and Egr-1 (Figure 4A and B). To test the hypothesis that Egr-1 and Sp1 co-operatively transactivate the PDGF-C promoter, we performed transient transfection analysis with Egr-1 and Sp1 expression vectors. Transfection with low amounts of Egr-1 results in modest activation of the PDGF-C promoter (Figure 5B, compare columns 2 and 3 to 1), but in the presence of Sp1, the same amounts of Egr-1 produced co-operative activation (Figure 5B, compare columns 5 and 6 to 4).
Figure 5.
Egr-1 and Sp1 cooperatively bind and transactivate the PDGF-C promoter. (A) EMSA was performed using 32P-PDGF-C(−560/−518) and human recombinant Egr-1 (100 ng), human recombinant Sp1 (100 ng) or both human recombinant Egr-1 and Sp1 (100 ng each). A major nucleoprotein complex formed (arrow) only when recombinant Egr-1 and recombinant Sp1 were both incubated with 32P-PDGF-C(−560/−518). The data are representative of three or more independent determinations. (B) WKY12-22 SMCs were transfected with 0, 1 and 2 µg of pCB6-Egr-1 (made up to 4 µg total with its backbone pCB6+), or identical amounts of pCB6-Egr-1 with 2 µg CMV-Sp1 (made up to 4 µg total with its backbone pCB6+), all with pPDGF-C-797 (5 µg) and pRL-null (0.5 µg, to normalize for transfection efficiency). ‘Firefly’ luciferase activity was normalized to ‘Renilla’. The data are representative of three or more independent determinations. Error bars represent the mean ± SE. Asterisk (*) denotes P < 0.001.
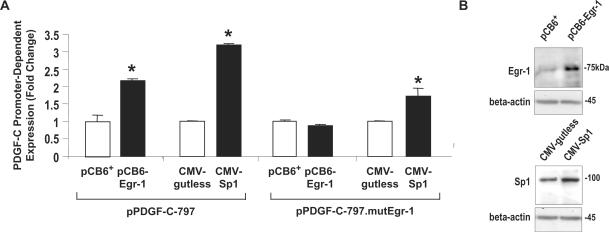
To determine the nucleotides critical for Egr-1/Sp1 binding to this region of the PDGF-C promoter, a transverse mutation was introduced into 32P-PDGF-C(−560/−518), changing the putative Egr-1 element located −543/−535 bp from 5′-CGCCCCCGC-3′ to 5′-ATAAAAATA-3′. EMSA was performed using this mutant form of the oligonucleotide [32P-mEgr1-PDGF-C(−560/−518)] with nuclear extracts from WKY12-22 SMCs treated with or without ATII (10−7 M) for 1 h (Figure 4). Neither Ni nor Nii formed under basal or ATII-stimulated conditions, demonstrating that the integrity of −543/−535 bp is critical for the interaction of Egr-1 and Sp1 with this region of the PDGF-C promoter.
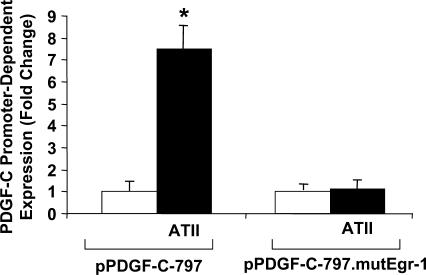
To establish the functional significance of this novel element in the PDGF-C promoter, the same transverse mutation was introduced into pPDGF-C-797 (pPDGF-C-797.mutEgr-1) and transient transfection was performed with both wild type and mutant PDGF-C promoter reporter construct. Transfectants were stimulated with or without ATII (10−7 M) for 24 h and PDGF-C promoter activation was analyzed using a luminometer. ATII activated the wild-type PDGF-C promoter by over 7-fold (Figure 6, consistent with Figure 1C), however, no significant difference was observed in PDGF-C promoter activity in cells transfected with pPDGF-C-797.mutEgr-1 treated with ATII or left untreated (Figure 6).
Figure 6.
Novel Egr-1 element in the PDGF-C promoter mediates ATII-inducible PDGF-C expression. Transient transfection analysis was performed on WKY12-22 SMCs transfected with pPDGF-C-797 (5 µg), or pPDGF-C-797.mutEgr-1 (5 µg) and pRL-null (0.5 µg, to normalize for transfection efficiency) and treated with or without ATII (10−7 M) for 24 h. ‘Firefly’ luciferase activity was normalized to ‘Renilla’. The data are representative of three or more independent determinations. Error bars represent the mean ± SE. Asterisk (*) denotes P < 0.05.
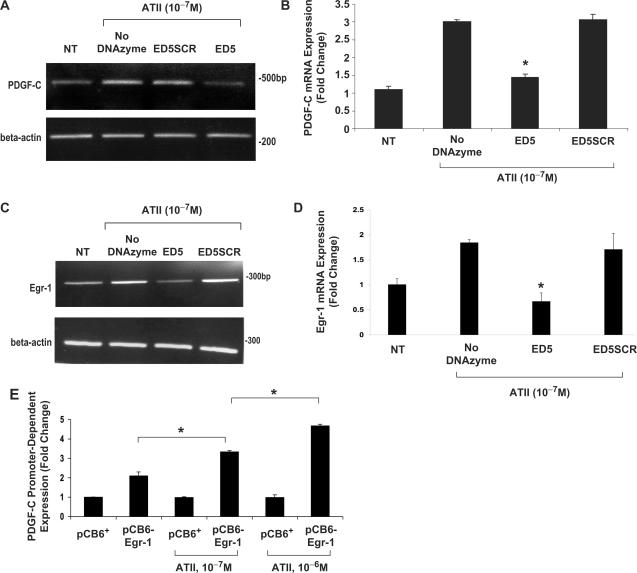
The preceding data demonstrate that Egr-1 inducibly binds to a novel upstream element in the PDGF-C promoter. To directly establish that Egr-1 mediates ATII-inducible PDGF-C expression, we assessed PDGF-C mRNA levels in neonatal SMCs that had been transfected with the Egr-1 DNAzyme, ED5 (25) or its arm-scrambled counterpart, ED5SCR, and stimulated with ATII for 2 h. DNAzymes are sequence-specific ‘gene knockdown’ agents that bind to and cleave the mRNA of their target gene (28,29). We have previously used DNAzymes to inhibit gene expression in SMCs in vitro and neointima formation in rat carotid arteries after balloon injury (25) and in pig coronary arteries following stenting (30). Using both conventional RT-PCR (Figure 7A) and real-time PCR (Figure 7B), our findings demonstrate that ED5, but not ED5SCR, perturbed the stimulatory effect of ATII on PDGF-C mRNA expression. To support these observations, we evaluated the effect of ED5 on levels of Egr-1 either by conventional or quantitative real-time PCR. ED5 inhibited the induction of Egr-1 by ATII, whereas ED5SCR had no effect (Figure 7C and D). These findings together demonstrate the dependence of ATII induction of PDGF-C expression on Egr-1. To demonstrate whether exogenous Egr-1 enhances the induction of PDGF-C by ATII, we co-transfected pCB6-Egr-1 or pCB6+ with pPDGF-C-797 and luciferase activity was assessed with or without exposure of the cells to ATII. We observed that a fixed amount of Egr-1 potentiated ATII-inducible PDGF-C transcription in an ATII dose-dependent manner (Figure 7E). Taken together, these data demonstrate that Egr-1 is critical for ATII-inducible PDGF-C expression.
Figure 7.
DNAzymes targeting Egr-1 block ATII-inducible PDGF-C expression. WKY12-22 SMCs were transfected twice with or without the DNAzyme ED5 (0.4 µM) or its scrambled counterpart ED5SCR (0.4 µM) and stimulated with ATII (10−7 M) for 2 h. (A) Conventional RT-PCR and (B) real-time PCR demonstrates the reliance on Egr-1 for ATII-inducible PDGF-C expression. The real-time data represents PDGF-C expression relative to the expression of GAPDH. (C) Conventional RT-PCR and (D) real-time PCR shows that ED5 (0.4 µM) inhibits ATII-inducible Egr-1 expression. The real-time data represents Egr-1 expression relative to the expression of GAPDH. (E) Growth-quiescent WKY12-22 SMCs were transfected with 2 µg pCB6+ or pCB6-Egr-1, together with pPDGF-C-797 (5 µg) and pRL-null (0.5 µg, to normalize for transfection efficiency), then exposed to ATII (10−7 or 10–6 M) for 24 h. ‘Firefly’ luciferase activity was normalized to ‘Renilla’. The data are representative of three or more independent determinations. Error bars represent the mean ± SE. Asterisk (*) denotes P < 0.05.
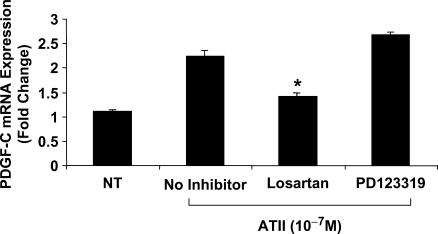
We next investigated the ATII receptor subtype mediating ATII-inducible PDGF-C expression using pharmacologic inhibitors. mRNA was isolated from WKY12-22 SMCs treated with Losartan (14), a competitive inhibitor of AT1R, or the AT2R antagonist PD123319 (14) for 1 h (both at 1 µM) prior to stimulation with ATII for 2 h. mRNA was reverse-transcribed to cDNA and real-time PCR was performed for PDGF-C. Losartan blocked ATII-inducible PDGF-C expression demonstrating that ATII employs the AT1 receptor to effect changes in PDGF-C gene expression (Figure 8).
Figure 8.
ATII-inducible PDGF-C expression acts through the AT1 receptor. WKY12-22 SMCs cells were treated with the AT1R inhibitor Losartan (1μM) or AT2R inhibitor, PD123319 (1 µM) for 1 h prior to stimulation with ATII (10−7 M) for 2 h. mRNA was isolated and real-time PCR was performed simultaneously for PDGF-C and GAPDH, a housekeeping gene. The graph represents PDGF-C expression relative to the expression of GAPDH to correct for cDNA concentration between samples. The data are representative of three or more independent determinations. Error bars represent the mean ± SE. Asterisk (*) denotes P < 0.05.
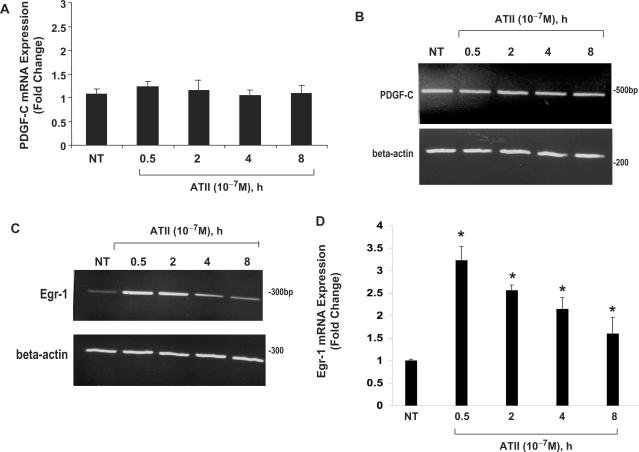
SMCs are well known for their phenotypically heterogeneity, particularly during development and arterial remodeling in atherosclerosis and restenosis in adults. Previous studies examining ATII-inducible PDGF-B expression revealed SMC subtype-specific responsiveness (15). ATII regulation of PDGF-A (14) or PDGF-D (16) expression has only been studied in adult rat SMCs. To determine whether ATII regulation of PDGF-C expression is SMC differentiation state-dependent, we isolated mRNA from adult rat aortic SMCs exposed to ATII (10−7 M) for various times and assessed PDGF-C levels (Figure 9A and B). In contrast to results obtained using neonatal SMCs (Figures 1A–C, 5B, 6, 7A, B, E and 8), ATII did not influence PDGF-C mRNA expression in adult SMCs (Figure 9A and B), indicating that PDGF-C expression in SMC is differentiation state-specific. We explored whether the inability of ATII to influence PDGF-C expression in adult SMCs is due to the lack of induction of Egr-1 by ATII in this cell type. ATII stimulated Egr-1 mRNA expression in adult SMCs (Figure 9C and D) as it did together with protein in neonatal cells (Figure 2B and C). These observations therefore indicate Egr-1-dependent and -independent transcriptional control of PDGF-C between these SMC subtypes. To determine the functional importance of this upstream element in relation to Egr-1 and Sp1, transient co-transfection analysis was performed in WKY12-22 SMCs using wild-type (pPDGF-C-797) and mutant (pPDGF-C-797.mutEgr1) PDGF-C promoter together with expression vectors for Egr-1 and Sp1. Both transcription factors, as expected, activated the wild-type PDGF-C promoter (Figure 10A). However, whereas mutation of the distal element (−543/−535) blocked Egr-1 activation of the PDGF-C promoter (Figure 10A), disruption of this element did not alter Sp1's ability to activate the promoter (Figure 10A). That Egr-1 and Sp1 were overexpressed in this context was demonstrated by immunoblot analysis (Figure 10B). These findings demonstrate that Sp1's transactivation of the PDGF-C promoter is not mediated by −543/−535, although Sp1 binds this element (Figure 4A–C). SMCs of distinct differentiation state respond to changes in the extracellular environment through different pathways (31). The present study demonstrates that ATII-inducible PDGF-C expression in SMCs is dissimilarly regulated in cells of neonatal and adult origin, by providing evidence for a novel ATII-AT1R-Egr-1-PDGF-C axis in SMCs of neonatal origin, but not in adult SMCs, where ATII induces Egr-1 but not PDGF-C. This study also demonstrates that ATII is a positive regulator of PDGF-C gene expression in neonatal SMCs, and for the first time, defines ATII as an agonist of all four known PDGF ligand chains (14–16). Using nuclear extracts and recombinant proteins, we discovered this novel upstream element cooperatively binds both Egr-1 and Sp1, and mediates ATII-inducible PDGF-C expression. DNAzymes targeting Egr-1 mRNA established that Egr-1 plays a critical role in ATII-inducible PDGF-C expression. Additionally, by using pharmacologic inhibitors of each ATII receptor, AT1R and AT2R, we demonstrated that ATII acts through AT1R to induce PDGF-C expression. Element −543/−535 plays a critical role in both Egr-1- and ATII-activation of the PDGF-C promoter. Thus, although Egr-1 is a key regulator of ATII-inducible PDGF-C expression, this present study invites further investigation to rationalize differences in the transcriptional regulation of PDGF-C between SMC subtypes. This information will help develop a more detailed understanding of SMC heterogeneity.
Figure 9.
ATII does not induce PDGF-C expression in rat adult SMCs. mRNA was isolated from adult rat aortic SMCs treated with or without ATII (10−7 M) for 0.5, 2, 4 or 8 h. (A) Real-time PCR and (B) conventional RT-PCR were performed simultaneously for PDGF-C and GAPDH, a housekeeping gene, as described in the Materials and Methods section. The graph represents PDGF-C expression relative to the expression of GAPDH to correct for cDNA concentration between samples. The data are representative of three or more independent determinations. (C) Conventional RT-PCR or (D) real-time PCR was performed simultaneously for Egr-1 as described in the Materials and Methods section. The graph represents Egr-1 expression relative to the expression of beta-actin to correct for cDNA concentration between samples. The data are representative of three or more independent determinations. Error bars represent the mean ± SE. Asterisk (*) denotes P < 0.05
Figure 10.
Mutation of the upstream element abrogates Egr-1, but not Sp1, activation of the PDGF-C promoter in neonatal SMCs. (A) Transient co-transfection analysis was performed in WKY12-22 SMCs with pPDGF-C-797 (5 µg) or pPDGF-C-797.mutEgr-1 (5 µg) and pCB6-Egr-1 (2 µg), pCB6-Egr-1 (2 µg), CMV-gutless (2 µg) or CMV-Sp1 (2 µg). The data are representative of three or more independent determinations. Error bars represent the mean ± SE. Asterisk (*) denotes P < 0.05. (B) Western blot analysis demonstrates that Egr-1 and Sp1 levels are increased in SMCs transfected with the respective expression vectors.
ACKNOWLEDGEMENTS
The authors are indebted to Fernando Santiago for technical assistance and other members of the Gene Targeting and Transcription Laboratory at UNSW. Funding to pay the Open Access publication charges for this article was provided by the Centre for Vascular Research. This work was supported by grants from the NHMRC, ARC and NHF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Li X, Ponten A, Aase K, Karlsson L, Abramsson A, Uutela M, Backstrom G, Hellstrom M, Bostrom H, et al. PDGF-C is a new protease-activated ligand for the PDGF alpha-receptor. Nat. Cell Biol. 2000;2:302–309. doi: 10.1038/35010579. [DOI] [PubMed] [Google Scholar]
- 2.Raines EW, Bowen-Pope DF, Ross R. In: Handbook of Experimental Pharmacology: Peptide Growth Factors and their Receptors. Sporn MB, Roberts, editors. Berlin: Springer; 1990. pp. 173–262. [Google Scholar]
- 3.Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 4.Campbell JS, Hughes SD, Gilbertson DG, Palmer TE, Holdren MS, Haran AC, Odell MM, Bauer RL, Ren HP, et al. Platelet-derived growth factor C induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc. Natl Acad. Sci. USA. 2005;102:3389–3394. doi: 10.1073/pnas.0409722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponten A, Li X, Thoren P, Aase K, Sjoblom T, Ostman A, Eriksson U. Transgenic overexpression of platelet-derived growth factor-C in the mouse heart induces cardiac fibrosis, hypertrophy, and dilated cardiomyopathy. Am. J. Pathol. 2003;163:673–682. doi: 10.1016/S0002-9440(10)63694-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao R, Brakenhielm E, Li X, Pietras K, Widenfalk J, Ostman A, Eriksson U, Cao Y. Angiogenesis stimulated by PDGF-CC, a novel member in the PDGF family, involves activation of PDGFR-alphaalpha and -alphabeta receptors. FASEB J. 2002;16:1575–1583. doi: 10.1096/fj.02-0319com. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Tjwa M, Moons L, Fons P, Noel A, Ny A, Zhou JM, Lennartsson J, Li HA, et al. Revascularization of ischemic tissues by PDGF-CC via effects on endothelial cells and their progenitors. J. Clin. Invest. 2005;115:118–127. doi: 10.1172/JCI19189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eitner F, Ostendorf T, Kretzler M, Cohen CD, Eriksson U, Grone HJ, Floege J. PDGF-C expression in the developing and normal adult human kidney and in glomerular diseases. J. Am. Soc. Nephrol. 2003;14:1145–1153. doi: 10.1097/01.asn.0000062964.75006.a8. [DOI] [PubMed] [Google Scholar]
- 9.Aase K, Abramsson A, Karlsson L, Betsholtz C, Eriksson U. Expression analysis of PDGF-C in adult and developing mouse tissues. Mech. Dev. 2002;110:187–191. doi: 10.1016/s0925-4773(01)00560-3. [DOI] [PubMed] [Google Scholar]
- 10.Hamada T, Ui-Tei K, Imaki J, Takahashi F, Onodera H, Mishima T, Miyata Y. The expression of SCDGF/PDGF-C/fallotein and SCDGF-B/PDGF-D in the rat central nervous system. Mech. Dev. 2002;112:161–164. doi: 10.1016/s0925-4773(01)00625-6. [DOI] [PubMed] [Google Scholar]
- 11.Ding H, Wu X, Bostrom H, Kim I, Wong N, Tsoi B, O’Rourke M, Koh GY, Soriano P, et al. A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling. Nat. Genet. 2004;36:1111–1116. doi: 10.1038/ng1415. [DOI] [PubMed] [Google Scholar]
- 12.Fang L, Yan Y, Komuves LG, Yonkovich S, Sullivan CM, Stringer B, Galbraith S, Lokker NA, Hwang SS, et al. PDGF C is a selective alpha platelet-derived growth factor receptor agonist that is highly expressed in platelet alpha granules and vascular smooth muscle. Arterioscler. Thromb. Vasc. Biol. 2004;24:787–792. doi: 10.1161/01.atv.0000120785.82268.8b. [DOI] [PubMed] [Google Scholar]
- 13.Berk BC, Corson MA. Angiotensin II signal transduction in vascular smooth muscle: role of tyrosine kinases. Circ. Res. 1997;80:607–616. doi: 10.1161/01.res.80.5.607. [DOI] [PubMed] [Google Scholar]
- 14.Day FL, Rafty LA, Chesterman CN, Khachigian LM. Angiotensin II (ATII)-inducible platelet-derived growth factor A-chain gene expression is p42/44 extracellular signal-regulated kinase-1/2 and Egr-1 dependent and modulated via the ATII type 1 but not type 2 receptor – induction by ATII antagonized by nitric oxide. J. Biol. Chem. 1999;274:23726–23733. doi: 10.1074/jbc.274.34.23726. [DOI] [PubMed] [Google Scholar]
- 15.Deguchi J-O, Masatoshi M, Nakaoka T, Collins T, Takuwa Y. Angiotensin II stimulates platelet-derived growth factor-B chain expression in newborn rat vascular smooth muscle cells through Ras, extracellular signal-regulated protein kinase, and c-Jun N-terminal protein kinase mechanisms. Circ. Res. 1999;85:565–574. doi: 10.1161/01.res.85.7.565. [DOI] [PubMed] [Google Scholar]
- 16.Liu MY, Eyries M, Zhang C, Santiago FS, Khachigian LM. Inducible platelet-derived growth factor D-chain expression by angiotensin II and hydrogen peroxide involves transcriptional regulation by Ets-1 and Sp1. Blood. 2006;107:2322–2329. doi: 10.1182/blood-2005-06-2377. [DOI] [PubMed] [Google Scholar]
- 17.Griendling KK, Ushio-Fukai M, Lassegue B, Alexander RW. Angiotensin II signaling in vascular smooth muscle. New concepts. Hypertension. 1997;29:366–373. doi: 10.1161/01.hyp.29.1.366. [DOI] [PubMed] [Google Scholar]
- 18.Hao H, Gabbiani G, Bochaton-Piallat ML. Arterial smooth muscle cell heterogeneity: implications for atherosclerosis and restenosis development. Arterioscler. Thromb. Vasc. Biol. 2003;23:1510–1520. doi: 10.1161/01.ATV.0000090130.85752.ED. [DOI] [PubMed] [Google Scholar]
- 19.Lemire JM, Covin CW, White S, Giachelli CM, Schwartz SM. Characterization of cloned aortic smooth muscle cells from young rats. Am. J. Pathol. 1994;144:1068–1081. [PMC free article] [PubMed] [Google Scholar]
- 20.Rafty LA, Khachigian LM. Zinc finger transcription factors mediate high constitutive PDGF-B expression in smooth muscle cells derived from aortae of newborn rats. J. Biol. Chem. 1998;273:5758–5764. doi: 10.1074/jbc.273.10.5758. [DOI] [PubMed] [Google Scholar]
- 21.Khachigian LM, Williams AJ, Collins T. Interplay of Sp1 and Egr-1 in the proximal PDGF-A promoter in cultured vascular endothelial cells. J. Biol. Chem. 1995;270:27679–27686. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 22.Midgley VC, Khachigian LM. Fibroblast growth factor-2 induction of platelet-derived growth factor-C chain transcription in vascular smooth muscle cells is ERK-dependent but not JNK-dependent and mediated by Egr-1. J. Biol. Chem. 2004;279:40289–40295. doi: 10.1074/jbc.M406063200. [DOI] [PubMed] [Google Scholar]
- 23.Delbridge GJ, Khachigian LM. FGF-1-induced PDGF A-chain gene expression in vascular endothelial cells involves transcriptional activation by Egr-1. Circ. Res. 1997;81:282–288. doi: 10.1161/01.res.81.2.282. [DOI] [PubMed] [Google Scholar]
- 24.Khachigian LM, Anderson KA, Halnon NJ, Resnick N, Gimbrone M.A., Jr,, Collins T. Egr-1 is activated in endothelial cells exposed to fluid shear stress and interacts with a novel shear-stress response element in the PDGF A-chain promoter. Arterioscl. Thromb. Vasc. Biol. 1997;17:2280–2286. doi: 10.1161/01.atv.17.10.2280. [DOI] [PubMed] [Google Scholar]
- 25.Santiago FS, Lowe HC, Kavurma MM, Chesterman CN, Baker A, Atkins DG, Khachigian LM. New DNA enzyme targeting Egr-1 mRNA inhibits vascular smooth muscle proliferation and regrowth factor injury. Nat. Med. 1999;5:1264–1269. doi: 10.1038/15215. [DOI] [PubMed] [Google Scholar]
- 26.Nelson JD, Denisenko O, Sova P, Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic Acids Res. 2006;34:e2. doi: 10.1093/nar/gnj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin JX, Leonard WJ. The immediate-early gene product Egr-1 regulates the human interleukin-2 receptor beta-chain promoter through noncanonical Egr and Sp1 binding sites. Mol. Cell. Biol. 1997;17:3714–3722. doi: 10.1128/mcb.17.7.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khachigian LM. Catalytic DNA as therapeutic agents and molecular tools to dissect biological function. J. Clin. Invest. 2000;106:1189–1195. doi: 10.1172/JCI11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santiago FS, Khachigian LM. Nucleic acid based strategies as potential therpeautic tools: mechanistic considerations and implications to restenosis. J. Mol. Med. 2001;79:695–706. doi: 10.1007/s001090100272. [DOI] [PubMed] [Google Scholar]
- 30.Lowe HC, Fahmy RG, Kavurma MM, Baker A, Chesterman CN, Khachigian LM. Catalytic oligodeoxynucleotides define a key regulatory role for early growth response factor-1 in the porcine model of coronary in-stent restenosis. Circ. Res. 2001;89:670–677. doi: 10.1161/hh2001.097867. [DOI] [PubMed] [Google Scholar]
- 31.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]