Abstract
Alus, primate-specific retroelements, are the most abundant repetitive elements in the human genome. They are composed of two related but distinct monomers, left and right arms. Intronic Alu elements may acquire mutations that generate functional splice sites, a process called exonization. Most exonizations occur in right arms of antisense Alu elements, and are alternatively spliced. Here we show that without the left arm, exonization of the right arm shifts from alternative to constitutive splicing. This eliminates the evolutionary conserved isoform and may thus be selected against. We further show that insertion of the left arm downstream of a constitutively spliced non-Alu exon shifts splicing from constitutive to alternative. Although the two arms are highly similar, the left arm is characterized by weaker splicing signals and lower exonic splicing regulatory (ESR) densities. Mutations that improve these potential splice signals activate exonization and shift splicing from the right to the left arm. Collaboration between two or more putative splice signals renders the intronic left arm with a pseudo-exon function. Thus, the dimeric form of the Alu element fortuitously provides it with an evolutionary advantage, allowing enrichment of the primate transcriptome without compromising its original repertoire.
INTRODUCTION
Alternative splicing is a mechanism by which transcriptomic diversity is generated from a relatively low number of human protein-coding genes (1–3). An average human gene is 28 000-nt long and is composed of 8.8 exons of ∼130 nucleotides each, separated by 7.8 introns of ∼3000 nt (3). Bioinformatic analyses estimate that >70% of all human genes generate more than one isoform by alternative splicing, which contributes significantly to human proteome complexity (4).
The splicing reaction involves the recognition of exon–intron junctions by the spliceosome and intron excision through a two-step transesterification reaction (5–7). The spliceosome is a large and dynamic complex assembled from five small nuclear ribonucleoproteins (snRNPs) and over 200 additional proteins (8–11). The spliceosome recognizes four conserved sequences located at both ends of introns: the 5′ and 3′ splice sites (5′ss and 3′ss), the polypyrimidine tract (PPT) and the branch point sequence (BPS) (5). In Metazoans, these four splicing signals alone are not sufficient to direct splicing. Cis-acting exonic and intronic splicing regulatory elements (ESRs/ISRs), recognized by trans-acting factors such as SR and hnRNPs proteins, modulate exon selection and regulate alternative splicing (6,12).
With 1.07 million copies, Alu elements are the most abundant repetitive element in the human genome, composing more than 10% of the genome (3,13). Alus belong to the SINE (short interspersed elements) family of repetitive elements and are unique to primates (14,15). The genomic distribution of Alu elements is non-uniform, with a strong bias towards gene-rich regions and a tendency to reside within intronic sequences (there are 718 460 intronic Alus within human protein-coding genes) (13,16). The first Alu sequence is thought to have been formed by dimerization of two distinct Alu monomers (Fossil Left and Right Alu Monomers, FLAM and FRAM) that arose from a 7SL RNA gene (17,18). A typical Alu is ∼300 nt in length, consisting of two similar, but distinct monomers—the left and the right arms—joined by an A-rich linker and followed by a poly(A) tail. The right arm contains a 31-bp insert that is not present in the left arm, whereas the left arm contains an internal promoter for RNA polymerase III (A and B boxes) that is absent from the right arm (19,20) (see also Figure 1A). These internal promoter elements significantly differ from the consensus sequence, therefore efficient transcription of Alu elements is dependent on sequences flanking their site of insertion and the state of Alu methylation (21–25). Although dimeric Alu elements are unique to primates, similar elements, called B1, exist in rodent genomes. B1 elements are monomeric Alu sequences that originated from FLAM and have reached a copy number of 500 000 in the mouse genome (13).
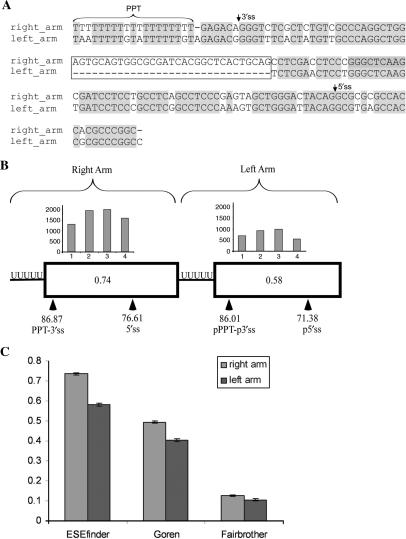
Figure 1.
Characteristics of Alu right arm exons and their counterpart intronic left arms. (A) Alignment of the right and left arm of AluJ consensus sequence (gi551536) in its antisense orientation (relative to the mRNA) using the MAVID alignment server (73). The PPT of the right arm was extended to 19 nt as, on average, the PPT length in exonized Alus is 19 bases ±3 (28) and is marked by horizontal brackets. The major 3′ss and 5′ss that are selected by Alu exons are indicated by arrows. Identical sequences are highlighted in gray. The 31-nt sequence that is present only in the right arm is indicated in a box. (B) Comparison of splice signals in the right and the left arms of 330 Alus that have undergone exonization originating from the right arm in the antisense orientation. Boxes indicate right and left arms. Locations of the real and putative splice sites are marked by arrowheads. Distribution of ESEs, based on ESEfinder, throughout the exon and the putative splice sites in the left arm are shown by a histogram divided into four bins, each of which represents a quartile of the exon length. The number in the center of the boxes represents the mean ESE density score based on ESEfinder. (C) Comparison of ESR densities between the right and left Alu arms. ESR densities are shown for three ESR datasets: ESEfinder (38), Goren et al. (39) and Fairbrother et al. (40).
Approximately 5% of the alternatively spliced internal exons in the human genome (exons that are flanked by an intron on both ends) are derived from Alu elements (26,27). Alu-derived exons originate from intronic Alu sequences by a process denoted ‘exonization’ (13,28–30). Throughout evolution, some of these intronic Alus accumulated mutations that led the splicing machinery to select them as internal exons. Previous studies have indicated that the majority of the Alu-derived exons are alternatively spliced (13,31). This enriches the human proteome with new isoforms without compromising its integrity (32). However, mutations leading to constitutive activation of Alu exons are prone to be deleterious due to the loss of the transcript encoding the original form of the protein and may result in a genetic disorder such as Alport and Sly syndromes or OAT deficiency (33–35).
Alus can undergo insertion into introns in the sense or antisense orientation relative to the coding sequence; in the human genome, 55% are in the antisense and 45% in the sense orientation (13,29). However, the majority of Alu exons (∼85%) originate from Alus in the antisense orientation and in most cases the right arm rather than the left arm is the one selected for exonization (13,26). We have recently shown that the exonization level of Alu elements is almost three times higher than that of other retroelements (e.g. LINE, MIR and CR1) in the human genome and that it is also higher than that of the B1 retroelement (the monomeric form of Alu) in the mouse genome (13). Therefore, retroelements have a greater effect on the human transcriptome in comparison to the mouse transcriptome, due to the exonization of Alu elements as internal exons that are alternatively spliced.
We therefore set out to examine why the right arm of the dimeric Alu element is prone to alternative splicing. Here we show that the left arm, the one that remains intronic, is involved in the splicing regulation of the Alu right arm exon. Deletion of the intronic left arm of the Alu element resulted in exonization that was constitutive; this can be detrimental as the original protein isoform is no longer produced. The effect of the left arm is independent of the sequence of the right arm. Insertion of the left arm downstream of another exon confers upon it alternative splicing. The selective advantage of alternative splicing of Alu exons by keeping the original transcript as a major product is discussed.
MATERIALS AND METHODS
Compilation of right arm exonization dataset
A typical exonization of an Alu element occurs from a right arm in the antisense orientation, as observed for the ADAR Alu exon. We have compiled a dataset of Alu elements that have undergone exonization in the antisense orientation by querying the TranspoGene database (36). This query yielded 744 exonized Alu sequences that overlap with EST sequences. Since a typical Alu is ∼300 nt in length, we next filtered out all Alus shorter than 250 nt, leaving 459 sequences. We used a simple algorithm to detect the border between the two Alu arms: We searched for the first stretch of four consecutive Ts (TTTT) between positions 120 and 200 of the Alu sequence. Visual inspection established that this algorithm correctly found the border between the left and right arms. We next demanded that both the 5′ and 3′ exon borders, as indicated by the supporting ESTs, were upstream of this polyT stretch, thereby creating our initial right arm dataset containing 342 sequences. Finally, we searched for putative splicing signals in the left arm and filtered out 12 cases in which either a putative 5′ss or a putative 3′ss (or both) could not be found in this arm. Our final dataset contained 330 Alu sequences.
The presented scores of the 5′ss and 3′ss were calculated using Analyzer Splice Tool (http://ast.bioinfo.tau.ac.il/SpliceSiteFrame.htm), which is based on procedures described by Shapiro and Senapathy (37). Splicing signals in the left arm were found using the following steps: (i) beginning 8 nt prior to the detected polyT, 30 running windows of 15 nt in length were given 3′ss scores. The value of 8 nt was chosen so as to allow the identified 4-nt polyT stretch to serve as the last four positions preceding the 3′ss AG dinucleotide. The highest scoring 15-nt window was chosen as the 3′ss. (ii) Beginning at a distance of 23-nt downstream to the detected 3′ss until the end of the Alu sequence, running windows of 9 nt were given 5′ss scores. The value of 23 was chosen to force the minimal distance between the 3′ss and the 5′ss to be 25, which is the minimal exon length in Alu right arm. The highest scoring window was chosen as the 5′ss.
ESR densities
We searched for ESRs in right and left arms, between the 3′ss and 5′ss. Three sets of ESRs were used in this analysis: (i) those obtained from the position-specific weight matrices in ESEfinder (38) using the default thresholds; (ii) all ESRs obtained by Goren et al. (39); and (iii) all ESEs obtained by Fairbrother et al. (40). For each right and left Alu arm in each ESR dataset, ESR density scores were calculated. The ESR density score was calculated as the number of nucleotides within a sequence covered by an ESR, divided by the length of the sequence. Mann–Whitney tests were used to compare the distributions of ESR densities between the right and left arms.
Plasmid construction
The desired minigenes were generated by amplifying human genomic fragments using primers containing an additional sequence-encoding restriction enzyme sites. The PCR products were restriction digested and inserted into the pEGFP-C3 plasmid (Clontech), which contains the coding sequence for Green Fluorescent Protein (GFP). The ADAR2 minigene (adenosine deaminase), containing the human genomic sequence of exons 7, 8 and 9 (2.2 kb), and the IMP minigene (IGF-II m-RNA-binding protein) with the human genomic sequence of four constitutive exons 10–13 (2.6 kb) were previously cloned (28,41). For the sequences of the minigene inserts see Supplementary Data.
Minigene mutagenesis
Site-directed mutagenesis was carried out to introduce mutations into the wild-type minigene by PCR using oligonucleotide primers containing the desired mutations. Mutations creating deletions in wild-type minigenes were performed by PCR using 5′ phosphorylated primers flanking the sequence to be deleted (see Supplementary Data for list of primers). PCR was performed using Pfuturbo DNA polymerase (Stratagene) with an elongation time corresponding to 2 min for each kilo base. The PCR products were treated with DpnI (20 U, New England BioLabs) at 37°C for 1 h. Plasmid mutants were ligated using T4 DNA Ligase (New England BioLabs) at 37°C for 2 h. Mutant plasmids were transformed into Escherichia coli XL1-competent cells. DNA was extracted from selected colonies by mini-prep extraction (Promega). All plasmid sequences were confirmed by standard sequencing.
Transfection, RNA isolation and RT–PCR amplification
Culturing of 293T cells in Dulbecco's Modification of Eagle medium was supplemented with 4.5 g/ml glucose (Biological Industries, Inc.), 10% fetal calf serum (FCS), 100 U/ml penicillin, 0.1 mg/ml streptomycin and 1 U/ml nystatin (Biological Industries, Inc.). Cells were cultured in 6-well plates under standard conditions at 37°C in 5% CO2. Cells were grown to 60% confluence and transfection was performed using 3 μl FuGENE6 (Roche) with 1 μg of plasmid DNA. RNA was isolated and harvested after 48 h. Total RNA was extracted using Trizol Reagent (Sigma), followed by treatment with 1 U RNase-free DNase (Ambion). RT–PCR amplification was preformed for 1 h at 42°C, using an oligo dT reverse primer and 2 U reverse transcriptase of avian myeloblastosisvirus (AMV, Roche). The spliced cDNA products derived from the expressed minigenes were detected by PCR. For both minigenes (ADAR2 and IMP), we used the pEGFP-specific reverse primer (5′CGCTTCTAACATTCCTATCCAAGCGT3′) for amplification. For the forward primers, we used an ADAR2 exon 7 primer (5′CCCAAGCTTTTGTATGTGGTCTTTCTGTTCTGAAG3′) and an IMP exon 9 primer (5′AATCTTACTCATGTTACTA3′). Amplification was performed for 28 cycles to maintain a linear relationship between the RNA input and signal (39). Each cycle consisted of 30 s at 94°C, 45 s at 61°C and 1.5 min at 72°C. The RT–PCR products were separated on a 2% agarose gel and confirmed by sequencing. Using the TOPO TA Cloning kit (invitrogen), we found that both the proximal and the distal AG of the Alu right arm exon were selected (28). The relative ratios of RNA products were measured using ImageJ software (http://rsb.info.nih.gov/ij/index.html), as we found previously that ImageJ quantification for ADAR2 RT–PCR products correlates with real-time RT–PCR quantification produced by the Roche LightCycler PCR and detection system (28). The level of mRNA of the housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase, was used as the internal control for each transfection.
RESULTS
Alu exons are stronger in splicing potential than their counterpart intronic arms
The right and left arms of Alu are highly similar, sharing ∼80% sequence identity after excluding the 31-bp insertion in the right arm (Figure 1A). Despite this sequence similarity, most exonizations occur in right arms of Alu elements inserted in the antisense orientation relative to the mRNA precursor. We first set to examine the presence and strength of the splice signals in the left and right arms. We thus compiled a dataset of Alus that have undergone exonization originating from the right arm and searched for potential splicing signals and ESR densities within the left arm. Specifically, we searched for optimal 3′ and 5′ splice sites with a minimum of 25 nt between them, chosen because the shortest Alu exon in this dataset is 25 nt. Following these filtrations, 330 Alus remained in this dataset (see ‘Materials and Methods’ section for details).
We found that the left arms, although not exonized, contained putative PPT-3′ss and putative 5′ss. However, the 5′ and the 3′ splice sites in the exonized right arms were stronger, in a statistically significant manner, than in the nonexonized counterpart (Figure 1B). Based on the Shapiro and Senapathy scoring method (42), the mean 3′ss in the right arm exon was 86.87, compared to 86.01 in the left arm (Mann–Whitney, P = 0.0485). The mean 5′ss in the right arm exon was 76.71, compared to 70.99 in the left arm (Mann–Whitney, P = 2.7e–019). Using different scoring methods for measuring the strength of the splice sites did not alter our conclusions.
We next compared the ESR densities in the two Alu arms. In this analysis we used three different sets of ESRs: (i) those obtained from the position-specific weight matrices in ESEfinder using the default thresholds (38); (ii) all ESRs compiled by Goren et al. (39); and (iii) all ESEs obtained by Fairbrother et al. (40). For each right and left Alu arm in each of the three ESR datasets, we calculated the ESR densities. The ESR density score was calculated as the number of nucleotides within a sequence covered by an ESR, divided by the length of the sequence. Figure 1C shows that all three analyses revealed that the right arm was enriched with ESRs compared to the left arm. Namely, the ESR densities of ESEfinder, Goren et al. and Fairbrother et al. were 0.74, 0.49 and 0.13, respectively, in the right arm, but only 0.58, 0.40 and 0.11 in the left arm, respectively, with these differences being highly significant (Mann–Whitney, P ≈ 0, P ≈ 0 and P = 6.43e–004, respectively). To examine whether any of the two Alu arms has a better potential to serve as an intron, we compiled a dataset of 10 experimentally validated binding sites, comprising binding sites of nine hnRNP proteins and Fox1 protein (extracted from http://ast.bioinfo.tau.ac.il/ESR.htm). We found a significantly higher density in the left arm compared to the right arm (density in right arm: 0.0531, density in left arm: 0.0628; Mann–Whitney P-value: 4.11e–5). These findings may suggest that not only does the right arm have a better potential to serve as an exon, the left arm is more suitable to serve as an intron, as well. However, comparison of the densities of ISRs in both Alu arms based on two computationally predicted datasets of Yeo et al. (43) and Voelker et al. (44) yielded contradictory results: the former indicated higher ISR densities in the right arm, while according to the latter the ISR density was higher in the left arm (data not shown).
These results indicate that the left arm has a lower potential for exonization, as these arms are characterized by weaker splice sites and lower ESR densities than the exonized right arms. Thus, in cases of Alu exonizations, the right arm Alu exon is accompanied by an intronic Alu arm that has the characteristics of an exon (potential splice sites) but is a weaker candidate for exonization.
The intronic left arm of the Alu element promotes alternative splicing of upstream exons
To determine whether the intronic left arm of the Alu elements has a role in regulating the exonization of the right arm we deleted the entire intronic left arm of an Alu element in ADAR2 (adenosine deaminase) minigene. Exon 8 of the ADAR2 gene is an Alu exon that is alternatively spliced. This exon adds 40 amino acids in frame to the protein (45). Exon 8 of the ADAR2 gene is representative of most Alu exonizations events reported to date, that is, exonizations of the right arm of an antisense Alu. Also, the left arm of this Alu element is generally weaker than the right exonized arm in terms of splice site strengths and ESR density, although the 5′ss of the exonized arm is weaker than that of the unexonized arm (Figure 2A). We therefore generated a minigene containing exons 7–9, and the introns in between, of the human ADAR2 gene (13,26). The minigene was transfected into 293T cells, total cytoplasmatic RNA was extracted and the splicing pattern was examined by RT–PCR using specific primers to the minigene mRNA.
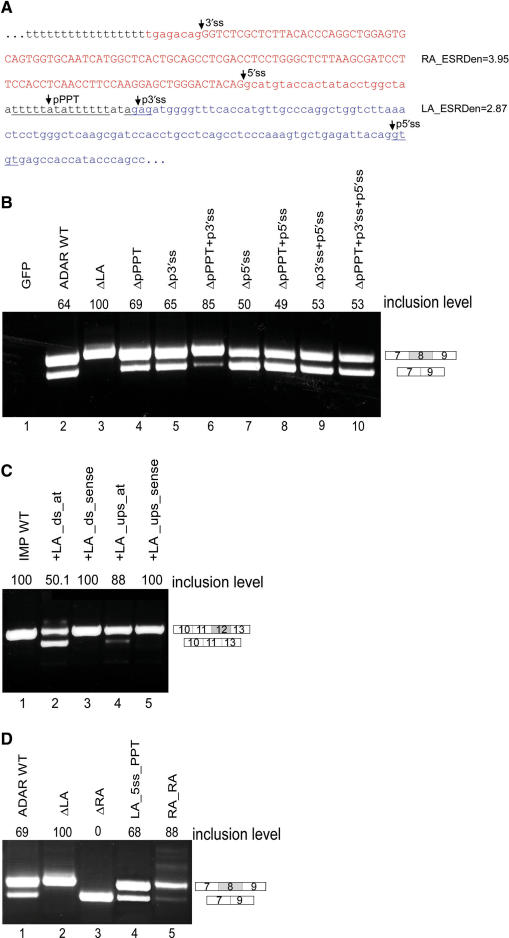
Figure 2.
The intronic left arm of the Alu element is essential for the alternative splicing of an upstream exon. (A) The sequence of an Alu retroelement in its antisense orientation from which exon 8 of ADAR2 is exonized. The right arm (in red) undergoes exonization while the left arm (in blue) is intronic. The potential PPT, 3′ss, and 5′ss in the left arm (unexonized) are underlined. ESE densities (according to ESEfinder) for each arm are indicated on the right. (B) Plasmids containing the indicated mutants were transfected into 293T cells. Total cytoplasmic RNA was extracted, and splicing products were separated in 2% agarose gel after reverse transcription polymerase chain reaction (RT–PCR). Lane 1, vector only (pEGFP); lane 2, splicing products of wild-type ADAR2; and lanes 3–10, splicing products of ADAR2 minigene mutated at the left arm of the Alu element in different combinations. ΔLA: deletion of the entire intronic left arm; ΔpPPT: deletion of the 14-bp pPPT sequence of the left arm; Δp3′ss: elimination of two potential 3′ss in the intronic left arm (AGAG to ACCG); Δp5′ss: elimination of two potential 5′ss in the intronic left arm (GTGT to GAAT). The two possible minigene mRNA isoforms are shown on the right. The numbers above the lanes indicate exon 8 inclusion level. (C) Insertion of an intronic left arm of an Alu element downstream to a constitutive exon results in a shift toward alternative splicing. The sequence of the intronic left arm of the Alu element, from which exon 8 of ADAR2 gene undergoes exonization (in its antisense orientation), was cloned downstream to constitutive exon 12 in the IMP minigene. The splicing assay was performed as described above. Lane 1, splicing product of wild-type IMP; lanes 2–3, splicing products of IMP minigene in which the intronic left arm of the Alu element from ADAR2 was inserted downstream to exon 12 in antisense and sense orientations, respectively; lanes 4–5, splicing products of IMP minigene in which the intronic left arm of the Alu element from ADAR2 was inserted upstream to exon 12 in antisense and sense orientations, respectively. The two possible minigene mRNA isoforms are shown on the right. The numbers above the lanes indicate exon 12 inclusion levels. (D): Exonization of the intronic left arm. Splicing assays were performed as described above. Lane 1, splicing products of wild-type ADAR2; lane 2, splicing products of ADAR2 mutant harboring a deletion of the intronic left arm; lane 3, splicing products of ADAR2 mutant harboring a deletion of the exonized right arm; lane 4, splicing products of ADAR2 mutant after strengthening of the p5′ss and pPPT of the intronic left arm; lane 5, splicing products of ADAR2 mutant harboring a replacement of the intronic left arm with the sequence of an additional right arm. The two minigene mRNA isoforms are shown on the right and were confirmed by sequencing.
The wild-type Alu exon was included in 64% of the transcripts (Figure 2B, lane 2). However, deletion of the intronic left arm resulted in a shift from alternative to constitutive inclusion of the Alu exon (Figure 2B, compare lanes 2 and 3). This suggests that the left arm in the antisense orientation is essential for alternative selection of the right arm.
To examine if the left arm of the Alu element promotes alternative splicing, we inserted it downstream of a non-Alu exon, exon 12 of IMP, that is known to be constitutively spliced (41). Remarkably, this insertion resulted in a shift from constitutive to alternative splicing of this exon (Figure 2C, compare lanes 1 and 2). As a control, the sequence was inserted in the sense orientation and had no effect on splicing of exon 12 (Figure 2C, lane 3). This indicates that the sequence of the left arm itself, and not a specific sequence that was interrupted by the insertion, was responsible for the shift from constitutive to alternative splicing. Insertion of the left arm upstream of the constitutive exon had a much weaker effect on the pattern of splicing and caused a slight shift from constitutive to alternative splicing. This slight effect was abolished when the sequence was inserted in the sense orientation (Figure 2C, lanes 4 and 5). Taken together, these results imply that the left arm of the Alu element in the antisense orientation, when located downstream of an exon, enhances alternative selection of the upstream exon. This allows expression of both the original transcript as well as that including the Alu exon, thus providing an advantage for Alu elements, making them more favorable candidates for exonization (see ‘Discussion’).
To identify sequence determinants in the intronic left arm that are responsible for the shift from constitutive to alternative splicing of the Alu right arm exon, we began by eliminating the three main potential splice signals within the intronic left arm: the putative PPT (pPPT), putative 3′ss (p3′ss) and putative 5′ss (p5′ss) (Figure 2A). The mutations were made such that the two consecutive potential 3′ss and 5′ss that exist in the left arm were eliminated (AGAG to ACCG and GTGT to GAAT; see Supplementary Data for primers). Deletion of the pPPT sequence or the elimination of the two potential 3′ss in the intronic left arm (AGAG to ACCG) had a minor effect on the level of alternative splicing of the Alu exon (Figure 2B, lanes 4 and 5). However, the combination of both mutations resulted in a strong shift toward constitutive inclusion (Figure 2B, lane 6). Mutations of the p5′ss alone or in combination with other putative splicing signals yielded puzzling, yet consistent results. Mutation of the p5′ss consistently did not affect the alternative selection of the upstream Alu exon. Moreover, the shift toward constitutive splicing that was demonstrated by combined mutations of the pPPT and p3′ss was abolished when we further mutated the p5′ss, leading to alternative splicing (Figure 2B, lanes 7–10). The above results are suggestive of a complex regulatory role for the intronic left arm. The most dramatic effect occurred when the potential pPPT and p3′ss were both eliminated. However, even in this case the inclusion rate increased only to 85% and not to the 100% inclusion rate as obtained after deletion of the entire left arm, suggesting that additional sequences along the left arm are also involved in the regulation of alternative splicing preventing constitutive inclusion of the Alu right arm exon.
The intronic left arm can exonize
The high sequence similarity between the two arms (Figure 1A) led us to examine whether the intronic left arm can undergo exonization. Deletion of the exonized right arm did not bring about exonization of the intronic left arm (Figure 2D, lane 3). This indicates that even in the absence of the competing right arm, the splicing signals in the left arm are insufficient to direct splicing. In the majority of pathological pseudo-exon inclusion events, a 5′ss creation or activation occurs (46). We thus hypothesized that a stronger 5′ss would be less sensitive to the negative context that prevents the left arm from being exonized (47). We strengthened the p5′ss of the left arm by mutating it from CAG/gtgtgt to CAG/gtaagt (a U1/5′ss ΔG change from −4.6 to −8.2). Strengthening the p5′ss alone was not sufficient to switch Alu exon selection from the right to the left arm (data not shown). Previous studies have shown that pseudo-exon activation or Alu exonization are not achieved by the combined strengthening of the 5′ss and 3′ss alone (48,49). We therefore strengthened both the pPPT (to 17 consecutive uridines, similar to the functional PPT of the right arm) and the p5′ss. These combined changes enabled selection of the left arm rather than the right arm and facilitated exclusive, albeit alternative, exonization of the left arm (Figure 2D, lane 4; RT–PCR products were confirmed by sequencing and revealed that the proximal p3′ss, TAG, was selected). This exonization did not occur when the pPPT was strengthened alone (data not shown). This result suggests that the intronic left arm is a few steps away from splicing competence, and thus might act as a pseudo-exon that competes with the right arm.
Our bioinformatic analysis showed that in most cases of Alu exonizations, the Alu exon is accompanied by an Alu arm that has the characteristics of a weak exon. To examine whether the splicing signals in the right arm must be stronger than those in the left arm for alternative exonization of the Alu exon, we substituted the intronic left arm with a right arm, thus producing a mutant minigene in which two identical arms of equal strength were located one after the other. We inserted a point mutation in right arm duplicate (the left position) in order to identify by sequencing which arm underwent exonization (see Supplementary Data). Competition between two exons resulted in a reduction in the total amount of mature mRNA (Figure 2D, lane 5). Sequencing of the PCR products revealed that each exon was selected separately, with a slight preference for the downstream exon.
The intronic left arm of the Alu element functions as a unit
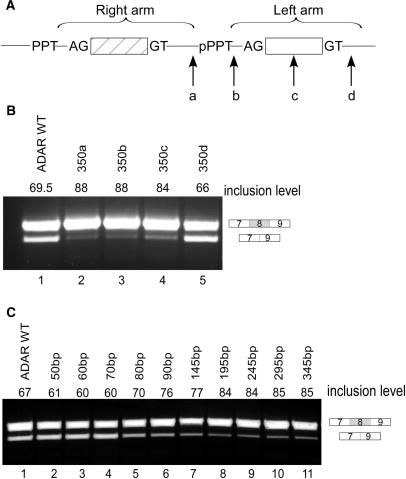
We then examined the effect of the distance between the two arms on alternative selection of the right arm. We inserted a 350-bp sequence originating from intron 11 of the IMP gene into four different positions: (a) between the exonized right arm and the intronic left arm; (b) between the pPPT and the p3′ss of the intronic left arm; (c) in the center of the intronic left arm; and (d) downstream of the intronic left arm (Figure 3A). Insertion into site ‘a’ separates the left from the right arm as a complete unit; insertion into sites ‘b’ and ‘c’ leaves part of the intronic left arm in the same distance from the Alu exon and separates only the downstream parts of the left arm from the right while insertion into site ‘d’ leaves the left arm intact. Insertion of a 350-bp fragment into sites ‘a’, ‘b’ or ‘c’ resulted in higher levels of exon inclusion than in the wild-type construct (Figure 3B, lanes 2–4), whereas insertion downstream of the left arm (site d) had a minor effect, if any, on splicing (Figure 3B, lane 5).
Figure 3.
Increasing the distance between the left and right arms of the Alu element results in a shift toward constitutive splicing. (A) Illustration of the Alu element in its antisense orientation. The right and left arms are marked by horizontal brackets. An intronic sequence of 350 bp was inserted in four different positions along the intronic Alu's left arm: (a) between the exonized right arm and the intronic left arm; (b) between the pPPT and the p3′ss of the intronic left arm; (c) in the center of the intronic left arm; (d) downstream of the intronic left arm. (B) Splicing assays were performed as described in Figure 2. Lane 1, splicing products of wild-type ADAR2; lanes 2–5, splicing products of ADAR2 minigene mutated as indicated in A. (C) Intronic sequences of growing lengths (10–300 bp) were inserted between the exonized right arm and the intronic left arm. Lane 1, splicing products of wild-type ADAR2; lanes 2–11, splicing products of ADAR2 minigene mutated as indicated.
The two arms are separated by a sequence of 40 bp (including the pPPT of the left arm). To further evaluate the importance of the distance between the two arms for the splicing of the right arm, we increased this distance by inserting sequences of growing lengths between the right and left arms. We used ESEfinder to ensure that the inserted sequence did not contain putative ESEs [(38) see also Supplementary Data]. As shown in Figure 3C, lanes 1–4, increasing the distance between the two arms from 40 to 70 bp had a minor effect on the inclusion level. However, when the distance between the two arms was 80 bp or longer there was a shift toward higher levels of inclusion (Figure 3C, lanes 5–11). We thus concluded that the effect of the intronic left arm on the splicing of the right arm is distance dependent. Once the distance between the two arms increased to over 70 bp, the effect of the left arm on the inclusion level of the right arm decreased considerably. Taken together, the data in Figures 2 and 3 indicate that the intronic left arm functions as a unit and different elements along the left arm must be present within a certain distance from the right arm to promote alternative splicing.
The effect of ESEs located in the left arm on exonization of the right arm
Mutations in the three main splice signals (pPPT, p3′ss and p5′ss) located in the left arm were not sufficient to induce constitutive splicing of the right arm, whereas deletion of the entire left arm shifted splicing from alternative to constitutive (Figure 2B). This implies that other sequences in the left arm affect splicing of the right arm.
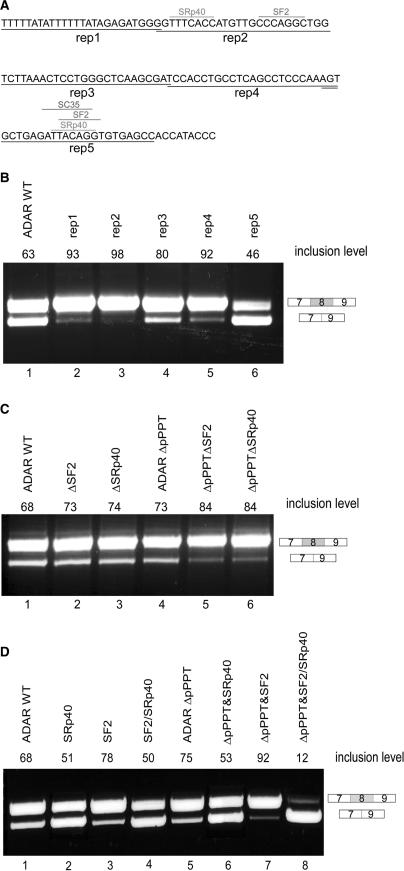
Previous analysis which compared the full-length Alu sequence of right-arm exonizing Alus to nonexonizing Alus did not reveal any regulatory positions along the intronic left arm (29). To identify such sequences, we systematically replaced segments of 25 bp along the left arm with a 25-bp sequence that did not contain any potential splice signals or SR-binding sites (Figure 4A; shown by underlined reps 1–5; see also Supplementary Data). Replacement of segments 1, 2, 3 and 4 resulted in a higher level of inclusion of the right arm than observed in the wild-type construct (Figure 4B, lane 3, 4 and 6, respectively), whereas replacement of the last segment, which contains the p5′ss, was similar to mutation of the p5′ss alone (compare Figure 4B, lane 6 with Figure 2B, lane 7). Thus it appears that the elements involved in regulation of the splicing of the Alu right arm exon are spread throughout the entire intronic left arm.
Figure 4.
The effect of pseudo splice signals and putative SR-binding sites within the left arm on the splicing pattern of the right arm. (A) Segments along the left arm 25 bp in length (rep1–rep5) were replaced with a designed sequence that did not contain any potential splice signals or SR-binding sites. The examined putative ESRs are shown. (B) Splicing assays were performed as described in Figure 2. Lane 2, splicing products of wild-type ADAR2; lanes 2–6, splicing products of ADAR2 minigene with replaced segments (rep1–rep5). (C) Lane 1, splicing products of wild-type ADAR2; lanes 2–6, splicing products of ADAR2 minigene mutated at the indicated binding sites in segments 2, with and without deletion of pPPT. (D) Changes in the binding capacities of putative SR proteins in segment 5 (near the potential 5′ss of the intronic left arm). Lane 1, splicing products of wild-type ADAR2; lanes 2–8, splicing products of ADAR2 minigene with the indicated mutations (see Supplementary Data).
To determine if the potential binding sites of SF2, SRp40 located in segments 2 are involved in alternative splicing of the Alu exon, we mutated each of the indicated sites. Figure 4A illustrates the locations of the putative binding sites along the intronic left arm that were mutated (for a list of all ESRs that are present along the left arm see Table S2 in the Supplementary Data). Elimination of the potential SF2 and SRp40 sites in segment 2, without creating a new putative SR-binding sites (according to ESEfinder) or putative ESR sequences [according to Goren et al. (39), Fairbrother et al. (40), Wang et al. (50) and Zhang et al. (51)], did not affect splicing (Figure 4C, lanes 2 and 3). However, when these mutations were combined with a deletion of the pPPT, which by itself had a minor effect on the splicing of the Alu exon (see Figure 2B, lane 4), there was an increase in the level of the Alu exon inclusion (Figure 4C, lanes 5 and 6). These results indicate that some of the potential SR-binding sites are functional ISRs that affect splicing of the upstream exon when combined with a deletion of the pPPT.
In segment 5 there are three overlapping potential binding sites for SF2 and SRp40 and SC35 located immediately upstream of the potential 5′ss. A high concentration of ESRs upstream of a 5′ss has been shown to be important for splicing and exon selection (39,52,53). Since the above results indicated that this region was involved in the regulation of the exonization of the upstream exon, we subjected the ESRs in this region to further analysis. We designed mutations that affected the ESRs but not the p5′ss (CAG/GTGTGA). Mutation that eliminated SC35 alone had no effect on the splicing pattern of the Alu right arm exon (data not shown). We therefore focused on SF2 and SRp40 putative binding sites. We created two mutant minigenes that harbor only SF2 or only SRp40-binding sites near the p5′ss with scores different from those in the wild-type sequence and an additional mutant minigene in which the SRp40-binding site was weaker and the SF2 binding was stronger than the original potential binding sites (see Supplementary Data). Strengthening the SF2-binding site alone resulted in a minor elevation of the inclusion level of the Alu exon (Figure 4D, lane 3). This effect was elevated when the mutation was combined with the deletion of the pPPT (Figure 4D, lane 7). When the binding affinity of SF2 site was strengthened and the potential SRp40 site was weakened, the right arm exon was more frequently skipped than in the wild-type construct (Figure 4D, lane 4). This effect was further enhanced when the mutation was combined with the deletion of the pPPT (Figure 4D, lane 8). In contrast, a mutation that slightly decreased the binding affinity of the SRp40 site, with and without deletion of the pPPT, had no effect on alternative splicing of the Alu exon (Figure 4D, lanes 2 and 6). These results further support our observations that ISRs in the intronic left arm are functional, although unlike those in segment 2, the exact roles of the putative SF2 and SRp40 sites in enhancing or repressing splicing of the Alu exon were not conclusive. Altogether, these results show an interplay between the pPPT and the functional ISRs.
We also examined whether the 31-bp sequence that is absent in the left arm (see Figure 1A) can affect Alu exon selection or the pattern of splicing of the Alu exon. Addition of this sequence to the left arm in a comparable position to that in the right and deletion of it from the right arm did not switch the Alu exon selection from the right arm to the left arm nor did it affect the pattern of splicing of the Alu exon (see Supplementary Data Figure 1S, lanes 2–4).
DISCUSSION
We have recently shown that the exonization level of Alu elements is significantly higher compared with other retroelements in the human genome. In addition, we have also shown that the exonization level of Alu elements is almost three times higher than that of B1 retroelement (the monomeric form of Alu) in the mouse genome even though they share high levels of sequence similarity and the same ancestral origin (7SL RNA) (13). Although Alu elements are not enriched with pseudo-splice sites (13,54), they exhibit a higher exonization rate compared to other repetitive elements in the human genome. This raises the intriguing question of what makes Alu elements more potent for exonization. In this report we set out to examine whether the high exonization capability of Alu elements, which is mostly observed in the antisense orientation (13,26), can be attributed to their unique dimeric structure.
The first indication that the nonexonized arm of Alu maintains alternative rather than constitutive exon selection was observed upon its deletion, leading to a shift from alternative to constitutive Alu exon selection (Figure 2B). Moreover, when we placed the nonexonized arm downstream to a constitutive exon, this exon was endowed with alternative splicing. Taken together, these results demonstrated that the left arm in the antisense orientation enabled alternative rather than constitutive selection of an upstream exon. In the case of newly emerged Alu exons, this observation is most significant since selection of an Alu exon in a constitutive manner will generate only a new transcript that may be deleterious, by causing a frameshift or introducing a premature stop codon, as documented in OAT deficiency and Alport and Sly syndromes (33–35). However, splicing of an Alu exon in an alternative manner can make such an exonization tolerable since it allows the new variant to be evolutionarily tested while the original transcript is expressed. Indeed, most of the Alu exons maintain a low inclusion level, thus maintaining the evolutionary conserved isoform as the major mRNA template generated from the gene (13,31). In this way, alternative splicing of Alu exons allows to increase transcriptomic diversity with new isoforms without compromising their original gene function as would occur if Alu exons were exonized constitutively. For this reason, newly born exons, and specifically newly born Alu exons, are almost always alternatively spliced (13,32,55,56).
A large-scale analysis of transposed elements in human and mouse genomes revealed a 3-fold higher exonization level of Alu elements in human, with respect to their murine counterpart B1 (13). The most prominent feature distinguishing Alu from B1 elements is the dimeric structure of Alu, as opposed to the monomeric structure of B1. Our results provide a possible explanation for the higher exonization rate found in human, namely that the intronic left arm of the Alu element facilitates alternative rather than constitutive splicing of the exonic right arm. This would decrease the evolutionary selection pressure against Alu exons, thus enabling a nondeleterious enrichment of the transcriptome.
By what mechanism does the left arm enable alternative splicing of the upstream exon? One possible explanation is that this region forms a secondary structure that impairs the ability of the splicing machinery or other regulatory proteins to correctly identify the exon (57,58). Alu RNAs are known to have a distinct secondary structure (59). Indeed, we found that deletion of the left arm caused considerable change in the minimal free energy of the predicted secondary structure of the ADAR Alu according to RNAfold program [(60) see Supplementary Data, Table S3]. However, we could not find a general correlation between strength of predicted secondary structure and splicing patterns. For example, upon insertion of the Alu left arm downstream of the IMP exon in either sense or antisense orientation, the predicted minimal free energy of the region decreased considerably. However, the splicing pattern of the IMP exon following insertion of left arm in sense or antisense orientations was completely different (constitutive versus alternative, respectively). We also visually examined the predicted secondary structures of the various mutants and could not observe a consistent correlation between changes in secondary structures and in splicing patterns. These results imply that while the secondary structure of the left arm of the Alu element may play a role in mediating alternative splicing of the IMP exon, it is not an exclusive factor.
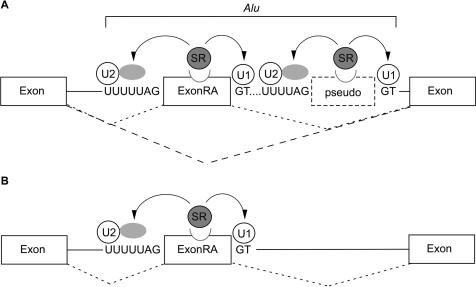
Another possible explanation for the ability of the left arm to weaken the selection of the Alu exon, manifested by the shift towards alternative splicing, is that the left arm acts as a ‘pseudo-exon’ competing with the right arm for spliceosome components. This model was motivated by results showing that tandemly duplicated exons are mostly spliced in a mutually exclusive fashion. Work by Letunic et al. (61) showed that when two exons are located in close proximity, one exon suppresses the selection of the other. Indeed, our bioinformatic analysis showed that the exonized arm tends to be characterized by stronger splicing signals than the intronic counterpart. These results are in line with the finding of Sironi et al. (54) who showed that pseudo-exons display, on average, weaker splice site consensuses and lower ESE frequencies than real exons and are suggestive of such a model in which the exonized (in our case right) arm contains sufficient signals required for its selection, while the nonexonized arm (in this case left) contains most, but not all, splicing signals. Figure 5 illustrates this possible model of competition between the two arms: When the Alu right arm exon competes with a weaker ‘pseudo-exon’ for spliceosome components, it reduces selection of the right arm as an exon and the right arm is alternatively spliced; however, when the Alu right arm exon is free of left arm regulation it is constitutively spliced.
Figure 5.
Why are Alu exons alternatively spliced? (A) The splicing pattern of exonization events originating from the right arm of Alu elements (ExonRA) is alternative due to the presence of a downstream arm that presumably acts as a pseudo-exon. Thus, competition between two highly similar arms shifts the splicing pattern of the stronger arm to alternative. Although the weaker left arm does not undergo exonization, it nonetheless participates in the regulation of the stronger right arm by reducing the affinity of the splicing machinery for the exonized arm, thereby causing it to be alternatively spliced. (B) However, when the exonized right arm (ExonRA) is free of left arm regulation it is constitutively spliced. Such events are presumably deleterious, and therefore may be selected against.
Our findings, however, indicate that a simple model of competition cannot fully account for the regulation exerted by the left arm on the alternative splicing of the Alu exon. This was shown when all three main potential splice signals in the left arm were mutated, and the alternative splicing pattern of the Alu exon was unaffected. However, a combination of mutations of the pPPT and the p3′ss had a dramatic effect on splicing, shifting exon selection from alternative to almost constitutive inclusion. This almost constitutive inclusion was reversed to alternative splicing when pPPT and p3′ss mutations were combined with a p5′ss mutation. This combination between two sites to exert an effect on splicing that was stronger than the effect of each splice signal alone was also observed when the putative SR-binding sites in the left arm were mutated in combination with the pPPT. This indicates that the shift toward constitutive splicing observed upon deletion of the entire intronic left arm was not due to the effect of each splice signal alone but rather due to interplay among several signals located along the left arm.
The synergistic activity of the PPT and the 3′ss is not surprising. Indeed, in the course of the splicing reaction, U2AF65 and U2AF35 bind cooperatively to the polypyrimidine tract and to the 3′ss AG dinucleotide, respectively (62,63). This process is highly regulated and recent work by Soares et al. (64) demonstrated that phosphorylated DEK, a chromatin and RNA associated protein, facilitates 3′ss recognition by U2AF35 and prevents binding of U2AF65 to pyrimidine tracts not followed by AG. Thus, our results provide a further example for the joint function of the PPT and the 3′ss.
How does the PPT mediate its effect? It has been reported that pyrimidine-rich motifs have an inhibitory effect on splicing in the human genome (65). This, however, was shown to be the case when the pyrimidine-rich motifs were inserted into a central exon of a three-exon minigene model, whereas in our case the pPPT is located downstream of the affected exon. PPTs, located in introns, are also known to regulate inclusion levels of upstream exons. Different proteins are known to bind such intronic PPTs, including U2AF65, TIA-1 and PTB (66–68). U2AF and TIA-1 both promote the recruitment of U1 snRNP to weak 5′ss that are followed by uridine-rich sequences, whereas PTB is known to antagonize the action of TIA-1 and U2AF and induce exon skipping (67,69–71). In the Alu exon, the pPPT of the intronic left arm appears downstream of the 5′ss. This raises the possibility that factors such as U2AF65, TIA-1 and PTB may play a role in splicing regulation of the Alu exon. In this context, two points are noteworthy: (i) The pPPT does not contain a consensus PTB-binding site [consensus UCUU/C within a pyrimidine-rich context (66,72)]. However, due to the degeneracy of the PTB-binding sites, PTB may nonetheless be involved in the Alu exon regulation. (ii) The effect of U2AF and TIA-1 on 5′ss usage has been reported in cases in which the PPT was located immediately downstream of the 5′ss (69,70). However, interaction of TIA-1 was also shown to occur even when the PPT was more than 30-bp downstream of the 5′ss (71). A secondary structure formed in the pre-mRNA or involvement of other splicing factors may facilitate the action of TIA-1 or the RS domain of U2AF65 in cases in which the PPT is not located immediately after the 5′ss. In the present case of Alu exonization, we find it very unlikely that the pPPT functions solely to mediate binding of TIA-1 or PTB; rather it is more likely to function in conjunction with the downstream p3′ss to facilitate pseudo-exon recognition, in which the downstream p5′ss and several SR-binding sites are involved.
We also showed that when two identical arms are located adjacent to each other and both have the potential to exonize, the overall effect is to decrease the selection of the exon and the levels of cytoplasmic mRNA. In our assay, either exon could be selected with a slight preference for the downstream exon. Thus, competition between two active exons of identical strength presumably disrupts mRNA splicing. A similar mechanism was suggested for mutually exclusive spliced exons that originate from exon duplication, in which only one exon is selected within a given mRNA, but never are both spliced into the mRNA (61). Our results experimentally validate this hypothesis and further imply that such competition between adjacent, identical exons imposes a problem for the splicing machinery and presumably leads to lower levels of splicing and hence to lower levels of mature mRNA.
To summarize, we have shown that the right arm Alu exons are accompanied by weaker competitors on the left arm and that the dimeric structure of the Alu element enables alternative rather then constitutive splicing of the Alu exon, thereby leading to the enrichment of the human transcriptome with new isoforms while preserving the original transcriptomic repertoire.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank Prof. David Givol, Eddo Kim, Hadas Keren and Noa Sela for reading the manuscript. This work was supported by a grant from the Israel Science Foundation (1449/04 and 40/05), MOP Germany-Israel, GIF and DIP. S.S. is a fellow of the Edmond J. Safra bioinformatics program at Tel Aviv University. Funding to pay the Open Access publication charges for this article was provided by DIP.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brett D, Pospisil H, Valcarcel J, Reich J, Bork P. Alternative splicing and genome complexity. Nat. Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- 2.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 5.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 6.Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell. Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 7.Brow DA. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 8.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 9.Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 10.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell. Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 12.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, Ast G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quentin Y. Emergence of master sequences in families of retroposons derived from 7sl RNA. Genetica. 1994;93:203–215. doi: 10.1007/BF01435252. [DOI] [PubMed] [Google Scholar]
- 15.Quentin Y. A master sequence related to a free left Alu monomer (FLAM) at the origin of the B1 family in rodent genomes. Nucleic Acids Res. 1994;22:2222–2227. doi: 10.1093/nar/22.12.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grover D, Mukerji M, Bhatnagar P, Kannan K, Brahmachari SK. Alu repeat analysis in the complete human genome: trends and variations with respect to genomic composition. Bioinformatics. 2004;20:813–817. doi: 10.1093/bioinformatics/bth005. [DOI] [PubMed] [Google Scholar]
- 17.Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Mighell AJ, Markham AF, Robinson PA. Alu sequences. FEBS Lett. 1997;417:1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- 19.Fuhrman SA, Deininger PL, LaPorte P, Friedmann T, Geiduschek EP. Analysis of transcription of the human Alu family ubiquitous repeating element by eukaryotic RNA polymerase III. Nucleic Acids Res. 1981;9:6439–6456. doi: 10.1093/nar/9.23.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willis IM. RNA polymerase III. Genes, factors and transcriptional specificity. Eur. J. Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 21.Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- 22.Khanam T, Rozhdestvensky TS, Bundman M, Galiveti CR, Handel S, Sukonina V, Jordan U, Brosius J, Skryabin BV. Two primate-specific small non-protein-coding RNAs in transgenic mice: neuronal expression, subcellular localization and binding partners. Nucleic Acids Res. 2007;35:529–539. doi: 10.1093/nar/gkl1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li TH, Schmid CW. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001;276:135–141. doi: 10.1016/s0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 24.Makalowski W, Mitchell GA, Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 25.Muller J, Benecke BJ. Analysis of transcription factors binding to the human 7SL RNA gene promoter. Biochem. Cell. Biol. 1999;77:431–438. [PubMed] [Google Scholar]
- 26.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gotea V, Makalowski W. Do transposable elements really contribute to proteomes? Trends Genet. 2006;22:260–267. doi: 10.1016/j.tig.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 29.Sorek R, Lev-Maor G, Reznik M, Dagan T, Belinky F, Graur D, Ast G. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 30.Hasler J, Strub K. Alu elements as regulators of gene expression. Nucleic Acids Res. 2006;34:5491–5497. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorek R. The birth of new exons: Mechanisms and evolutionary consequences. RNA. 2007;13:1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 33.Knebelmann B, Forestier L, Drouot L, Quinones S, Chuet C, Benessy F, Saus J, Antignac C. Splice-mediated insertion of an Alu sequence in the COL4A3 mRNA causing autosomal recessive Alport syndrome. Hum. Mol. Genet. 1995;4:675–679. doi: 10.1093/hmg/4.4.675. [DOI] [PubMed] [Google Scholar]
- 34.Vervoort R, Gitzelmann R, Lissens W, Liebaers I. A mutation (IVS8 + 0.6kbdelTC) creating a new donor splice site activates a cryptic exon in an Alu-element in intron 8 of the human beta-glucuronidase gene. Hum. Genet. 1998;103:686–693. doi: 10.1007/pl00008709. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell GA, Labuda D, Fontaine G, Saudubray JM, Bonnefont JP, Lyonnet S, Brody LC, Steel G, Obie C, Valle D. Splice-mediated insertion of an Alu sequence inactivates ornithine delta-aminotransferase: a role for Alu elements in human mutation. Proc. Natl Acad. Sci. USA. 1991;88:815–819. doi: 10.1073/pnas.88.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levy A, Sela N, Ast G. TranspoGene and microTranspoGene: transposed elements influence on the transcriptome of seven vertebrates and invertebrates. Nucleic Acids Res. 2007;36:D47–52. doi: 10.1093/nar/gkm949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences–The complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 41.Kol G, Lev-Maor G, Ast G. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum. Mol. Genet. 2005;14:1559–1568. doi: 10.1093/hmg/ddi164. [DOI] [PubMed] [Google Scholar]
- 42.Carmel I, Tal S, Vig I, Ast G. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004;10:828–840. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeo GW, Nostrand EL, Liang TY. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voelker RB, Berglund JA. A comprehensive computational characterization of conserved mammalian intronic sequences reveals conserved motifs associated with constitutive and alternative splicing. Genome Res. 2007;17:1023–1033. doi: 10.1101/gr.6017807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai F, Chen CX, Carter KC, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol. Cell. Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buratti E, Dhir A, Lewandowska MA, Baralle FE. RNA structure is a key regulatory element in pathological ATM and CFTR pseudoexon inclusion events. Nucleic Acids Res. 2007;35:4369–4383. doi: 10.1093/nar/gkm447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson KK, Green MR. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988;2:319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- 48.Lei H, Day IN, Vorechovsky I. Exonization of AluYa5 in the human ACE gene requires mutations in both 3′ and 5′ splice sites and is facilitated by a conserved splicing enhancer. Nucleic Acids Res. 2005;33:3897–3906. doi: 10.1093/nar/gki707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun H, Chasin LA. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 51.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fairbrother WG, Holste D, Burge CB, Sharp PA. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004;2:E268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Majewski J, Ott J. Distribution and characterization of regulatory elements in the human genome. Genome Res. 2002;12:1827–1836. doi: 10.1101/gr.606402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sironi M, Menozzi G, Riva L, Cagliani R, Comi GP, Bresolin N, Giorda R, Pozzoli U. Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res. 2004;32:1783–1791. doi: 10.1093/nar/gkh341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alekseyenko AV, Kim N, Lee CJ. Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes. RNA. 2007;13:661–670. doi: 10.1261/rna.325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang XH, Chasin LA. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc. Natl Acad. Sci. USA. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA Secondary Structures Influence Exon Recognition. PLoS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinnett D, Richer C, Deragon JM, Labuda D. Alu RNA secondary structure consists of two independent 7 SL RNA-like folding units. J. Biol. Chem. 1991;266:8675–8678. [PubMed] [Google Scholar]
- 60.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Montash. Chem. 1994;125:167–188. [Google Scholar]
- 61.Letunic I, Copley RR, Bork P. Common exon duplication in animals and its role in alternative splicing. Hum. Mol. Genet. 2002;11:1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- 62.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 63.Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- 64.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 65.Fairbrother WG, Chasin LA. Human genomic sequences that inhibit splicing. Mol. Cell. Biol. 2000;20:6816–6825. doi: 10.1128/mcb.20.18.6816-6825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 67.Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P, Valcarcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 68.Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Forch P, Merendino L, Martinez C, Valcarcel J. U2 small nuclear ribonucleoprotein particle (snRNP) auxiliary factor of 65 kDa, U2AF65, can promote U1 snRNP recruitment to 5′ splice sites. Biochem. J. 2003;372:235–240. doi: 10.1042/BJ20021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zuccato E, Buratti E, Stuani C, Baralle FE, Pagani F. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 2004;279:16980–16988. doi: 10.1074/jbc.M313439200. [DOI] [PubMed] [Google Scholar]
- 72.Perez I, Lin CH, McAfee JG, Patton JG. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 73.Bray N, Pachter L. MAVID: constrained ancestral alignment of multiple sequences. Genome Res. 2004;14:693–699. doi: 10.1101/gr.1960404. [DOI] [PMC free article] [PubMed] [Google Scholar]