Abstract
Fully modified 4′-thioDNA, an oligonucleotide only comprising 2′-deoxy-4′-thionucleosides, exhibited resistance to an endonuclease, in addition to preferable hybridization with RNA. Therefore, 4′-thioDNA is promising for application as a functional oligonucleotide. Fully modified 4′-thioDNA was found to behave like an RNA molecule, but no details of its structure beyond the results of circular dichroism analysis are available. Here, we have determined the structure of fully modified 4′-thioDNA with the sequence of d(CGCGAATTCGCG) by NMR. Most sugars take on the C3′-endo conformation. The major groove is narrow and deep, while the minor groove is wide and shallow. Thus, fully modified 4′-thioDNA takes on the A-form characteristic of RNA, both locally and globally. The only structure reported for 4′-thioDNA showed that partially modified 4′-thioDNA that contained some 2′-deoxy-4′-thionucleosides took on the B-form in the crystalline form. We have determined the structure of 4′-thioDNA in solution for the first time, and demonstrated unexpected differences between the two structures. The origin of the formation of the A-form is discussed. The remarkable biochemical properties reported for fully modified 4′-thioDNA, including nuclease-resistance, are rationalized in the light of the elucidated structure.
INTRODUCTION
Synthetic short oligonucleotides are utilized to control gene expression. A variety of modified nucleoside units have been synthesized and incorporated into oligonucleotides with the aim of developing useful applications. Walker, Imbach and their collaborators (1–5) have reported the synthesis and properties of 4′-thioDNAs, i.e. oligonucleotides comprising of 2′-deoxy-4′-thionucleosides. They indicated that 4′-thioDNAs preferably hybridize to complementary RNA (1,3,5). They investigated the nuclease sensitivity of 4′-thioDNAs (3,5). No significant resistance to degradation by the exonuclease, snake venom phosphodiesterase (SVPD), was observed when compared with the corresponding unmodified oligodeoxynucleotide. However, 4′-thioDNAs were found to be highly resistant to degradation by the endonuclease S1 (3). These features of 4′-thioDNA are suitable for its application as functional DNA such as an antisense DNA. It was also demonstrated that 4′-thioDNAs elicit Escherichia coli RNase H hydrolysis of the RNA target only at high enzyme concentration (3).
The DNAs investigated by Walker's group were partially modified 4′-thioDNAs in which 2′-deoxy-4′-thionucleosides were incorporated just at several specific positions. Then, we reported the synthesis and properties of fully modified 4′thioDNA which comprises 2′-deoxy-4′-thionucleosides exclusively (6,7). Fully modified 4′-thioDNA also exhibited preferable hybridization to complementary RNA and resistance to an endonuclease. It was unexpectedly found that fully modified 4′-thioDNA behaves like an RNA molecule in terms of hybridization properties and structural aspects monitored by circular dichroism (CD). Fully modified 4′-thioDNA exhibited a CD spectrum characteristic to RNA. It was also shown that fully modified 4′-thioDNA interacts with an RNA major groove binder, lividomycin A, resulting in an increase in thermal stability, but not with DNA groove binders. Moreover, the interaction of fully modified 4′-thioDNA with RNase V1 was also revealed. These findings have suggested that fully modified 4′-thioDNA takes on an RNA-like structure, i.e., the A-form.
The structure of partially modified 4′-thioDNA, d(CGCGAA4′ST4′STCGCG)2, in which 4ST is 2′-deoxy-4′-thiothymidine has been reported (8). The two 4′ST residues took on the C3′-exo conformation, which belongs to the category of the B-form. Although some structural differences were noticed between the partially modified 4′-thioDNA and the native non-modified DNA, the partially modified 4′-thioDNA basically took on the B-form, both locally and globally (8). Thus, the crystal structure of partially modified 4′-thioDNA does not coincide with our experimental results, implying the A-form for fully modified 4′-thioDNA. The crystal structure of partially modified 4′-thioRNA, r(CC4′SCCGGGG)2, in which 4′SC is 4′-thioribocytidine, has also been reported (9). The A-form structure was seen, as expected for RNA. No other structure has been reported for either 4′-thioDNA or 4′-thioRNA. Thus, detailed structural analysis of fully modified 4′-thioDNA beyond CD analysis has not been reported yet.
The implication of the RNA-like structure, together with the promising features as a functional DNA, has encouraged us to determine the structure of fully modified 4′-thioDNA. Here, we present the structure of fully modified 4′-thioDNA with the sequence of d(CGCGAATTCGCG) determined by NMR. This is the first structure determined in solution for 4′thioDNA or 4′-thioRNA. It was revealed that, in contrast to partially modified 4′-thioDNA taking on the B-form in the crystalline form, fully modified 4′-thioDNA takes on the A-form with the C3′-endo sugar conformation, which is characteristic of RNA. This is surprising because the base sequence itself is identical for fully and partially modified 4′-thioDNAs. The reasons for the unexpected formation of the A-form of this DNA are discussed. Then, remarkable features reported for fully modified 4′-thioDNA, including resistance to nucleases, are explained rationally on the basis of the elucidated structure.
MATERIALS AND METHODS
Sample preparation
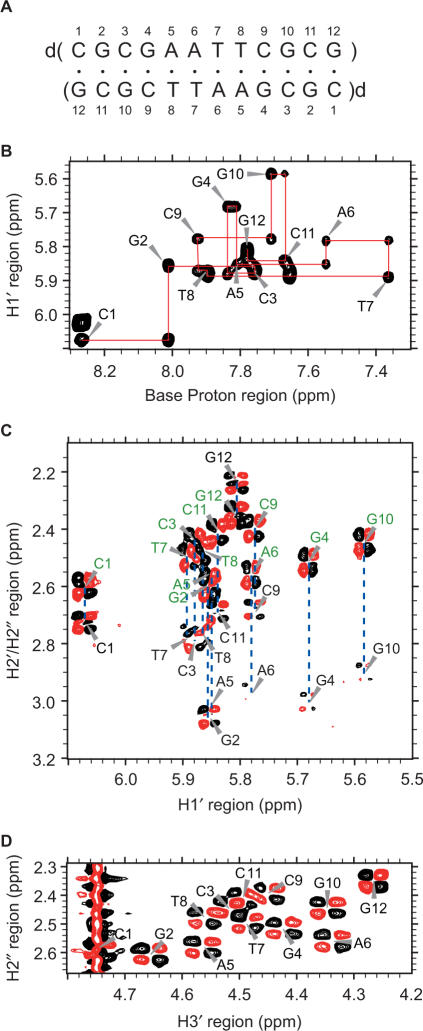
Fully modified 4′-thioDNA with the sequence of d(CGCGAATTCGCG), which exclusively comprises 2′-deoxy-4′-thionucleosides (Figure 1A), was synthesized and purified as described previously (6,7). 4′-thioDNA was dissolved in a solution containing 10 mM NaCl and 10 mM Na-phosphate (pH 6.5). The concentration of the 4′-thioDNA strand was 0.4 mM. 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS) was used as an internal chemical shift reference. The sample was heated at 85°C for 5 min, followed by gradual cooling to room temperature prior to the measurement.
Figure 1.
(A) The sequence and numbering of fully modified 4′-thioDNA. (B) The H6/H8-H1′ region of the NOESY spectrum of 4′-thioDNA. The H1′ (i − 1)–H6/H8(i)–H1′ (i) connectivities are traced, the intraresidue correlation peaks being labeled with the residue numbers. The H1′–H2′ (labeled in black) and H1′–H2″(labeled in green) regions (C) and the H2″–H3′ region (D) of the DQF-COSY spectrum of 4′-thioDNA.
NMR spectroscopy
NMR spectra were recorded with Bruker DRX800 and DRX600 spectrometers each equipped with a cryoprobe with a Z-gradient. The following NMR experiments were performed to assign the resonances, and to obtain distance and dihedral angle constraints: NOESY, TOCSY, DQF-COSY and 1H-13C HSQC. Spectra were processed and analyzed with XWIN-NMR (Bruker), NMRPipe (10), Capp/Pipp/Stapp (11) and Sparky (12).
Distance and dihedral angle constraints
Interproton distances were obtained from a NOESY spectrum with a mixing time of 50 ms in 2H2O and one with a mixing time of 80 ms in H2O, supplemented by ones with a mixing time of 230 ms in 2H2O and H2O, as described previously (13–15). In total, 786 distance constraints were obtained per duplex.
The dihedral angle constraints for the δ, and endocyclic ν0, ν1, ν2, ν3 and ν4 torsion angles were derived from the 3JH1′-H2′, 3JH2″-H3′ and 3JH3–H4′ couplings (16,17), as described previously (13–15). The sugar pucker was determined as follows: the C3′-endo conformation for the C3, G4, A5, A6, T7, T8, C9 and G10 residues, the C4′-exo conformation for the G2 and C11 residues, and a conformation between the C4′-exo and O4′-endo ones for the C1 and G12 residues. The δ and endocyclic ν0–ν4 torsion angles of the residues taking on the C3′-endo conformation were moderately constrained, leaving the sugar free to take on any conformation without an energy penalty between C2′-exo and C4′-exo including C3′-endo in the pseudorotation cycle. In the same way, those of the residues taking on the C4′-exo conformation were constrained between C3′-endo and O4′-endo including C4′-exo, and those of C1 and G12 were also moderately constrained similarly.
Structure calculations
Structure calculations were carried out using distance and dihedral angle constraints with a simulated annealing protocol supplied with X-PLOR v. 3.851 (18), CNS v. 1.1 (19), and Xplor-NIH v. 2.18 (20). Hydrogen-bonding constraints and planarity constraints for Watson–Crick base pairs were also included.
As described above and also under ‘Results’ section, the NMR data revealed that all sugars of our 4′thioDNA duplex took on the N-type pucker, typically C3′-endo, not the S-type pucker. Crystal structures have been reported for partially modified 4′-thionucleic acids: 4′-thiosugars of r(CC4′SCCGGGG)2 took on the N-type pucker, C3′-endo (9), while those of d(CGCGAA4′ST4′STCGCG)2 took on the S-type pucker, C3′-exo (8). Thus, the conformation of 4′-thiosugars is the same for our 4′thioDNA and r(CC4′SCCGGGG)2. Therefore, geometrical parameters regarding 4′-thiosugars, i.e. bond lengths for S4′–C1′ and S4′–C4′, and bond angles for C4′–S4′–C1′, S4′–C1′–C2′, C1′–C2′–C3′, C2–C3′–C4′ and C3′–C4′–S4′, were extracted from the crystal structure of r(CC4′SCCGGGG)2 (PDB IB: 2a0p), and added to the data base that was referred to during the structure calculation.
Ten final structures were selected from 100 calculations on the basis of the criteria of the smallest residual energy values. None of them violated the distance constraints by more than 0.5 Å or the dihedral angle constraints by more than 5°. The structures of the grooves were analyzed with Curves (21), and the stacking was analyzed with 3DNA (22).
Coordinates
The coordinates of a representative structure with the lowest energy have been deposited in the Protein Data Bank (PDB ID code is 2rmq).
RESULTS
Resonance assignments
Non-exchangeable 1H resonances were assigned using standard methods (16), as described for other DNAs and RNAs (13–15) (Table 1S). As an example, Figure 1B shows expansion of the NOESY spectrum allowing the sequential assignment of the H1′ and H6/H8 through H1′ (i−1)–H6/H8(i)–H1′ (i) connectivities. The H2′, H2″ H3′, H4′ and H5′/H5″ resonances were assigned by means of NOESY, TOCSY and DQF-COSY spectra. The exchangeable 1H resonances were also assigned with standard methods.
It was noted that an H2′ resonance appeared in the lower field in comparison to an H2″ resonance for all residues except for G12. The discrimination of H2′ and H2″ resonances was unequivocally accomplished on the basis of the fact that an H1′–H2″ correlation peak should be stronger than an H1′–H2′ one in a NOESY spectrum because of the vicinity of H1′–H2″ for any sugar conformation. Usually, the H2′ resonance appeared in the higher field not only for the B-form, but also for the A-form (23). Therefore, the reversal of the resonance positions may be a characteristic of a 2′-deoxy-4′-thionucleoside.
Appearance of the N-type sugar conformation characteristic to the A-form
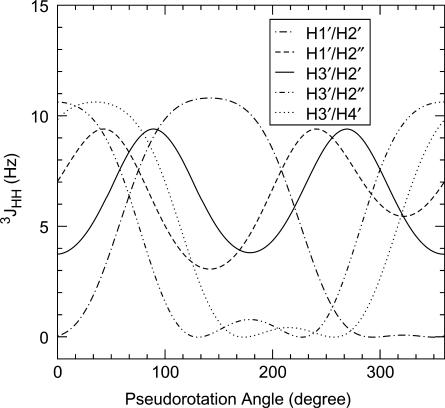
The parameters used to describe the relationship between the pseudorotation angle, P, and the torsion angle between two vicinal sugar protons, ϕHH, for 2′-deoxy-4′-thioribose were optimized and validated by Walker and Chattopadhyaya, and their collaborators (24). On the basis of the reported parameters, the dependence of five vicinal 1H–1H coupling constants, 3JHH, on the pseudorotation angle was calculated using the simple Karplus equation (25,26), 3JHH = 10.2cos2ϕHH − 0.8cosϕHH, and is illustrated in Figure 2. The maximum puckering amplitude was set to 45° (24). The simple Karplus equation was used, because it had been reported that the simple Karplus equation and a more complicated Karplus equation (27,28) give reasonably similar results regarding the dependence of 3JHH on P as to 2′-deoxyribose (29). The dependence obtained for 2′-deoxy-4′thioribose (Figure 2) is basically the same as that reported for 2′-deoxyribose (29). However, some differences are noticeable. For example, 3JH2′-H3′ for the C3′-endo sugar conformation is ∼4 Hz for 2′-deoxy-4′-thioribose (Figure 2), while the corresponding coupling constant is 6–7 Hz for 2′-deoxyribose (29).
Figure 2.
Dependency of five vicinal 1H–1H coupling constants, 3JHH, on the pseudorotation angle, P, for 2′-deoxy-4′-thioribose.
The sugar conformation of fully modified 4′-thioDNA was deduced from Figure 2. It was remarkable that the strong H2″H3′ correlation peaks in the DQF-COSY spectrum were observed for all twelve residues of fully modified 4′-thioDNA (Figure 1D). When Figure 2 was referred to, this indicated that all residues took on the N-type sugar conformation characteristic of the A-form, not the S-type sugar conformation characteristic of the B-form, for which the absence of the corresponding correlation peaks was expected. Observation of the strong H3′–H4′ correlation peaks for all residues (data not shown) is also consistent with the N-type sugar conformation.
The sugar conformation of the central eight residues, C3–G10, was further limited to around C3′-endo, on the basis of the weakness of the H1′–H2′ correlation peaks (Figures 1C and 2). It should be added that the H2′–H3′ correlation peaks were weak for these residues (data not shown). This is expected from Figure 2, because the corresponding J-value is relatively small, 4 Hz, for C3′-endo. On the other hand, the J-value expected for C3′-endo of 2′-deoxyribose is not small, 6–7 Hz, (29), as mentioned above, which cannot explain the weakness of the correlation peaks. This fact seems to confirm the validity of Figure 2 calculated specifically for 2′-deoxy-4′-thioribose.
At this stage, the sugar conformation of G2 and C11 was presumed to be around C4′-exo on the basis of the medium intensity of the H1′–H2′ correlation peaks, and that of C1 and G12 to be around a conformation between C4′-exo and O4′-endo, on the basis of the relatively strong intensity of these peaks. In order to take account of the experimental errors and resulting uncertainties, the δ and endocyclic ν0–ν4 torsion angles were moderately constrained during the structure calculation, as described in ‘Materials and methods’ section.
Structure of fully modified 4′-thioDNA
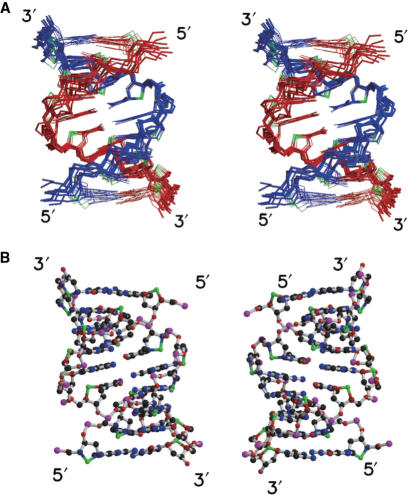
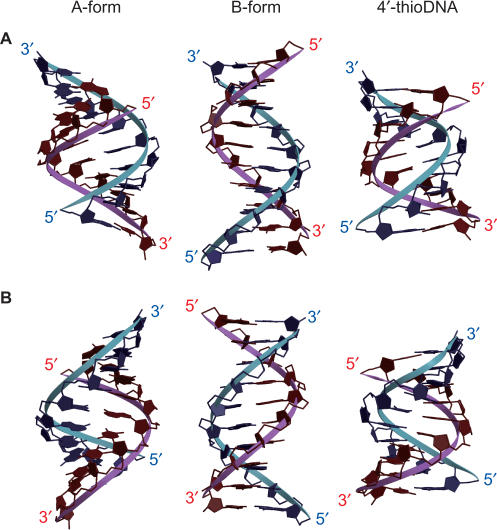
The applied constraints and structural statistics for the 10 final structures are summarized in Table 1. The root mean square deviations (RMSD) of the 10 final structures versus the mean structure for all heavy atoms, terminal residues being excluded, were 0.83 ± 0.10 Å. Figure 3A shows a stereo view of the superposition of the 10 final structures. A representative structure with the lowest energy is shown in Figure 3B. The canonical A-form and B-form of the sequence of d(CGCGAATTCGCG)2 were constructed on the basis of the reported coordinates and parameters (30,31). The structure of fully modified 4′-thioDNA is shown with the canonical A-form and B-form for comparison (Figure 4). The widths and depths of the major and minor grooves were calculated for each structure with Curves (20) and are summarized in Table 2. For fully modified 4′-thioDNA, the values averaged for the whole of the representative structure are indicated.
Table 1.
NMR constraints and structural statistics for fully modified 4′-thioDNA
| A. NMR constraints | |
| Distance constraints | 786 |
| Intraresidue distance constraints | 452 |
| Sequential (i, i + 1) distance constraints | 228 |
| Medium-to-long range ≥(i, i + 2) distance constraints | 106 |
| Dihedral angle constraints | 120 |
| ν0–ν4 | 120 |
| Planarity constraints for base pairs | 12 |
| Hydrogen bonding constraints | 64 |
| B. Structural statistics for 10 final structures | |
| CNS energies (kcal/mol) | |
| Etotal | 585 ± 11 |
| Ebond | 24 ± 1 |
| Eangle | 128 ± 4 |
| Eimproper | 45 ± 3 |
| Evdw | 117 ± 5 |
| Enoe | 0 ± 0 |
| Ecdih | 6 ± 1 |
| RMSD from idealized geometry | |
| Bond lengths (Å) | 0.004 ± 0.000 |
| Bond angles (deg.) | 0.71 ± 0.01 |
| Impropers (deg.) | 0.8 ± 0.5 |
| NOE violations | |
| Number of violations >0.5 Å | 0 ± 0 |
| RMSD of violations (Å) | 0.07 ± 0.001 |
| Dihedral angle violations | |
| Number of violations >5° | 0 ± 0 |
| RMSD of violations (deg.) | 0.81 ± 0.37 |
| RMSD of 10 final structures versus mean structure | 0.83 ± 0.1 |
| for all heavy atoms (Å) (terminal residues excluded) |
Figure 3.
(A) A stereo view of the superposition of the 10 final structures of fully modified 4′-thioDNA. The two strands are colored red and blue, respectively. The S4′ atoms are colored green. (B) The representative structure with the lowest energy, viewed from the major groove (left) and the minor groove (right).
Figure 4.
Comparison of the structure of fully modified 4′-thioDNA (right) with those of the canonical A-form (left) and B-form (middle), viewed from the major groove (A) and the minor groove (B).
Table 2.
Widths and depths of the grooves of A-form, B-form and 4′-thioDNA
| A-form (Å) | B-form (Å) | 4′-thioDNA (Å) | |
|---|---|---|---|
| Major groove | |||
| Width | Narrow (3.4) | Wide (11.5) | Narrow (3.5) |
| Depth | Deep (10.0) | Intermediate (3.3) | Deep (9.7) |
| Minor groove | |||
| Width | Wide (10.0) | Narrow (5.0) | Wide (8.0) |
| Depth | Shallow (1.0) | Intermediate (4.5) | Shallow (1.1) |
Pseudorotation angle, P, was calculated for each residue for 10 final structures; C1(P = 39.6 ± 5.1°), G2(19.9 ± 2.9°), C3(14.5 ± 4.6°), G4(4.3 ± 3.0°), A5(1.7 ± 6.1°), A6(−0.4 ± 7.0°), T7(−0.1 ± 3.6°), T8(5.7 ± 5.8°), C9(4.2 ± 5.2°), G10(35.1 ± 4.8°), C11(22.7 ± 4.3°) and G12(25.1 ± 2.5°). Thus, the structure calculation indicated that the sugar conformations of all residues are C3′-endo (0° < P < 36°) or very close to C3′-endo. The appearance of the N-type sugar conformation, characteristic of the A-form, for all residues of fully modified 4′-thioDNA is quite in contrast to the finding that all residues of partially modified 4′-thoDNA, d(CGCGAA4′ST4′STCGCG)2, took on the S-type sugar conformation, characteristic of the B-form, including 4′-thionucleosides.
The major groove of fully modified 4′thioDNA is narrow (3.5 Å) and deep (9.7 Å) (Figure 4 and Table 2), and the minor groove is wide (8.0 Å) and shallow (1.1 Å) (Figure 4 and Table 2). These characteristics of the shapes of the two grooves of 4′-thioDNA coincide with those of the A-form (Figure 4 and Table 2). In the case of the B-form, in contrast, the major groove is wide and of intermediate depth, and the minor groove is narrow and of intermediate depth (Figure 4 and Table 2). These characters of the B-form are quite different from those of fully modified 4′-thioDNA. It was revealed in the previous section that the local sugar conformation of fully modified 4′-thioDNA is the N-type, which is characteristic of the A-form. Here, it is concluded that fully modified 4′-thioDNA takes on the A-form not only locally but also globally.
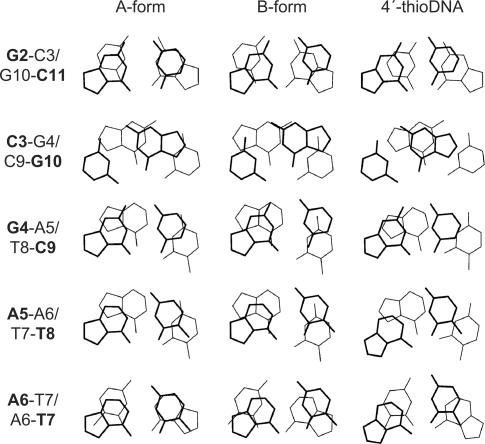
The stacking pattern observed for fully modified 4′-thioDNA is shown in Figure 5. Those of the A-form and B-form are also shown, respectively, for comparison. One remarkable difference between the A-form and B-form is the interstrand stacking. As to a 5′-pyrimidine-purine-3′ step such as C3–G4/C9–G10, interstrand stacking between two purine bases is seen for the A-form, but not for the B-form (Figure 5). It was found that the interstrand stacking characteristic of the A-form is also present in 4′-thioDNA, being even more intense than that in the A-form (Figure 5). As to 5′-purine-purine-3′ steps such as G4-A5/T8-C9 and A5-A6/T7-T8, the six-membered ring of the upper purine stacks on both the five-membered and six-membered rings of the lower purine for the B-form, but just on the five-membered ring for the A-form. The situation for 4′-thioDNA is the same as that for the A-form (Figure 5).
Figure 5.
The stacking pattern observed for fully modified 4′-thioDNA (right), with those for the canonical A-form (left) and B-form (middle), respectively, the terminal part being excluded. The views are perpendicular to base pairs, not along the helix axes. The upper bases are indicated by bold lines and characters.
Usually, DNA takes on the B-form and RNA takes on the A-form (31). Additionally, it has been reported that partially modified 4′-thioDNA, d(CGCGAA4′ST4′STCGCG)2, also took on the B-form (8). The base sequence of this DNA is identical to that of fully modified 4′-thioDNA. Therefore, it is remarkable that fully modified 4′-thioDNA takes on the A-form, not the B-form. This finding is consistent with our previous suggestion that fully modified 4′-thioDNA takes on an RNA-like structure (7).
DISCUSSION
DNA usually takes on the B-form in solution, although a structural change to the A-form occurs under dehydrating conditions (31,32). Structure determination by means of NMR has demonstrated that fully modified 4′-thioDNA unexpectedly takes on the A-form in the solution of moderate salt and neutral pH conditions. The B-form of DNA is stabilized through interactions with hydration spines located in the minor groove (31,33). The O4′ atoms of 2′-deoxynucleosides are critically involved in these interactions through a series of hydrogen bonds, and thus contribute to stabilization of the B-form. The S4′ atoms of 2′-deoxy-4′-thionucleosides are more hydrophobic than the O4′ atoms of 2′-deoxynucleosides. Thus, the hydration spines essential for stabilization of the B-form would not be formed for fully modified 4′-thioDNA, which could account for the resultant formation of the A-form in solution.
It has been reported that a 2′-deoxy-4′-thiothymidine monomer prefers the S-type sugar conformation linked to the B-form to the N-type sugar conformation linked to the A-form, the population ratio being ∼7 : 3 (24). It is assumed that the stacking, particularly the interstrand stacking, is more effective in the A-form than in the B-form (31,34). Therefore, in terms of merely stacking, the formation of the A-form might be more favorable than that of the B-form. In addition to the destruction of the hydration spine, this may account to some extent for the formation of the A-form for a 4′-thio-oligomer, in spite of the preference of a 4′-thiomonomer for the B-form sugar conformation. The finding that the stacking pattern, including interstrand stacking, for fully modified 4′-thioDNA is, in fact, similar to that for the A-form would be consistent with this idea.
The gauche effect for O(S)–C–C–O fragments and the O4′ (S4′)–C1′–N9/N1 anomeric effect are affected by the introduction of sulfur atom in the deoxyribose ring. Some work in this respect has been done on nucleosides (35,36). The changes of these effects may also contribute to the formation of the A-form for fully modified 4′-thioDNA, although the rational interpretation is not available at this moment.
Partially modified 4′-thioDNA, d(CGCGAA4′ST4′STCGCG)2, took on the B-form in the crystalline form, including 4′-thionucleosides (8). This may be due to the fact that the hydration spines, which stabilize the B-form, still persist to some extent for this DNA containing only four 4′-thionucleosides out of 24 residues per duplex. In fact, inspection of the crystal structure deposited in the Protein Data Bank (PDB ID: 233D) revealed that many water molecules are present in the minor groove and that the hydration spine-like structure seems to be present in the minor groove, although the hydration spine-like structure was not discussed in the original paper (8).
On the basis of the elucidated structure, the remarkable properties reported for fully modified 4′-thioDNA can be rationally interpreted. First, it was revealed that this DNA exhibits a CD spectrum characteristic of the A-form (7), although DNA usually gives a CD spectrum characteristic of the B-form. It is dangerous to come to any conclusion regarding the structure of DNA merely from its CD spectrum, because the correlation between the structure and the CD spectrum is sometimes perturbed and not clear (37). Structure determination by means of NMR has now firmly revealed the formation of the A-form, which is consistent with the results of CD analysis.
Second, the results regarding the interactions with several groove binders can be rationalized. It was found that fully modified 4′-thioDNA unexpectedly interacts with lividomycin A, resulting in an increase in thermal stability (7). Lividomycin A is known to be an RNA major groove binder. The shape of the major groove of RNA is supposed to be suitable for accommodating lividomycin A. RNA usually takes on the A-form, and 4′-thioDNA was demonstrated to take on the A-form. Thus, the shape of the minor groove of 4′-thioDNA is similar to that of RNA, being suitable for the accommodation. Thus, the unexpected interaction of 4′-thioDNA with lividomycin A is rationalized. It was also found that 4′-thioDNA interacts with neither distamycin A nor methyl green (7). Distamycin A is known to be a DNA minor groove binder preferring the AATT sequence, and methyl green to be a DNA major groove binder. The shapes of the minor and major grooves of DNA are supposed to be suitable for accommodating distamycin A and methyl green, respectively. DNA usually takes on the B-form, while fully modified 4′-thioDNA takes on the A-form. Therefore, the shapes of the grooves of 4′-thioDNA are quite different form those of usual DNA of the B-form (Figure 4 and Table 2), being not suitable for accommodating either distamycin A or methyl green. Thus, the absence of the interactions with these binders is explained.
Third, it was found that fully modified 4′-thioDNA is resistant to cleavage by DNase I, a DNA-specific endonuclease (7). This is consistent with the report by Walker and collaborators, who first revealed the endonuclease-resistance (1,3). It was further demonstrated that the addition of fully modified 4′-thioDNA does not inhibit the cleavage of usual DNA by DNase I, which indicates that fully modified 4′-thioDNA is not even recognized, i.e. not even bound, by DNase I (7). The crystal structure of a DNase I–DNA complex has been reported, and the importance of the minor groove width and depth of the B-form was suggested for achievement of the interactions (38). The minor groove width and depth of fully modified 4′-thioDNA are quite different from those of the B-form (Figure 4 and Table 2). This accounts for the finding that fully modified 4′-thioDNA was not recognized by DNase I and exhibited resistance to cleavage by DNase I.
A crystal of d(CGCGAA4′ST4′STCGCG)2 exhibited the B-form, but it exhibited a small, but detectable, distortion of the conformation of the sugar-phosphate backbone in the regions at and adjacent to the positions of the 4′-thionucleosides (8). It was suggested that an endonuclease binds less well to this partially modified 4′-thioDNA due to the distortion, resulting in endonuclease-resistance. In the case of fully modified 4′-thioDNA, the structural change is more drastic and it is clearly understood why an endonuclease, DNase I, does not bind to this DNA, resulting in endonuclease-resistance. Because of this, fully modified 4′-thioDNA can be applied in a direct way as a nuclease-resistant functional DNA.
The interaction of a restriction endonuclease, EcoRV, with DNA containing a single 4′-thionucleoside has been examined (1). It was revealed that this DNA is bound by EcoRV as strongly as native non-modified DNA, but that this DNA is cleaved much more slowly than the native DNA. It was also indicated that EcoRV methylase does not methylate DNA, in which one or two Ts are substituted by 2′-deoxy-4′-thiothymidine in the sequence d(GACGATATCGTC) (1). The reason for the reduced cleavage rate was not elucidated, as the crystal structure of the complex between EcoRV and this DNA showed that the conformation of DNA had not significantly changed (1,8,39). The mechanism of endonuclease resistance seems to differ between this DNA and fully modified 4′-thioDNA. In any case, fully modified 4′-thioDNA is not recognized, i.e. not bound, by endonuclease, DNase I, due to the formation of the A-form, which inevitably results in endonuclease resistance.
Fourth, the competition assay surprisingly indicated that fully modified 4′-thioDNA is recognized, i.e. bound, by RNase V1 (7), which is an RNA-specific endonuclease (40). Although the structure of RNase V1 has not been reported, it is suggested that RNase V1 interacts with the minor groove of RNA (41). It is also suggested that the interaction of RNase V1 with the 2′-hydroxyl group of RNA is not necessary for binding (42). Therefore, the fact that the shape of the minor groove of fully modified 4′-thioDNA is similar to that of RNA (Figure 4 and Table 2) allows rationalization of the unexpected recognition of fully modified 4′-thioDNA by RNase V1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
M.K. was supported by Grants-in-Aid for Scientific Research (Nos. 18011008, 18370046, 19036026 and 19657034) and the Protein 3000 Project of MEXT, by a PRESTO grant from JST, and by a grant for the Strategic Research Project (No. K19014) of YCU. A.M. was supported by the post-doctoral fellow program of JSPS. Funding to pay the Open Access publication charges for this article was provided by Grants-in-Aid for Scientific Research of MEXT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hancox EL, Connolly BA, Walker RT. Synthesis and properties of oligodeoxynucleotides containing the analogue 2′-deoxy-4′-thiothymidine. Nucleic Acids Res. 1993;21:3485–3491. doi: 10.1093/nar/21.15.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar S, Horton JR, Jones GD, Walker RT, Roberts RJ, Cheng X. DNA containing 4′-thio-2′-deoxycytidine inhibits methylation by HhaI methyltransferase. Nucleic Acids Res. 1997;25:2773–2783. doi: 10.1093/nar/25.14.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones GD, Lesnik EA, Owens SR, Risen LM, Walker RT. Investigation of some properties of oligodeoxynucleotides containing 4′-thio-2′-deoxynucleotides: duplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1996;24:4117–4122. doi: 10.1093/nar/24.21.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones GD, Altmann K.-H, Husken D, Walker RT. Duplex- and triplex-forming properties of 4′-thio-modified oligodeoxynucleotides. Bioorg. Med. Chem. Lett. 1997;7:1275–1278. [Google Scholar]
- 5.Leydier C, Bellon L, Barascut JL, Morvan F, Rayner B, Imbach JL. 4′-Thio-RNA: synthesis of mixed base 4′-thio-oligoribonucleotides, nuclease resistance, and base pairing properties with complementary single and double strand. Antisense Res. Dev. 1995;5:167–174. doi: 10.1089/ard.1995.5.167. [DOI] [PubMed] [Google Scholar]
- 6.Inoue N, Kaga D, Minakawa N, Matsuda A. Practical synthesis of 2′-Deoxy-4′-thioribonucleosides: substrates for the synthesis of 4′-ThioDNA. J. Org. Chem. 2005;70:8597–8600. doi: 10.1021/jo051248f. [DOI] [PubMed] [Google Scholar]
- 7.Inoue N, Minakawa N, Matsuda A. Synthesis and properties of 4′-ThioDNA: unexpected RNA-like behavior of 4′-ThioDNA. Nucleic Acids Res. 2006;34:3476–3483. doi: 10.1093/nar/gkl491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boggon TJ, Hancox EL, McAuley.-Hecht. KE, Connolly BA, Hunter WN, Brown T, Walker RT, Leonard GA. The crystal structure analysis of d(CGCGAASSCGCG)2, a synthetic DNA dodecamer duplex containing four 4′-thio-2′-deoxythymidine nucleotides. Nucleic Acids Res. 1996;24:951–961. doi: 10.1093/nar/24.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haeberli P, Berger I, Pallan PS, Egli M. Syntheses of 4′-thioribonucleosides and thermodynamic stability and crystal structure of RNA oligomers with incorporated 4′-thiocytosine. Nucleic Acids Res. 2005;33:3965–3975. doi: 10.1093/nar/gki704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 11.Garrett DS, Powers R, Gronenborn AM, Clore GM. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Goddard TD, Kneller DG. SPARKY 3,. San Francisco: University of California; 2006. [Google Scholar]
- 13.Matsugami A, Kobayashi S, Ouhashi K, Uesugi S, Yamamoto R, Taira K, Nishikawa S, Kumar PKR, Katahira M. Structural basis of the highly efficient trapping of the HIV Tat protein by an RNA aptamer. Structure. 2003;11:533–545. doi: 10.1016/s0969-2126(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 14.Matsugami A, Ouhashi K, Kanagawa M, Liu H, Kanagawa S, Uesugi S, Katahira M. An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J. Mol. Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]
- 15.Matsugami A, Okuizumi T, Uesugi S, Katahira M. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J. Biol. Chem. 2003;278:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- 16.Varani G, Aboul-ela F, Allain FHT. NMR investigation of RNA structure. Prog. Nucl. Magn. Reson. Spect. 1996;29:51–127. [Google Scholar]
- 17.Smith JS, Nikonowicz EP. Phosphorothioate substitution can substantially alter RNA conformation. Biochemistry. 2000;39:5642–652. doi: 10.1021/bi992712b. [DOI] [PubMed] [Google Scholar]
- 18.Brünger AT. X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. New Haven: Yale University Press; 1993. [Google Scholar]
- 19.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 21.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 22.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishizaki T, Iwai S, Ohkubo T, Kojima C, Nakamura H, Kyogoku Y, Ohtsuka E. Solution structures of DNA duplexes containing a DNA x RNA hybrid region, d(GG)r(AGAU)d(GAC) x d(GTCATCTCC) and d(GGAGA)r(UGAC) x d(GTCATCTCC) Biochemistry. 1996;35:4016–4025. doi: 10.1021/bi9519821. [DOI] [PubMed] [Google Scholar]
- 24.Koole LH, Plavec J, Liu H, Vincent BR, Dyson MR, Coe PL, Walker RT, Hardy GW, Rahim SG, Chattopadhyaya J. Conformations of two 4′-thio-2′-deoxynucleoside analogs studied by 500-MHz 1H NMR spectroscopy and X-ray crystallography. J. Am. Chem. Soc. 1992;114:9936–9943. [Google Scholar]
- 25.Davies DB. Conformations of nucleosides and nucleotides. Prog. Nucl. Magn. Reson. Spectrosc. 1978;12:135–225. [Google Scholar]
- 26.Hosur RV, Ravikumar M, Chary KVR, Sheth A, Govil G, Zu-Kun T, Miles HT. Solution structure of d-GAATTCGAATTC by 2D NMR. A new approach to determination of sugar geometries in DNA segments. FEBS Lett. 1986;205:71–76. doi: 10.1016/0014-5793(86)80868-7. [DOI] [PubMed] [Google Scholar]
- 27.Haasnoot CAG, de Leeuw FAAM, Altona C. The relationship between proton-proton NMR coupling constants and substituent electronegativities-I: an empirical generalization of the Karplus equation. Tetrahedron. 1980;36:2783–2792. [Google Scholar]
- 28.Van de Ven FJ, Hilbers CW. Nucleic acids and nuclear magnetic resonance. Eur. J. Biochem. 1988;178:1–38. doi: 10.1111/j.1432-1033.1988.tb14425.x. [DOI] [PubMed] [Google Scholar]
- 29.Kim SG, Lin LJ, Reid BR. Determination of nucleic acid backbone conformation by 1H NMR. Biochemistry. 1992;31:3564–3574. doi: 10.1021/bi00129a003. [DOI] [PubMed] [Google Scholar]
- 30.Arnott S, Hukins DW. Optimised parameters for A-DNA and B-DNA. Biochem. Biophys. Res. Commun. 1972;47:1504–1509. doi: 10.1016/0006-291x(72)90243-4. [DOI] [PubMed] [Google Scholar]
- 31.Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag Inc.; 1983. [Google Scholar]
- 32.Johnson CW., Jr. Determination of the Conformations of Nucleic Acids by Electronic CD. In: Fasman GD, editor. Circular Dichroism and the Conformational Analysis of Biomolecules. NY: Plenum Press; 1996. pp. 433–468. [Google Scholar]
- 33.Kopka ML, Fratini AV, Drew HR, Dickerson RE. Ordered water structure around a B-DNA dodecamer. A quantitative study. J. Mol. Biol. 1983;163:129–146. doi: 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- 34.Arnott S, Chandrasekaran R, Hukins DW, Smith PJ, Watts L. Structural details of double-helix observed for DNAs containing alternating purine and pyrimidine sequences. J. Mol. Biol. 1974;15:523–533. doi: 10.1016/0022-2836(74)90499-9. [DOI] [PubMed] [Google Scholar]
- 35.Crnugelj M, Dukhan D, Barascut JL, Imbach JL, Plavec J. How S-C-N anomeric effects and energetic preference across [S-C-C-O] fragments steer conformational equilibria in 4′-thionucleosides. 1H NMR and ab initio MO study. J. Chem. Soc., Perkin Trans. 2000;2:255–262. [Google Scholar]
- 36.Crnugelj M, Plavec J. NMR conformational study reveals that S-C-N anomeric effect in thionucleosides is weaker than O-C-N anomeric effect in natural nucleosides. Croatica Chim. Acta. 2001;74:381–398. [Google Scholar]
- 37.Sato M, Ono A, Higuchi H, Ueda T. CD spectra and some properties of deoxyoligonucleotide duplexes having a C:G terminus (nucleosides and nucleotides. Part 69) Nucleic Acids Res. 1986;14:1405–1416. doi: 10.1093/nar/14.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weston SA, Lahm A, Suck D. X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3. J. Mol. Biol. 1992;226:1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]
- 39.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. The crystal structures of EcoRV endonuclease, and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lockard RE, Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981;9:5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auron PE, Weber LD, Rich A. Comparison of transfer ribonucleic acid structures using cobra venom and S1 endonucleases. Biochemistry. 1982;21:4700–4706. doi: 10.1021/bi00262a028. [DOI] [PubMed] [Google Scholar]
- 42.Wyatt JR, Walker GT. Deoxynucleotide-containing oligoribonucleotide duplexes: stability and susceptibility to RNase V1 and RNase H. Nucleic Acids Res. 1989;17:7833–7842. doi: 10.1093/nar/17.19.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]