Abstract
Following DNA damage, mRNA levels decrease, reflecting a coordinated interaction of the DNA repair, transcription and RNA processing machineries. In this study, we provide evidence that transcription and polyadenylation of mRNA precursors are both affected in vivo by UV treatment. We next show that the polyadenylation factor CstF, plays a direct role in the DNA damage response. Cells with reduced levels of CstF display decreased viability following UV treatment, reduced ability to ubiquitinate RNA polymerase II (RNAP II), and defects in repair of DNA damage. Furthermore, we show that CstF, RNAP II and BARD1 are all found at sites of repaired DNA. Our results indicate that CstF plays an active role in the response to DNA damage, providing a link between transcription-coupled RNA processing and DNA repair.
INTRODUCTION
The cellular response to DNA damage involves changes in the properties of a number of nuclear proteins, resulting in coordinated control of gene expression and DNA repair. One example is provided by the transient decrease in mRNA levels following UV irradiation (1,2). Although the mechanism underlying this response is still unresolved, it has been suggested that the UV-induced inhibition of transcription, reflecting turnover of the RNAP II largest subunit (RNAP II LS), is responsible for the decrease (3). This indeed is likely a significant part of the mechanism. However, those studies have not considered the important effect of RNA processing on mRNA levels. Indeed, it has been shown that processing of mRNA precursors, and specifically 3′ end formation, is also affected by DNA damage. Our previous data indicated that mRNA polyadenylation in cell extracts is strongly but transiently inhibited following treatment of cells with DNA damage-inducing agents (4). These results suggested a functional interaction between RNA processing and DNA repair.
The poly(A) tail found on almost all eukaryotic mRNAs plays important roles in regulation of mRNA stability, translation and RNA transport from the nucleus (5–7). The polyadenylation reaction consists of an endonucleolytic cleavage followed by synthesis of the poly(A) tail (reviewed in 8–10). While a relatively simple signal sequence in the mRNA precursor is required for the reaction, a surprisingly large number of protein factors are necessary for 3′ processing. Cleavage stimulation factor (CstF) is one of the essential 3′ processing factors. Genetically modified chicken B cells deficient in CstF-64, a CstF subunit, undergo cell cycle arrest and apoptotic death (11). Another subunit, CstF-50, has been shown to interact with the C-terminal domain of the RNAP II LS (CTD), likely facilitating the RNAP II-mediated activation of 3′ processing (12,13). The stimulatory role of RNAP II in polyadenylation highlights the link between RNA processing and transcription. This link is supported by a variety of chromatin immunoprecipiation experiments documenting the association of polyadenylation factors with transcribed genes (e.g. 14–16).
As part of our efforts to characterize links between mRNA 3′ processing and other nuclear events, we uncovered and characterized an association between CstF and the BRCA1/BARD1 tumor suppressor complex. We showed that this association was mediated by a direct interaction between CstF-50 and BARD1, and inhibits 3′ processing in vitro (17). The complex is increased transiently in concentration following DNA damage-inducing treatments, and results in inhibition of 3′ processing in extracts from the treated cells (4). It has also been shown that DNA damage-induced BARD1 phosphorylation is critical for inhibition of polyadenylation and RNAP II LS degradation (18). After UV treatment, a fraction of RNAP II LS is phosphorylated, ubiquitinated and degraded by the proteasome (reviewed by 19,20). While UV-induced turnover of RNAP IIA, the form engaged at promoters, occurs by phosphorylation and conversion to RNAP IIO, turnover of RNAP IIO, which functions in elongation, occurs by ubiquitination and degradation (21–26). We have shown that degradation of RNAP IIO in fact contributes to inhibition of 3′ processing in response to DNA damage (27), suggesting the existence of another, possibly redundant, mechanism to explain the inhibitory effect of UV irradiation. Significantly, both BRCA1 and BARD1 are necessary for ubiquitination of RNAP II LS and its turnover in response to UV treatment (27,28).
UV-induced turnover of RNAP II is part of the transcription-coupled repair (TCR) response (reviewed by 19,20). TCR is a pathway that operates on certain types of DNA damage found in the transcribed strand of expressed genes. Accumulated evidence suggests that the blockage of elongating RNAP II at sites of DNA damage is an early event that initiates TCR. Levels of mRNA are transiently decreased, and normal recovery depends on TCR (1,2,29,30). One of the earliest indications of the existence of TCR was the key observation that when mammalian cells are exposed to UV light, RNA synthesis resumes before any significant amount of UV-induced damage is removed from the bulk of the genome by global genome repair (31). One reason for this may be that TCR serves to repair transcription-blocking lesions and, therefore, to facilitate a rapid recovery of transcription. Transcription complexes can be extremely stable when they are stalled at endogenous pause sites or at sites of damage (32). It has been suggested that RNAP II stalled at sites of DNA damage could respond in either of two ways. If the lesion is repaired rapidly, RNAP II re-engages and continues transcription, but if the lesion persists, RNAP II is ubiquitinated and degraded (26,33,34). Stalling and/or degradation of RNAP II have another potential function: to prevent transcription across sites of DNA repair and thereby prevent formation of potentially deleterious proteins. However, this could result in release of prematurely terminated transcripts, and inhibition of the 3′ processing machinery would then function to prevent polyadenylation and stabilization of such RNAs.
In this article, we describe studies extending the links between 3′ processing and DNA repair. We first provide evidence that UV treatment in fact affects both transcription and polyadenylation of nascent mRNAs in vivo. We then show that depletion of CstF in DT40 cells enhances sensitivity to UV treatment, reduces UV-induced ubiquitination of RNAP II and, significantly, causes a delay in TCR or a related pathway. Extending these results, we provide evidence that following UV treatment BRCA1/BARD1, RNAP II and CstF associate at sites of repaired DNA. Taken together, our results indicate that CstF plays active roles not only in 3′ processing but also in DNA repair, providing a link between transcription-coupled RNA processing and DNA repair.
MATERIALS AND METHODS
Tissue culture methods and DNA damaging agents
HeLa cells were cultured in Dulbecco's modified Eagles medium (DMEM)-10% fetal bovine serum (FBS), 10 mg/ml Penicillin–Streptomycin. Ninety percent confluent cultures of transiently transfected cells were exposed to UV and harvested after the times indicated. UV doses (20 Jm−2) were delivered in two pulses using a Stratalinker (Stratagene). Prior to pulsing, medium was removed and replaced immediately after treatment. DT40-64 cells (11) were cultured in RPMI 1640 media supplemented with 10% FBS, 1% chicken serum and Hygromycin B 100 µg/ml. DT40 cells were grown in the presence or absence of 10 µg/ml of tet as indicated.
Plasmids expressing actin
The human β-actin cDNA was amplified from a cDNA library using a sense primer which contains the coding sequence of HA-flu epitope next to a BamH I site, while the antisense primer contains another BamH I site (sense primer: 5′-ATGGATCCATGTACCCATACGATGTTCCAGATTACGCTCTTATGGATGATGATATCGCC-3′ and antisense primer: 5′-GAGGATCCCTAGAAGCATTTGCGGTGGA-3). The HA-actin was introduced into the BamH I site of either pcDNA 3.1(+) vector (Invitrogen) or the pAPSV-Zeo vector (35). pcDNA 3.1(+) vector has the CMV promoter and the BGH polyadenylation signal. pAPSV-Zeo contains the chicken β-actin promoter, the SV40 late poly(A) signal and the zeocin resistance gene. To synthesize the ssDNA probe used in the Southern blot analysis, the first 200 bp sequence from human actin gene was subcloned into the pBluescript KS(−) vector (Stratagene). This sequence was amplified from the actin cDNA using the forward primer (BF, 5′-ATGGATCCATGGATGATGATATCGCCGC-3′), which contains a BamH I recognition sequence, and the reverse primer (BR, 5′-GCGAATTCCAGGGTGAGGATGCCTCTCT-3′), which contains an EcoR I site. The amplified sequence was introduced into the multiple cloning site of pBluescript generating the pBprobe vector. The plasmid constructions were cloned into Escherichia coli DH5α for amplification and purified using Qiagen kits.
HeLa cell transfection with pcDNA3.1 (+)/HA actin
HeLa cells were grown in a 10-cm plate in complete DMEM at 50% confluence. The cells were then transfected with pcDNA3.1 (+)/HA-actin DNA using 30 μl TransIT-HeLa and 20 μl of Monster reagents (Mirus) according to the manufacturer's protocol. After culturing for 24 h, the cells were exposed to UV (20 Jm−2) and harvested 30 min, 2, 5 or 10 h later. Alternatively, HeLa cells were transfected with pcDNA3.1 (+)/HA-actin plasmid exposed to 900 J/m2 UV-C radiation to induce CPDs. Cells were harvested after 8 and 24 h after transfection. Cells were used for either RNA purification or western blot analysis.
Analysis of RNA by RT–PCR and real-time quantitative RT-PCR (RT-qPCR)
RNA was purified from cells transfected with the pcDNA 3.1(+)/HA-β-actin vector using the RNeasy (Qiagen). The purified RNA was used as a template to synthesize cDNA using oligo d(T) primers and MMLV reverse transcriptase (Promega) according to the manufacturer's protocol. PCR was performed in a 50 μl volume using the RT product and primers against GAPDH and HA-actin with Pfu DNA polymerase (Stratagene) according to the standard protocol provided by the manufacturer. PCR fragments were amplified from RT products using the forward primer HA (5′-CCTACGATGTTCCAGATTACGC-3′) and the following reverse primers: A (5′-GCCCATGTCGTCCCAGTTGGTG-3′), B (5′-GGGAAGCTGTAGCCGCGCTCGG-3′), C (5′-CCGAGGATCCCTAGAAGCATTTT-3′) and a modified oligo(dT) primer/adapter (36). Equal volumes of the PCR products were run on a 1% agarose gel and visualized by ethidium bromide staining. Real-time qPCR was performed for endogenous transcripts using the RT product described before and commercially available primers. Reactions were performed with the Applied Biosystems TaqMan Gene Expression Assays on 7500 RT-PCR detection system. The reaction was performed in triplicate in 25 μl reaction volume containing cDNA [2–20 ng of poly(A)+ RNA purified as described before], 300 nM primers, 12.5 μl Taqman MGB Gene expression system (Applied Biosystems) and water to complete. The RT products obtained from UV untreated cells were used as endogenous control.
NE preparation and immunoblotting
NEs were prepared from harvested cells essentially as described (4). Cells were lysed by douncing in 4 ml of 10 mM Tris pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT) and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). Lysates were centrifuged for 10 min at 6000g, and pellets were resuspended in 20 mM Tris pH 7.9, 1.5 mM MgCl2, 25% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF and 0.3 M NaCl. Preparations were rocked for 30 min at 4°C and centrifuged for 15 min at 10 000g. Supernatants were quick frozen and stored at −80°C. Sixty microgram of each NE was analyzed by immunoblotting with antibodies against RNAP II (H5, Covance), CSA (Santa Cruz H-266), CSB (Santa Cruz H-300), actin (Sigma A2066) and BARD1 (Santa Cruz, H-300).
ssDNA probe synthesis
32P-labeled single-strand DNA probes were synthesized according to the procedure described by Ruven et al. (37). Pure pBprobe plasmid was used as template in an asymmetric polymerase chain reaction (30 cycles at 94°C for 1 min, 54°C for 2 min, 72°C for 3 min and a last cycle at 72°C for 7 min). A total 48 ng of pBprobe vector digested with either BamH I or EcoR I was used in the reaction as templates with 20 pmol of either BR or BF primer, respectively.
Host cell recovery assays
HeLa cells were transfected with UV-damaged or non-damaged pcDNA3.1 (+)/HA-actin. To induce photolesions, plasmid DNA was treated with 900 J/m2 UV-C radiation with a Stratalinker (Stratagene) at room temperature. HeLa cells were grown in a 10-cm plate in complete DMEM at 50% confluence. Transfections were done with TransIT-HeLa Monster reagent (Mirus) according to the manufacturer's protocol. After transfection, cells were incubated at 37°C for the times specified. Cells were collected; RNA was purified and analyzed by RT-PCR as described before. Alternatively, DT40–64 cells grown in the presence or absence of tet were transfected with 10 µg of UV-damaged or non-damaged pAP-actin plasmids. Plasmids were damaged as described earlier. Twenty-four hours before the transfection, 2.4 107–3.4 107 DT40–64 cells were transferred to 10 ml fresh media with or without 10 µg/ml tet. Transfections were done with TransIT-LT1 reagent (Mirus) according to the manufacturer's protocol. After transfection, cells were incubated in fresh media with or without 10 µg/ml tet at 37°C for the times indicated. Cells were collected, lysed and plasmid DNA was purified using a Qiaprep Spin Miniprep kit (Qiagen). The presence of plasmid in those preparations was confirmed by PCR with the primers used to construct the pAP-actin plasmid. The rest of the plasmid preparation was subjected to Kpn I linearization. Half of the linearized plasmid preparation was treated with T4 endo V (Epicentre) and the other half was mock treated for 1 : 30 h. The material was electrophoresed on 1% agarose gel at 92 V. Southern blot transfer to a charged nylon membrane and hybridization with 32P ssDNA probe were done as described (38). The membranes were exposed to a Phosphor Imager Screen (Kodak) and scanned in a Molecular Dynamics Typhoon 9410 (General Electrics). Images were analyzed with the software ImageJ 1.37a (Wayne Rasband, NIH, USA).
Chromatin immunoprecipitation-type assays
We performed ChIP assays using a modification of previously published methods (39). Ninety percent confluent cultures of HeLa cells were exposed to UV (two pulses of 50 J/m2) using a Stratalinker (Stratagene), incubated with BrdU (10 μM) and FrdU (fluorodeoxyuridine, 1 μM) to label the repaired DNA, and then the cells were cross-linked with formaldehyde at the stated times. After formaldehyde treatment, NEs were prepared from HeLa cells as described (4) and samples were sonicated to produce soluble chromatin in the presence of proteinase inhibitors (Sigma, P2714). To obtain DNA fragmentation of average length of 2000 base pairs, sonications were done two times for 20 s each. Samples were then pre-cleared by treatment with protein-G Sepharose 4B beads (Sigma). DNA-protein complexes were immunoprecipitated by incubation with BrdU monoclonal antibody (Covance) coupled to blocked protein-G Sepharose 4B beads. Immunoprecipitations were carried out for 3 h at 4°C in 150 µl of sonication buffer (10 mM Tris pH 8.0, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 0.5 mM PMSF, 1 X protease inhibitor cocktail). Washing was with sonication buffer. Crosslinks were reversed by boiling the samples for 30 min. The protein complex bound to repair DNA was analyzed by western blot.
RESULTS
UV treatment affects transcription/3′ processing in vivo
The DNA damage response requires coordination between the gene expression and DNA repair machineries. We showed previously that following DNA damage, 3′ end formation is strongly but transiently inhibited in extracts from the damaged cells as a result of BRCA1/BARD1/CstF complex formation (4) and of proteasome-mediated degradation of RNAP II (27). We provided evidence that this complex (which we dub ‘the CBB checkpoint’) was active between 2 and 8 h post-UV, and proposed that this reflects a mechanism to prevent polyadenylation of prematurely terminated RNAs that could arise at sites of DNA damage (4).
An important question is whether DNA damage in fact induces formation of such transcripts, and if so whether it inhibits polyadenylation to prevent their accumulation in vivo. As such RNAs might be heterogeneous, unstable and therefore difficult to observe directly, we developed an assay to determine the effect of UV-induced DNA lesions on transcription and 3′ processing of RNAs produced from a reporter gene (Figure 1A). Briefly, the assay involves transient transfection of HeLa cells with a reporter plasmid, followed by analysis of expression and polyadenylation of transcripts by oligo(dT) selection and RT-PCR. Importantly, a collection of primers was designed with the potential to detect RNAs with 3′ ends located throughout the length of the reporter gene. Twenty-four hours after transfection, cells were exposed to UV light and poly(A)+ RNA was purified at different times after UV treatment (0 min, 30 min, 2, 5 and 10 h). Expression of the HA-β-actin reporter gene was analyzed by RT-PCR of the oligo(dT)-selected RNA using an oligo(dT) primer for the RT reaction and PCR was done with a common forward primer that anneals to the HA region of the tagged gene and reverse primers derived from different parts of the β-actin gene (Figure 1A). As a common forward primer was used, we expected to obtain differently sized PCR products in each sample. Elevated intensities of the shorter species would indicate the presence of prematurely polyadenylated forms of HA-β-actin RNA. For comparison, we also analyzed by RT-PCR transcripts from an endogenous gene (GAPDH).
Figure 1.
Effect of DNA damage on reporter gene expression is transfected in HeLa cells. (A) Diagram of the RT-PCR analysis. The reporter gene and the PCR primers are described in the diagram. (B) Analysis of the RT-PCR products generated from RNA samples obtained at different times after UV treatment from HeLa cells transfected with the pcDNA3.1 (+)/HA actin vector. RNA samples were purified from equal numbers of cells. Equivalent amounts of RNA, quantified by optical density at 260 nm, from each sample were used in the RT-PCR reactions, which were run for 20 cycles. Equal volumes of the PCR reactions were analyzed in agarose gels. The relative density of each PCR band was determined by the Image J program. (C) Poly(A)+ RNA was isolated from cells after UV exposure, and RT-PCR (top panel) and real-time RT-PCR (bottom panel) were carried out to measure expression of GAPDH. The RT products from cells not treated with UV were used as endogenous control. Data shown are the mean ± SEM from three independent experiments. (D) Preparation of damaged pcDNA3.1 (+)/HA actin vector. Plasmids were irradiated with the UV doses indicated (lanes 4–15), treated or not treated with T4 endo V and electrophoresed on a 1% neutral agarose gel. A plasmid digested with EcoRI was used as an indicator of the linear form (6.6 kb, lane 2). Molecular size markers are also included. Positions of the relaxed (R), linear (L) and supercoiled (S) forms are indicated. (E) Analysis of the RT-PCR products obtained from RNA samples of transfected HeLa cells with plasmids treated or not treated with UV. RNA samples were purified from equivalent numbers of cells. RT-PCR reactions were performed and analyzed as in (C). (F) Analysis of truncated/polyadenylated transcripts obtained from RNA samples of transfected HeLa cells with plasmids treated (D) or not treated (ND) with UV. The poly(A) transcripts were purified as in (E) and cDNA was synthesized using an oligo(dT) primer followed by a PCR amplification with the common forward/HA primer together with the oligo(dT) primer/adapter. PCR products were analyzed by agarose gel electrophoresis.
Our results suggest that UV treatment can indeed lead to transient formation of truncated, polyadenylated RNAs. A significant decrease in accumulation of full-length HA-β-actin mRNA (HA-C) was detected as early as 30 min after UV treatment (Figure 1B, top panel). Accumulation of this species reached its lowest levels at 2 h, and high levels were fully restored by 10 h. Similar changes in expression of the endogenous GAPDH gene were also observed by both RT-PCR (Figure 1C, top) and RT-qPCR assays (Figure 1C, bottom). A 2-fold decrease in GAPDH poly(A)+ mRNA was detected by RT-qPCR 30 min after UV treatment, reaching the lowest level (8-fold decrease) at 2 h. We also observed a transient decrease in the shorter RT-PCR products (HA-A and HA-B). Importantly however, the relative decrease at 30 min. was significantly less for these shorter products (∼80% of the untreated control; quantitation at right in Figure 1B) than that of the full-length mRNA (∼40% of the control), suggesting that some of these products arose from prematurely terminated, polyadenylated transcripts. At 2 h the longer products were almost undetectable, consistent with the idea that their production was blocked by activation of the CBB checkpoint. At the 5 and 10 h-time points, accumulation of the shorter products paralleled that of the full-length mRNA, suggesting that these RT-PCR products were derived predominantly from full-length HA-β-actin mRNA.
Our results indicated that levels of the endogenous GAPDH mRNA decrease somewhat after UV treatment. It is noteworthy that several studies have addressed the accuracy of traditional reference or housekeeping genes, such as GAPDH, as control for error between samples (40,41). Those studies have shown that GAPDH can show significant variation in RNA expression in different biological systems and in different conditions. Indeed, Kartasova et al. (42) described a change in the expression of GAPDH mRNA after UV irradiation.
If indeed the shorter RNAs described before resulted from DNA damage-induced premature termination and erroneous polyadenylation that occurs prior to activation of the CBB checkpoint, then accumulation of these RNAs would be expected to be greater if activation of the checkpoint could be avoided. To test this, we utilized a variation of the host cell reactivation (HCR) assay, which is a transfection-based approach in which cells repair damage localized to exogenous DNA (43,44). In our experimental approach, the HCR assay allowed us to study the effect of UV-induced DNA lesions on transcription and 3′ processing of RNAs produced from a reporter gene. Briefly, the assay involves transfection of HeLa cells with a UV-treated reporter plasmid followed by analysis of expression and polyadenylation of transcripts by oligo(dT) selection and RT-PCR exactly as before.
Photoproducts were first introduced into the reporter plasmid expressing HA-β-actin (Figure 1D) by exposing the plasmid to UV light (254 nm) for various doses. We determined the number of lesions per molecule by treating the plasmids with a damage-specific endonuclease, T4 endonuclease V (T4 endo V), that specifically cleaves DNA at sites of cyclobutane pyrimidine dimers (CPDs, 45). The extent of damage was determined by comparison of the intensity of bands corresponding to relaxed, linear and supercoiled forms of the plasmid (45). As T4 endo V introduces nicks where CPDs remain, we used the appearance of full–length linear forms as indicative of treatments inducing ∼1 lesion per plasmid. Based on the results shown in Figure 1B, we used UV doses of 900 Jm−2 per microgram of DNA to induce ≤3 lesions per plasmid.
HeLa cells were transiently transfected with the UV-treated or untreated plasmids. Preliminary results (data not shown) suggested that expression of HA-actin from either damaged or non-damaged plasmids was readily detectable and similar 24 h after transfection. Therefore, poly(A)+ RNA was purified at a very early time (8 h) and at 24 h after transfection and expression of the HA-β-actin gene analyzed by oligo(dT) selection and RT-PCR as before. For comparison, we also analyzed by RT-PCR transcripts from the endogenous GAPDH gene for each sample. As the cells were not exposed to UV, no changes in expression of GAPDH mRNA were observed (Figure 1E). The results of our RT-PCR analysis support the idea that DNA damage can indeed lead to aberrant 3′ processing. Expression of full-length mRNA (HA-C) was readily detected 8 h after transfection with the control plasmid (Figure 1E). Significantly, however, cells transfected with the damaged plasmid did not produce full-length HA-β-actin mRNA after 8 h, but shorter polyadenylated forms (HA-A and HA-B) were readily detected. These products likely reflect prematurely terminated and incorrectly polyadenylated transcripts arising specifically from the damaged plasmid. At 24 h, the ratios of full-length (HA-C) to shorter products (HA-A and HA-B) were similar in both cases, suggesting that most if not all of the shorter products at this time were amplified from the full-length cDNA, consistent with the idea that the damaged plasmid had been fully repaired and essentially all transcripts were now full length. It is important to point out that as the cells were not exposed to UV light, UV-induced inhibition of 3′ processing would not be expected to occur.
Although our results suggest that UV treatment leads to transient formation of truncated, polyadenylated RNAs, we could not rule out the possibility that the results could have been generated by blocking of the RT reaction by UV-induced lesions in the mRNA, having a stronger effect on longer mRNAs that are more likely to acquire those lesions. It is also possible that UV treatment induced destabilization and decay of cellular mRNAs, and that the TCR complexes stabilize the intermediates. This could generate the RT-PCR patterns observed for both the exogenous (HA-A>HA-B>HA-C) and the endogenous genes (GAPDH).
To determine whether UV treatment indeed generates truncated polyadenylated forms, we mapped the poly(A) sites in HA-actin transcripts present in HeLa cells transiently transfected with reporter plasmids under different conditions (samples from Figure 1B and E). For this analysis, we used an oligo(dT) primer for the synthesis of cDNA followed by PCR amplification with the common forward/HA primer together with the oligo(dT) primer adapter. The results with the samples from HeLa cells transiently transfected with the UV-treated or untreated plasmids are shown in Figure 1F. Similar results were obtained with samples from HeLa cells transiently transfected with the reporter plasmid and then exposed to UV treatment (samples from Figure 1B). The electrophoretic analysis of the undigested RT-PCR products showed broad bands larger than the HA-C full-length product for both the damaged and non-damaged samples (over 1163 nt, lanes 3–5). These bands, which encompassed fragments from 1163 nt to approximately 1263 nt, could have resulted from the different size of fully extended polyadenylated forms. The electrophoretic analysis also showed bands larger than the HA-B product only for the UV-damaged samples (over 628 nt, lanes 2–3). The appearance of this novel band only in the UV-damaged samples indicates that truncated polyadenylated forms could arise in vivo. The identity of the RT-PCR products shown in Figure 1F was confirmed by mapping these fragments using restriction digestion (data not shown). The pattern of bands in the UV-damaged samples provide evidence that prematurely terminated, aberrantly polyadenylated RNAs can arise from transcription of damaged DNA templates in the absence of the CBB checkpoint.
CstF functions in recovery from UV-treatment, in UV-induced ubiquitination of RNAP II and in TCR
The data presented above and previously has provided evidence that DNA damage can influence 3′ end formation, via interactions involving CstF. We next wished to investigate whether the converse might be true; i.e. whether CstF might function in DNA repair. To this end, we used genetically modified chicken DT40 cells in which the only source of CstF-64 is from a tet-repressible transgene (DT40–64, 11). These cells allow tet-dependent depletion of CstF-64, which destabilizes the entire CstF complex. CstF-64 became undetectable in DT40–64 cells treated with 10 μg/ml of tet for 48 h as measured by western blot (Figure 2B–C). While the cells were still viable at this time, they stopped growing after 3–4 days in tet-containing medium and started to die shortly thereafter (data not shown; 11). We used these cells first to determine whether the presence or absence of CstF affects the ability of the cells to recover following UV treatment. DT40–64 cells were treated with 10 μg/ml of tet for 48 h, exposed to UV light (20 Jm−2), and cell viability determined after 5 h, first measured simply by the appearance of cell death, which appeared significantly enhanced in the tet-treated cells compared to untreated controls (Figure 2A). Cell viability was quantitated by trypan blue staining, which showed that the cells with reduced levels of CstF indeed displayed enhanced sensitivity to UV treatment.
Figure 2.
Cells with reduced levels of CstF show enhanced sensitivity to UV and reduced ability to ubiquitinate RNAP II. (A) DT40–64 cells were grown in presence or absence of tet (10 μg/ml) for 48 h. Then the cells were exposed to UV irradiation (20 Jm−2) and allowed to recover for the times indicated. Viable cells are seen as bright dots by contrast microscopy, while non-viable cells are seen as condensed dark dots. The percentage of viable cells after UV treatment, determined by trypan blue staining, is also shown. (B) DT40–64 and DT40-ASF cells were treated with tet and UV as in A. One hundred microgram of each cell extract was analyzed by immunoblotting with antibodies against CstF-64, CSA, CSB and actin. The percentage of viable cells after UV and tet treatment is also shown. (C-D) CstF-64, RNAP IIO and actin protein levels in cell extracts were monitored by western blot of DT40–64 cells treated or not treated with UV. As indicated in the figure, cells were incubated (C) or not (D) with the proteasome inhibitor MG132 immediately after UV treatment.
We next wished to ask whether the heightened UV sensitivity was specific to CstF depletion, or might be a characteristic of DT40 cells poised to undergo cell death. To address this, DT40-ASF cells, which express the essential splicing factor ASF/SF2 under tet control (46), were analyzed as before (Figure 2B). Significantly, DT40-ASF cells did not show enhanced sensitivity to UV, supporting the idea that CstF has a specific role in recovery from exposure to UV. As CstF is a general polyadenylation factor that functions in the 3′ processing of many if not most mRNA precursors, it is conceivable in principle that the effect of CstF on DNA repair might be indirect. However, Takagaki and Manley (11) showed that depletion of CstF in DT40–64 cells did not detectably affect the steady-state levels of actin mRNA and several other less abundant transcripts, at least over time courses such as employed in our experiments. To determine whether this might also apply to mRNAs encoding proteins involved in DNA repair, we examined levels of CSA and CSB proteins, which are involved in TCR (47,48), following depletion of CstF and UV treatment. Tet-dependent depletion of CstF for 24 h and UV treatment did not significantly affect the expression levels of CSA and CSB (Figure 2B), supporting the idea that any effect of the depletion of CstF on DNA repair (see subsequently) is in fact due to a direct role of this RNA processing factor in this response. As the samples analyzed were obtained 2 h after UV treatment, our western blot analysis did not show the CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway that occurs 3–4 h after UV irradiation (48).
UV-induced degradation of RNAP IIO LS, initiated by BRCA1/BARD1 ubiquitination, contributes to inhibition of 3′ processing (27). We next wished to determine whether CstF functions in DNA damage-induced ubiquitination of RNAP II, and again used DT40–64 cells. Ubiquitination and degradation of RNAP IIO LS was examined by western blot using antibodies directed against the Ser 2-phosphorylated CTD epitope of RNAP II (H5), which reflects elongating RNAP IIO. The proteasome inhibitor MG132 was added to the cells immediately after UV exposure to prevent degradation of RNAP II, and cell extracts were prepared at different times after UV/MG132 treatment. With degradation blocked, we were able to observe apparent ubiquitinated forms of RNAP IIO in cells expressing normal levels of CstF (Figure 2C, lanes 2 and 3). Importantly, cells with reduced expression levels of CstF showed lower accumulation of ubiquitinated RNAP IIO. This was apparent after 24 h tet treatment and essentially complete after 48 h. In the absence of MG132, UV-induced degradation of RNAP IIO was observed in the presence of CstF, but strikingly, turnover was reduced (24 h) or completely blocked (48 h) when CstF was depleted (Figure 2D). Taken together these results indicate that CstF is required for UV-induced proteasomal degradation of RNAP II.
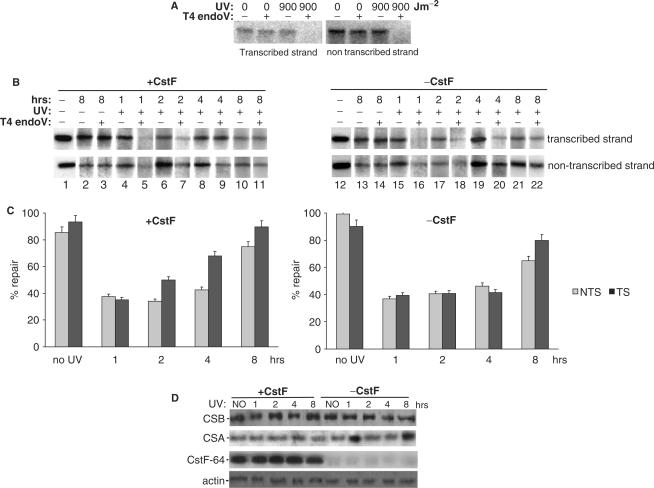
We next wished to determine the effect of reduced levels of CstF on DNA repair, and specifically on TCR. Although there is little direct information regarding the removal of lesions from reporter genes, and most of the published studies consider the relative level of expression of the reporter gene to be an indicator of the repair capacity of the host cell (43,44,47,48), we decided to use the HCR assay and DT40–64 cells to examine the possibility that CstF plays a direct role in the removal of such lesions. First, we prepared a plasmid expressing HA-β-actin using the chicken expression vector pAPSV-Zeo (pAP-actin). UV doses up to 900 Jm−2 were used to induce photoproducts. Damaged plasmids were linearized, treated or not treated with T4 endo V and analyzed by Southern blot with single strand-specific DNA probes produced by asymmetric PCR of a fragment of the β-actin gene. The autoradiogram (Figure 3A) shows that UV doses of 900 Jm−2 generated enough damage to be readily detected by this assay.
Figure 3.
CstF plays a role in DNA repair. (A) Southern blot analysis of the pAP-actin plasmids. Autoradiogram illustrating CPD damage of the plasmid. The plasmids were treated or not treated with UV, digested with Kpn I, treated or mock-treated with T4 endo V, electrophoresed on 1% agarose gel and transferred to nitrocellulose membrane. The transcribed and non transcribed strands of the cloned fragment of the HA-actin gene were detected by sequential hybridization with the indicated single strand-specific DNA probes. (B) Strand-specific DNA repair of CPDs from damaged pAP-actin plasmid purified 1, 2, 4 or 8 h after transfection from DT40-64 cells containing or lacking CstF. Purified plasmids from transfected cells were treated as in A. Plasmids not treated with UV were used as control. The frequency of induction of CPD and their rate of removal were determined by the appearance of the full-length restriction fragments in the T4 endo V-treated and mock-treated samples upon quantitation by Phosphorimager analysis. The images were analyzed by software Image J. (C) Graph of repair profile for removal of CPDs in cells containing or lacking CstF. The graph shows the mean values ± standard deviations for the percentage of repair of photolesions from the transcribed and non-transcribed strand. The values were obtained from Phosphorimager quantification of three independent experiments. The values of the autoradiogram shown in panel B were also included in the graph. (D) DT40–64 cells were treated with tet and transfected with damaged or untreated plasmid DNA. One hundred microgram of each cell extract was analyzed by immunoblotting with antibodies against CstF-64, CSA, CSB and actin.
DT40–64 cells were next grown in the presence or absence of tet for 24 h. Cells were then transfected with damaged or untreated plasmid DNA and grown with tet for another 1, 2, 4 and 8 h to allow repair to take place before isolation of plasmid DNA (Materials and Methods). To measure strand-specific repair, we linearized the purified plasmids with a restriction enzyme, then treated half the sample with T4 endo V to digest fragments that contain unrepaired lesions. For loading/purification control purposes, equal amounts of untreated and T4 endo V-treated plasmid DNA purified at each time point were analyzed by Southern blot with single strand-specific DNA probes as before (Figure 3B). The proportion of lesions that were repaired at different time-points was calculated by comparing T4 endo V-treated DNA with untreated DNA of each time point and quantifying the signal of full-length fragment by densitometry (Figure 3C). Cells containing CstF repaired the transcribed strand significantly more efficiently than the non-transcribed strand at the 2 and 4 h-time points (Figure 3B, lanes 6–7 and 8–9), indicative of activation of a TCR pathway. This preference was considerably reduced by 8 h, when both strands were repaired completely. Strikingly, the preference for the transcribed strand was not detected in the cells lacking CstF (Figure 3B, lanes 17–18 and 19–20); the rate of repair of the transcribed and non-transcribed strands was essentially the same. The presence or absence of CstF did not affect the rate of repair of the non-transcribed strand at any time point.
To determine whether the depletion of CstF followed by transfection with plasmid DNA affects the expression of proteins involved in DNA repair in DT40–64 cells, we examined levels of CSA and CSB proteins under these conditions (47,48). As shown in Figure 2B, neither tet-dependent depletion of CstF for 48 h nor transfection with damaged/untreated plasmid DNA affected the levels of CSA and CSB (Figure 3D), supporting the idea that any effect of CstF depletion on DNA repair in fact reflects a direct role of CstF in this response. As the cells were not exposed to UV treatment, our western blot analysis did not show the CSA-dependent degradation of CSB (48).
Supporting our results, Link and colleagues (49) showed that at earlier times the frequencies for photoproducts removal from the DHFR gene in CHO hamster cells are similar on the transcribed and non-transcribed strand. They also found that 62% of the lesions were repaired in the transcribed strand by 8 h compared with 43% in the non-transcribed strand, a 1.44 ratio that favors the repair of the transcribed strand. Although our study in DT40 cells containing CstF showed a similar difference between both strands (68% repair for transcribed versus 43% repair for non-transcribed), this difference was reached at an earlier time (by 4 h after UV treatment) than in the previous study. This differences could reflect the biological systems used in the two studies and/or technical reasons. In fact, several studies have indicated that it is difficult to make unambiguous conclusions from HCR studies, because the results are dependent upon many factors (43,50), such as the vector, the reporter gene, the promoter, the host cell and transfection methods. For example, the differences between our study and that of Link and colleagues (49) could reflect different transcriptional levels between reporter plasmids and endogenous genes, and/or differences in the repair mechanism between different cell types and organisms. In spite of these complications, it is apparent from our results that CstF plays a role in the TCR pathway, or in a related pathway, in DT40 cells.
RNAP II, CstF and BARD1 associate at sites of DNA repair
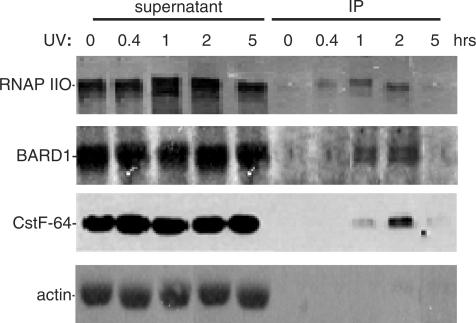
The data presented before provides evidence that CstF participates in ubiquitination of RNAP II in response to DNA damage, and in the TCR response itself. Based on this and on our previous data establishing an interaction between CstF and BRCA1/BARD1, we next wished to determine whether RNAP II, CstF and BRCA1/BARD1 all associate at sites of DNA damage. To this end, a variation of the chromatin immunoprecipitation (ChIP) assay (39,51) was employed. This method has been used largely to study chromatin associated factors, but has also been valuable in analysis of proteins apparently associated with elongating RNAP II (e.g. 52–54). In our experiments, BrdU was added to HeLa cells immediately after exposure to UV light to label repaired DNA. Cells were crosslinked with formaldehyde at different times after UV exposure. As ubiquitination of RNAP II occurs within 15 min of exposing cells to UV and persists for about 8–12 h (55), we performed our analysis in a period between 0–5 h after UV treatment. Extracts of these cells were prepared and following sonication DNA–protein complexes were IPed by incubation with an anti-BrdU monoclonal antibody. Following reversal of crosslinks, rather than analyzing DNA by PCR, we determined whether specific proteins were associated with the BrdU-containing DNA by western blot. Samples from cells not treated with UV were used as a control.
The data shows that RNAP II, CstF and BARD1 all associated with repaired/BrdU-containing DNA (Figure 4). These findings corroborate earlier observations that part of RNAP IIO does not dissociate from the damaged DNA during the assembly of the TCR complex (54). The presence of RNAP II supports the hypothesis proposed in yeast that RNAP II is not always degraded at sites of DNA damage and might re-engage and continue transcription (26,33,54). RNAP II associated with the repaired DNA was detected at the earliest time after UV irradiation (0.4 h), suggesting that the arrest of the RNAP II is an early event in TCR. Consistent with previous results, the western blot analysis also revealed that UV treatment decreased accumulation of RNAP II at later times (Figure 4, 2 h after UV treatment; 27,54), likely reflecting the turnover of stalled RNAP II shown in Figure 2C–D. Significantly, we also detected CstF-64 and BARD1 associated with the BrdU-containing DNA, with a time course very similar to that displayed by RNAP II. Together, this data supports the idea that RNAP II, CstF and BARD1 associate at sites of DNA damage and play a direct role in the DNA repair response.
Figure 4.
RNAP II, CstF and BARD1 localize to sites of repaired DNA. Analysis of protein complexes associated with BrdU-labeled DNA after UV treatment and the indicated recovery times. BrdU was added to HeLa cells immediately after exposure to UV light. Cells were cross-linked with formaldehyde after UV exposure at the times indicated. Sonicated cell extracts were IPed with anti-BrdU antibody. Equivalent amounts of the pellets (IP) and normalized amounts of the supernatants, which represents 7% of the input, were analyzed by immunoblotting with anti-RNAP II (H5), anti-CstF-64, anti-BARD1 and anti-actin antibodies.
DISCUSSION
Our previous work showed that polyadenylation is inhibited after DNA damage as a result of both BRCA1/BARD1/CstF complex formation (4) and proteasome-mediated degradation of RNAP II (27). As CstF-50 can interact with BARD1 to inhibit polyadenylation (17) and with the CTD of RNAP II to activate polyadenylation (13), we proposed that CstF plays an important role in the response to DNA damage. In this study, we provided evidence that prematurely terminated polyadenylated transcripts can be detected in vivo following DNA damage, especially under conditions when the CBB checkpoint is not activated. We also determined that cells with reduced levels of CstF displayed enhanced sensitivity to UV treatment. The depletion of CstF was found to correlate with decreases in both ubiquitination and turnover of RNAP IIO and repair of the transcribed DNA strand, which are events in the TCR response (22,23,25,54,55). Consistent with our model for CstF function, we also found that RNAP IIO, BARD1 and CstF were all transiently associated with sites of repaired DNA. This finding also suggests that a fraction of RNAP II elongation complexes arrested at sites of DNA damage are stable and remain associated with the DNA. Taken together, our results suggest that the polyadenylation machinery, specifically CstF, plays an important role in the response to DNA damage.
Based on the results presented here, we can confirm and extend the model that has been proposed in previous work (Figure 5; 4,17,54). Our data has provided evidence that DNA damage can induce premature transcription termination and polyadenylation, likely at sites of DNA damage and that accumulation of such species is blocked by activation of the CBB checkpoint. How might the checkpoint prevent such RNAs from accumulating? Milligan et al. (56) observed not only reduction in the levels of different mRNA species but also of truncated RNAs in yeast strains with a defective poly(A) polymerase. Defective polyadenylation of prematurely terminated transcripts is known to activate a nuclear surveillance pathway, eliminating those mRNAs by deadenylation and exosome-mediated degradation (56–58). Extending this idea, our ongoing work (our unpublished data) indicates that another CstF-50-interacting protein is the poly(A) specific ribonuclease (PARN; 59–61). PARN has been shown to co-purify with essential nonsense-mediated decay factors (62) and PARN down-regulation abrogates nonsense-mediated decay (63). Although more work is necessary to determine the functional relevance of the CstF/PARN interaction, the association of a polyadenylation factor and a deadenylation factor is mechanistically intriguing, and could contribute to the turnover of different RNA species by a nuclear quality control pathway after UV treatment.
Figure 5.
Model for the role of CstF in the DNA damage response. Coupling polyadenylation, transcription and DNA repair. After exposure to DNA damage-inducing agents, the elongating RNAP II-CstF holoenzyme complex stalls at sites of damage. A BRCA1/BARD1-containing complex is activated and recruited to sites of repair, inhibiting RNAP II and the associated polyadenylation machinery by ubiquitination followed by degradation of the RNAP IIO. This process facilitates repair by allowing access to the repair machinery, while simultaneously preventing polyadenylation of aborted nascent mRNAs, which are eliminated by exosome-mediated degradation in a nuclear surveillance pathway. Alternatively, the RNAP II complexes arrested at certain DNA lesions are not degraded, CSA and CSB proteins regulate the recruitment of chromatin remodeling and TCR factors (54), and then RNAP II re-engages and continues transcription once repair is completed. Given that CstF-50 can functionally interact with all the elements of this model, we propose an important role for this protein in the transcription-coupled DNA damage response.
Our results suggest that CstF is involved in DNA damage-induced ubiquitination of RNAP II LS, which is an important event in the TCR response (22,23,25,54). As CstF-50 can bind BARD1 (17), the CTD of RNAP II (13) and ubiquitin (Ub, our unpublished data), it is possible that it functions to help in the assembly or stabilization of the ubiquitination complex. In this scenario, CstF-50 might function as a cofactor for ubiquitination of RNAP II by BRCA1/BARD1 (27,28). Recent studies suggest that Ub-binding proteins may be critical in determining substrate specificity and substrate fate (64–66). In this respect CstF-50 could function to help the BRCA1/BARD1 heterodimer recognize some of its substrates, such as RNAP IIO. By this model, loss of CstF would have a negative effect on clearance of the stalled RNAP II from sites of damage, by preventing ubiquitination and degradation of RNAP IIO. This could in turn block access of repair enzymes to the DNA, thereby interfering with the repair process and enhancing cell death. Our results with DT40 cells depleted of CstF showing deficiencies not only in recovery from UV treatment but also in repair of UV-induced DNA damage support this idea.
Our data has indicated that CstF plays a role in the DNA repair response. As just discussed, CstF could affect DNA repair by inhibiting the erroneous processing of nascent, truncated RNAs, by inducing RNAP II ubiquitination, and/or by re-engaging and continuing transcription with stalled RNAP II complexes. It is also possible that CstF plays a more direct role in the repair process. Several observations support this hypothesis. First, CstF interacts with the DNA replication and repair factor PCNA (17). It has been shown that PCNA co-localizes with BRCA1/BARD1 at sites of DNA repair (67,68) and associates with DNA repair proteins as part of the TCR response (69). It is possible that PCNA is the repair factor that links the stalled RNAP II complex to the repair machinery during TCR. Second, several polyadenylation factors have been shown to interact with DNA repair factors. For example, cleavage factor CFIIm co-purifies with the BRCA1-associated protein hMre11 (70), which has been implicated in DNA repair and cancer predisposition (reviewed by 71). Additionally, the transcriptional co-activator PC4 interacts not only with CstF-64 (72) but also with the DNA repair protein XPG (73). XPG is known to function in multiple DNA repair pathways. XPG recruits PC4 to the bubble-containing DNA substrate, PC4 displaces XPG and forms a DNA-PC4 complex (73). PC4 can also interact with the elongating RNAP IIO through CstF-64, preventing premature termination during the elongating phase (72). It is thus possible that the interaction of PC4 with CstF-64 mediates the damage-induced association of the stalled RNAP II and the DNA repair machinery. In any case, our data have provided evidence that CstF plays a role in TCR, reinforcing the functional interaction between components of the transcription, 3′ processing and DNA repair machineries.
ACKNOWLEDGEMENTS
We thank A. Norris and N. Berezovskaya for technical assistance, and T. Hasson and Dr Kawamura for their assistance with real time-PCR. This work was supported by National Institute of General Medical Sciences-Score grant S06-60654 to F.E.K, Research Centers in Minority Institutions award RR-03037, from the National Center for Research Resources, which supports the infrastructure of Hunter College, and RO1 GM26983 to J.L.M. Funding to pay the Open Access publication charges for this article was provided by and NIH grant RO1 GM26983 to J.L.M.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hanawalt PC. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 2.Ljungman M, Zhang F, Chen F, Rainbow AJ, McKay BC. Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene. 1999;118:583–592. doi: 10.1038/sj.onc.1202356. [DOI] [PubMed] [Google Scholar]
- 3.Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc. Natl Acad. Sci. USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleiman FE, Manley JL. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/s0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- 5.Anderson JT. RNA turnover: unexpected consequences of being tailed. Curr. Biol. 2005;15:635–638. doi: 10.1016/j.cub.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223–237. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neugebauer KM. On the importance of being co-transcriptional. J. Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 8.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 9.Shatkin AJ, Manley JL. The ends of the affair: capping and polyadenylation. Nature Struct. Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, Hyman L, Moore C. Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 12.Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 13.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 14.Calvo O, Manley JL. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 2005;24:1009–1020. doi: 10.1038/sj.emboj.7600575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 16.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleiman FE, Manley JL. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science. 1999;285:1576–1579. doi: 10.1126/science.285.5433.1576. [DOI] [PubMed] [Google Scholar]
- 18.Kim HS, Li H, Cevher M, Parmelee A, Fonseca D, Kleiman FE, Lee SB. DNA damage-induced BARD1 phosphorylation is critical for the inhibition of messenger RNA processing by BRCA1/BARD1 complex. Cancer Res. 2006;66:4561–4565. doi: 10.1158/0008-5472.CAN-05-3629. [DOI] [PubMed] [Google Scholar]
- 19.Muratani M, Tansey W. How the ubiquitin-proteasome system controls transcription. Nature Revs. Mol. Cell. Biol. 2003;4:1–9. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 20.van den Boom V, Jaspers NGJ, Vermeulen W. When machines get stuck-obstructed RNA polymerase II: displacement, degradation or suicide. Bioessays. 2002;24:780–784. doi: 10.1002/bies.10150. [DOI] [PubMed] [Google Scholar]
- 21.Lee KB, Wang D, Lippard SJ, Sharp PA. Transcription-coupled and DNA damage-dependent ubiquitination of RNA polymerase II in vitro. Proc. Natl Acad. Sci. USA. 2002;99:4239–4244. doi: 10.1073/pnas.072068399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Z, Zheng J, Lu Y, Bregman DB. Ultraviolet radiation alters the phosphorylation of RNA polymerase II large subunit and accelerates its proteasome-dependent degradation. Mutat. Res. 2001;486:259–274. doi: 10.1016/s0921-8777(01)00097-0. [DOI] [PubMed] [Google Scholar]
- 23.McKay BC, Chen F, Clarke ST, Wiggin HE, Harley LM, Ljungman M. UV light-induced degradation of RNA polymerase II is dependent of Cockayne's syndrome A and B proteins but not p53 or MLH1. Mutat. Res. 2001;485:93–105. doi: 10.1016/s0921-8777(00)00064-1. [DOI] [PubMed] [Google Scholar]
- 24.Mitsui A, Sharp PA. Ubiquitination of RNA polymerase II large subunit signaled by phosphorylation of carboxyl-terminal domain. Proc. Natl Acad. Sci. USA. 1999;96:6054–6059. doi: 10.1073/pnas.96.11.6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratner JN, Balasubramanian B, Corden J, Warren SL, Bregman DB. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J. Biol. Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 26.Woudstra EC, Gilbert C, Fellows J, Jansen L, Brouwer J, Erdjument-Bromage H, Tempst P, Svejstrup JQ. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature. 2002;415:929–933. doi: 10.1038/415929a. [DOI] [PubMed] [Google Scholar]
- 27.Kleiman FE, Wu-Baer F, Fonseca D, Kaneko S, Baer R, Manley JL. BRCA1/BARD1 inhibition of mRNA 3′ processing involves targeted degradation of RNA polymerase II. Genes Dev. 2005;19:1227–1237. doi: 10.1101/gad.1309505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starita L, Horwitz AA, Keogh MC, Ishioka C, Parvin JD, Chiba N. BRCA1/BARD1 ubiquitinate phosphorylated RNA polymerase II. J. Biol. Chem. 2005;280:24498–24505. doi: 10.1074/jbc.M414020200. [DOI] [PubMed] [Google Scholar]
- 29.Derheimer FA, Chang CW, Ljungman M. Transcription inhibition: a potential strategy for cancer therapeutics. Eur. J. Cancer. 2005;41:2569–2576. doi: 10.1016/j.ejca.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Mullenders LHF. Transcription response and nucleotide excision repair. Mut. Res. 1998;409:59–64. doi: 10.1016/s0921-8777(98)00048-2. [DOI] [PubMed] [Google Scholar]
- 31.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 32.Svejstrup JQ. Mechanisms of transcription-coupled DNA repair. Mol. Cell. Biol. 2002;3:21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- 33.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 34.Tornaletti S, Patrick SM, Turchi JJ, Hanawalt PC. Behavior of T7 RNA polymerase and mammalian RNA polymerase II at site-specific cisplatin adducts in the template DNA. J. Biol. Chem. 2003;278:35791–35797. doi: 10.1074/jbc.M305394200. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, Manley JL. Core promoter elements and TAFs contribute to the diversity of transcriptional activation in vertebrates. Mol. Cell. Biol. 2003;23:7350–7362. doi: 10.1128/MCB.23.20.7350-7362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleiman FE, Ramírez AO, Dodelson de Kremer R, Gravel RA, Argaraña CE. A frequent TG deletion near the polyadenylation signal of the human HEXB gene: occurrence of an irregular DNA structure and conserved nucleotide sequence motif in the 3′ untranslated region. Hum. Mutat. 1998;12:320–329. doi: 10.1002/(SICI)1098-1004(1998)12:5<320::AID-HUMU5>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 37.Ruven HJ, Seelen CM, Lohman PH, Mullenders LH, van Zeeland AA. Efficient synthesis of 32P-labeled single-stranded DNA probes using linear PCR; application of the method for analysis of strand-specific DNA repair. Mutat. Res. 1994;315:189–195. doi: 10.1016/0921-8777(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Russell RW. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 39.Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 40.Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37:112–119. doi: 10.2144/04371RR03. [DOI] [PubMed] [Google Scholar]
- 41.Maccoux LJ, Clements DN, Salway F, Day PJ. Identification of new reference genes for the normalisation of canine osteoarthritic joint tissue transcripts from microarray data. BMC Mol. Biol. 2007;8:62–72. doi: 10.1186/1471-2199-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kartasova T, Cornelissen BJ, Belt P, van de Putte P. Effects of UV, 4-NQO and TPA on gene expression in cultured human epidermal keratinocytes. Nucleic Acids Res. 1987;15:5945–5962. doi: 10.1093/nar/15.15.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spivak G, Pfeifer GP, Hanawalt PC. In vivo assays for transcription-coupled repair. Methods Enzymol. 2006;408:23–246. doi: 10.1016/S0076-6879(06)08014-1. [DOI] [PubMed] [Google Scholar]
- 44.Boszko IP, Rainbow AJ. Removal of UV photoproducts from an adenovirus-encoded reporter gene following infection of unirradiated and UV-irradiated human fibroblasts. Somat. Cell. Mol. Genet. 1999;25:301–315. doi: 10.1023/a:1019916415806. [DOI] [PubMed] [Google Scholar]
- 45.Spivak G, Hanawalt PC. Determination of damage and repair in specific DNA sequences. Methods. 1995;7:147–161. [Google Scholar]
- 46.Wang J, Takagaki Y, Manley JL. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- 47.Laine JP, Egly JM. When transcription and repair meet: a complex system. Trends Genet. 2006;22:430–436. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, Kisselev AF, Harel-Bellan A, Nakatani Y. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 2006;20:1429–1434. doi: 10.1101/gad.378206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Link CJ, Jr, Mitchell DL, Nairn RS, Bohr VA. Preferential and strand-specific DNA repair of (6-4) photoproducts detected by a photochemical method in the hamster DHFR gene. Carcinogenesis. 1992;13:1975–1980. doi: 10.1093/carcin/13.11.1975. [DOI] [PubMed] [Google Scholar]
- 50.Ganesan AK, Hunt J, Hanawalt PC. Expression and nucleotide excision repair of a UV-irradiated reporter gene in unirradiated human cells. Mutat Res. 1999;433:117–126. doi: 10.1016/s0921-8777(98)00070-6. [DOI] [PubMed] [Google Scholar]
- 51.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 52.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 55.Bregman DB, Halaban R, van Gool AJ, Henning KA, Friedberg EC, Warren SL. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc. Natl Acad. Sci. USA. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milligan L, Torchet C, Allmang C, Shipman T, Tollervey D. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol. Cell. Biol. 2005;25:9996–10004. doi: 10.1128/MCB.25.22.9996-10004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 58.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 59.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 60.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 61.Wu M, Reuter M, Lilie H, Liu Y, Wahle E, Song H. Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 2005;24:4082–4093. doi: 10.1038/sj.emboj.7600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell. Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 63.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 64.Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 65.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat. Rev. Mol. Cell. Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 66.Kim I, Rao H. What's Ub chain linkage got to do with it? Sci. STKE. 2006;330:18. doi: 10.1126/stke.3302006pe18. [DOI] [PubMed] [Google Scholar]
- 67.Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 69.Balajee AS, May A, Dianova I, Bohr VA. Efficient PCNA complex formation is dependent upon both transcription coupled repair and genome overall repair. Mutat. Res. 1998;409:135–146. doi: 10.1016/s0921-8777(98)00051-2. [DOI] [PubMed] [Google Scholar]
- 70.de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petrini JH. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 72.Calvo O, Manley JL. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- 73.Wang JY, Sarker AH, Cooper PK, Volkert MR. The single-strand DNA binding activity of human PC4 prevents mutagenesis and killing by oxidative DNA damage. Mol. Cell. Biol. 2004;24:6084–6093. doi: 10.1128/MCB.24.13.6084-6093.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]