Abstract
Ribosomal protein L2 is a core element of the large subunit that is highly conserved among all three kingdoms. L2 contacts almost every domain of the large subunit rRNA and participates in an intersubunit bridge with the small subunit rRNA. It contains a solvent-accessible globular domain that interfaces with the solvent accessible side of the large subunit that is linked through a bridge to an extension domain that approaches the peptidyltransferase center. Here, screening of randomly generated library of yeast RPL2A alleles identified three translationally defective mutants, which could be grouped into two classes. The V48D and L125Q mutants map to the globular domain. They strongly affect ribosomal A-site associated functions, peptidyltransferase activity and subunit joining. H215Y, located at the tip of the extended domain interacts with Helix 93. This mutant specifically affects peptidyl–tRNA binding and peptidyltransferase activity. Both classes affect rRNA structure far away from the protein in the A-site of the peptidyltransferase center. These findings suggest that defective interactions with Helix 55 and with the Helix 65–66 structure may indicate a certain degree of flexibility in L2 in the neck region between the two other domains, and that this might help to coordinate tRNA–ribosome interactions.
INTRODUCTION
Even well after the discovery of RNA catalysis (1), it was thought that peptidyltransfer was catalyzed by a ribosomal protein. Indeed, a series of ribosome reconstitution and ribosomal protein omission experiments had narrowed the candidates to two ribosomal proteins required for this activity: L2 and L3 (2–4). Early chemical modification experiments particularly implicated L2 (5,6), and later studies focused on specific histidines thought to be involved in this process (7–9). In fact, only after high-resolution crystal structures of ribosomal large subunits revealed that no ribosomal proteins were close enough to the tRNA 3′ ends to participate in catalysis that it became clear that this essential activity is completely catalyzed by RNA (10,11).
Although it does not play a direct role in peptidyltransfer, ribosomal protein L2 remains an intriguing subject of study. L2 is so highly conserved that archaebacterial and even the human isoforms of the protein can be functionally reconstituted into Escherichia coli ribosomes (8). Along with L3, L4 and L16 (L10 in eukaryotes), L2 is a member of a group of core ribosomal proteins containing acidic globular domains at the periphery and highly basic extensions penetrating into the rRNA core of the ribosome that form an X-shaped protein scaffold encased by rRNA (Figure 1). L2 contacts multiple domains of the large subunit rRNA (12) and participates in the B7b intersubunit bridge with the 18S rRNA of the small subunit (13). Its SH3 β-barrel globular domain participates in the intersubunit bridge, and interacts mainly with rpL43, and helices 79 and 65. The middle bridge region of the protein L2 is sheathed in rRNA, making extensive contacts with many separate helices, including H33, H65, H66 and H67. The extension region closely approaches the peptidyl–tRNA in a particularly strong interaction with the major groove of helix 93 at the PTC (12).
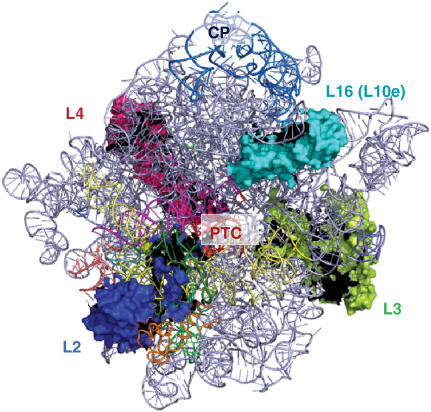
Figure 1.
Crown view of the large ribosomal subunit. Ribosomal proteins L2, L3, L4 and L16 (eukaryotic ribosomal protein L10) are indicated. PTC denotes the peptidyltransferase center. CP denotes the central protuberance. The image was generated with PyMol using the yeast cryo-EM-based ribosome mapped onto the H. marismoutui X-ray crystal structure (13).
Previous functional studies of L2 were primarily performed in prokaryotes (2–7,9,14–16), and very little research has been conducted in eukaryotes. These analyses were primarily performed using the yeast Saccharomyces cerevisiae two or more decades ago (15,17–22). Since then our knowledge and understanding of the relationship between ribosome structure and function has significantly matured, and the repertoire of molecular genetic, biochemical and computational tools that can be brought to bear on these issues has been greatly expanded. In the current study, the new tools have been used to revisit yeast ribosomal protein L2. Screening of a library of randomly generated L2 alleles identified three mutants unable to maintain the yeast ‘Killer’ dsRNA virus, a highly sensitive indicator of defects in the protein synthetic apparatus. Molecular genetic, biochemical and structural analyses revealed that these could be grouped into two general classes. One group, composed of the V48D and L125Q mutants, maps to the SH3 β-barrel globular domain of L2 and their biochemical and phenotypic effects appear to mainly be caused by 60S subunit biogenesis defects, possibly due to defective interactions with Helix 55, and the Helix 65–66 structure respectively. The other, H215Y lies near the tip of the basic extended domain where it interacts with Helix 93 along the P-site side of the PTC and affects peptidyl–tRNA binding. This initial foray into L2 mutagenesis has served to identify regions of the protein important for translational fidelity and lays the groundwork for deeper investigations.
MATERIALS AND METHODS
Strains, plasmids and media
The S. cerevisiae strains, and oligonucleotides (IDT, Coralville, IA, USA) used in this study are shown in Supplementary Tables S1 and S2. rpl2a and rpl2b gene-deletion strains were obtained from Invitrogen (Carlsbad, CA, USA), and L-A and M1 ‘killer’ viruses were introduced into these strains by cytoplasmic mixing by mating with kar1-1 strains of opposite mating types as previously described (23). Plasmids were amplified in E. coli strain DH5α. Escherichia coli were transformed using a calcium chloride method (24) and yeast were transformed with an alkali cation protocol (25). YPAD, synthetic complete (SC), synthetic dropout medium (H-), and 4.7 MB plates for testing the killer phenotype were used as previously described (26,27).
Construction of RPL2A clones and of rpl2A/rpl2B gene-deletion strains
In yeast, L2 is encoded by two identical genes (RPL2A and RPL2B). RPL2A was cloned in three segments (5′ UTR, 815 bp; ORF, 949 bp; and 3′ UTR, 270 bp) each separated by unique restriction sites which were added during PCR, and the resulting product was cloned into pRS316 (CEN6-URA3) (28) to make pRPL2A-URA3. The three-segment RPL2A cassette was also cloned into the pRS315 plasmid (CEN6-LEU2) to generate pRPL2A-LEU2. This is diagrammed in Supplementary Figure S1. In parallel, haploid strains of opposite mating type each harboring a deletion of one of the RPL2 isogenes (JD1205 and JD1207 transformed with pRPL2A-URA3) were mated, sporulated and tetrads dissected to obtain haploid rpl2A, rpl2B double knockout strains supported by a plasmid borne copy of the gene from pRPL2A-URA3. These were confirmed by their inability to grow in the presence of 5-fluoroorotic acid (5-FOA).
Generation of rpl2A mutants
Mutations in the RPL2A ORF were generated by both random mutagenesis and site-directed mutagenesis techniques. Site-directed mutagenesis was carried out using the Quikchange XL site-directed mutagenesis kit following the directions of the manufacturer (Stratagene, Madison, WI, USA). Random mutagenesis of the RPL2A ORF was performed using an error-prone PCR and gap-repair procedure (29). The Stratagene Genemorph II Random Mutagenesis Kit (Stratagene, La Jolla, CA, USA) was utilized and reaction conditions were optimized for 1–3 mutations/1000 nt. The primers (forward: RPL2ARMF and reverse: RPL2ARMR, see Supplementary Table S2) included the RPL2A start and stop codons and were complementary to the 5′ and 3′ untranslated regions of the gene. The PCR products were cotransformed into yeast along with BamHI/SpeI linearized pRPL2A-LEU2 that contained only the 5′ and 3′ UTR regions of RPL2A. Cells were then plated onto medium lacking leucine (–leu) to select for recombinant clones containing mutant rpl2A alleles. Colonies were subsequently replica plated onto media containing 5-FOA to select for cells that had lost pRPL2A-URA3, leaving colonies expressing pRPL2A-LEU2 clones. RPL2A was chosen for use instead of RPL2B because of prior data showing that expression of RPL2A as the only gene-encoding L2 resulted in apparently wild-type cells, while expression of RPL2B as the sole form of the protein promoted a slow growth phenotype and decreased accumulation of 60S subunits (21).
As an initial screen for defects in the translational apparatus, ∼10 000 colonies were tested for their inability to maintain the killer virus as previously described. Killer assays were performed by replica plating test colonies onto 4.7-MB plates seeded with a lawn of killer-susceptible 5 × 47 indicator cells (23). Plasmids were rescued from colonies with defective killer phenotypes (Killer loss, K−; or Killer reduction Kr), amplified in E. coli, reintroduced into JD1269, the wild-type pRPL2A-URA3 was eliminated using 5-FOA and killer assays were repeated.
Temperature response, drug sensitivity and growth curves
Dilution spot assays were used to qualitatively monitor cell growth at various temperatures and drug concentrations. For all conditions, yeast cells were grown to logarithmic growth phase and then diluted to 1 × 106 CFU/ml. Subsequently, 10-fold serial dilutions of each strain were spotted onto YPAD and incubated at 15°, 30° and 37°C, or onto YPAD containing paromomycin (5 mg/ml) or anisomycin (20 µg/ml). Sensitivity to sparsomycin was monitored using a filter disc assay (30). Yeast cells were grown overnight to saturation and 300 µl of cells diluted to OD595 = 0.2 were spread onto YPAD plates and allowed to dry, after which sterile 6-mm Whatman filter discs saturated with 30 µg of sparsomycin were placed onto the center of the plates. Growth curves were generated in quadruplicate with a Synergy HT micro-plate reader utilizing the KC4 software package (Bio-Tek Instruments, Inc., Winooski, VT). Yeast growth at 30° C was measured in 48-well plates beginning with 0.5-ml cultures of cells in –leu medium diluted to OD595 = 0.05. Cultures were subjected to constant high-intensity shaking and automatic OD595 readings were taken of each well at 20-min intervals for 40 h. Duplicate cultures were independently assayed twice and the four readings were averaged for each time point.
Translational fidelity assays
Effects of rpl2A mutants on translational fidelity were assayed by monitoring their effects on the ability to suppress an in-frame UAA codon (nonsense suppression), programmed −1, and +1 ribosomal frameshifting (PRF) using previously described methods (31). The reporter constructs are shown in Supplementary Figure S1. The test construct for −1 PRF includes the −1 PRF signal from the yeast L-A virus, and translation of the downstream firefly luciferase gene requires a −1 frameshift event. The +1 PRF test construct contains the +1 PRF signal from the Ty1 yeast retrotransposable element, and synthesis of the downstream firefly gene requires a +1 shift in reading frame. Nonsense suppression was measured by simply inserting an in-frame stop codon between the Renilla and firefly luciferase genes, so that a readthrough event is necessary for expression of the downstream firefly luciferase. The readthrough control plasmid supplied the baseline data to which the data from test constructs were compared. Reaction reagents were from the Dual-Luciferase® Reporter Assay System (Promega Corporation, Madison, WI, USA) and luminescence readings were measured by a TD20/20 luminometer (Turner Designs Inc. Sunnyvale, CA, USA). Each individual assay compares a ratio of two luciferase (firefly and Renilla) intensities from the test construct to a 0-frame control plasmid. The ratio of this test to the control was calculated and normalized to the wild-type value, with enough replicates for >95% confidence (minimum of nine replicates). All calculations and standard errors were determined as previously described (32).
Ribosome biochemistry
Lysates of cycloheximide arrested yeast cells were sedimented through 7–47% sucrose gradients and polysome profiles were determined by monitoring A254 nm as previously described (33). Yeast phenylalanyl–tRNA was purchased from Sigma (St Louis, MO, USA), charged with [14C]-Phe or Ac-[14C]-Phe and purified by HPLC as previously described (34). Ribosomes were isolated from isogenic wild-type and mutant cells, and puromycin reactions to monitor peptidyltransferase activity, and tRNA-binding studies were carried out as previously described (34). Purified ribosomes were chemically probed with dimethylsulfate (DMS), kethoxal or carbodiimide metho-p-toluenesulfonate (CMCT), followed by reverse transcriptase primer extension analyses of modified rRNAs as previously described (35) with the modifications that (i) reactions were performed at 30°C instead of 37°C and (ii) low concentrations of deoxyribonucleotides were employed to maximize reverse transcriptase strong stops (36). Primer 25-6 was used for analysis of helices 89–93, and primer 25-11 covered helices 73–74 of 25S rRNA. The molecular visualization tool Pymol (37) was used to view and examine available crystal structures of the ribosome. Images in this document are based primarily on structures from (13), in which atomic resolution crystal structures from Haloarcula marismortui and Thermus thermophilus were docked into cryo-EM structures from S. cerevisiae (pdb accession numbers 1s1h and 1s1i).
RESULTS
Generation and genetic characterization of rpl2A alleles
Random PCR mutagenesis was used to generate a library of rpl2a alleles. PCR products and linearized EcoRI/SpeI digested pPRL2A-LEU2 were introduced into JD1315 cells, transformants were initially selected for growth on –leu medium and then replica plated to medium containing 5-FOA. Preliminary analysis using 5-FOA revealed that ∼30% of the mutants were not viable as the sole form of rpl2a. All mutations created both in random mutagenesis and later site-directed mutagenesis are catalogued in Supplementary Table S3. Maintenance of the endogenous yeast M1 dsRNA satellite virus is sensitive to a wide variety of defects in the translational apparatus including changes in 60S ribosomal biogenesis (38) and to changes in rates of −1 PRF efficiency (23). The latter can in turn be caused by changes in tRNA binding, peptidyltransferase activity and interactions between ribosomes and numerous trans-acting factors (39). Thus, mutants were initially screened for ‘Killer’ phenotypic defects. We note however, that this screen cannot detect other translational defects, e.g. in translational initiation or mRNA decay (40). Of ∼10 000 candidate colonies screened, 79 initially displayed Killer loss (K−) or Killer reduced (Kr) phenotypes. Plasmids were rescued from these colonies, reintroduced into JD1315 cells, and taken again through the selection and Killer analysis protocols. This analysis revealed that 16 of the mutants conferred K−, and 20 promoted Kr phenotypes. These are summarized in Supplementary Table S3.
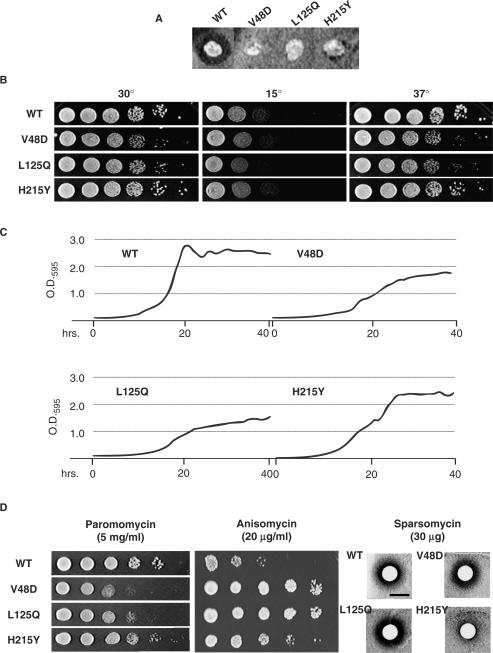
The majority of the Killer-defective rpl2A alleles contained multiple mutations and resulting amino acid changes, with 22 of the alleles having three or more mutations. Two alleles had mutations in their stop codons, resulting in C-terminal extensions containing an extra four amino acids (YIMY). Six strains had double mutations and six alleles contained single mutations. Based on the frequency with which individual mutations occurred, and their locations within the structure of L2, 16 alleles harboring single mutations were generated for further analysis using oligonucleotide site-directed mutagenesis. Three of the rpl2A single mutant alleles, V48D, L125Q and H215Y conferred K– phenotypes (Figure 2A) and were used for all subsequent analyses. Ten-fold serial dilution spot assays at 15°, 30° and 37°C revealed that the V48D and L125Q mutants grew slower than wild-type across all tested temperatures with the most pronounced effects at 37°C (Figure 2B). To obtain a higher-resolution perspective of the effects the mutants, cellular growth curves were generated at 30° over a period of 40 h (Figure 2C). This analysis revealed that the V48D and L125Q mutants promoted slower growth rates and failed to approach the density of the saturated wild type cultures. The H215Y mutant also promoted decreased growth rates in logarithmic phase, but was able to attain saturation concentration similar to wild-type cells. Growth curves of all 16 of the single mutant strains are shown in Supplementary Figure S2. Notably, all of the mutants showed changes in growth rates.
Figure 2.
The L2 mutants promote numerous phenotypic defects. (A) ‘Killer’ virus phenotypes. The Killer+ phenotype is scored by the presence of a halo of growth inhibition around wild-type colony. Lack of the halo around colonies expressing the V48D, L125Q and H215Y L2 mutants indicates the Killer− phenotype. (B) Ten-fold dilutions of indicated cells were spotted onto rich medium and incubated at the indicated temperatures. (C) Yeast cell growth was monitored for 40 h at 30°C with a Synergy HT micro-plate reader and growth curves were generated from four independent readings utilizing the KC4 software package. (D) Drug-sensitivity phenotypes. To monitor changes in sensitivity to paromomycin and anisomycin, 10-fold serial dilutions of each strain were spotted onto rich medium containing these drugs at the indicated concentrations. Sensitivity to sparsomycin was monitored using 6-mm Whatman filter discs saturated with 30 µg of sparsomycin placed onto the center of plates seeded with OD595 = 0.2 of yeast cells expressing the indicated L2 mutants.
The effects of three translational inhibitors were also assayed on the three mutants and compared to wild type. Dilution spot assays revealed that the V48D and L125Q were similar in their responses to all three drugs: hypersensitive to paromomycin, strongly resistant to anisomycin and having wild-type responses to sparsomycin (Figure 2D). In contrast, while H215Y was slightly anisomycin-resistant, it was significantly resistant to sparsomycin, while paromomycin had no effect relative to wild-type cells. The results of these genetic studies indicated that, although all three mutants promoted the K− phenotype, they could be divided into two classes: V48D and L125Q were similar to one another, while H215Y was unique.
The V48D, L125Q and H215Y mutants promote specific translational fidelity defects
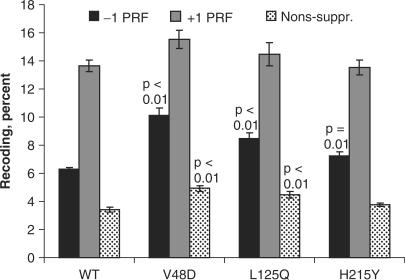
To test whether changes in rates of −1 PRF were responsible for the K− phenotype, isogenic cells expressing wild-type or mutant forms of L2 were transformed with either a −1 PRF reporter plasmid harboring the yeast L-A viral −1 PRF signal, or an in-frame control (Supplementary Figure S1), and rates of −1 PRF were monitored as described in the Materials and Methods section. The results of these experiments are summarized in Figure 3. The baseline −1 PRF rate in wild-type cells was 6.29 ± 0.15%. The V48D mutation promoted 10.10 + 0.52% PRF (1.6-fold wild-type, p = 8.5 × 10−5), L125Q promoted 8.49 + 0.42% (1.4-fold increase, p = 2.2 × 10−3), and −1 PRF in H215Q cells was 7.23 + 0.31 (1.2-fold WT, p = 0.01). Programmed +1 frameshifting was similarly monitored using a reporter harboring the Ty1 +1 PRF signal (Figure 3). The baseline rate of +1 PRF rate was 13.65 ± 0.43%. The V48D, L125Q and H215Y mutants did not significantly affect this aspect of translational recoding. Specifically, +1 PRF for V48D was 15.55 + 0.64% (1.1-fold WT); L125Q was 14.45 + 0.82% (1.0-fold WT), and H215Y was 13.52 + 0.53 (1.0-fold WT).
Figure 3.
The L2 mutants promote specific defects in translational fidelity. Isogenic yeast cells expressing either wild-type or mutant forms of L2 were transformed with the dual luciferase reporter and control plasmids shown in Supplementary Figure S1 and rates of translational recoding were determined. −1 PRF indicates percent of programmed −1 ribosomal frameshifting promoted by the yeast L-A virus frameshift signal. +1 PRF denotes programmed +1 ribosomal frameshifting directed by the frameshift signal of the Ty1 retrotransposable element. Nons-suppr. denotes the percentage of ribosomes able to suppress an in-frame UAA termination codon located between the Renilla and firefly luciferase reporter genes. p values are indicated above samples showing statistically significant changes. Error bars denote SE as previously described (32).
Hypersensitivity to paromomycin is often accompanied by changes in the ability of ribosomes to properly recognize and decode termination codons. Thus, a dual luciferase assay was used to monitor the effects of the L2 mutants on nonsense suppression. This reporter is diagrammed in Supplementary Figure S1. Consistent with their paromomycin hypersensitivity, the V48D and L125 mutants promoted increased rates of stop-codon readthrough (Figure 3). Specifically, nonsense suppression rates in cells expressing wild-type L2 were 3.41 + 0.15%, while these values were 4.92 + 0.20% (1.4-fold WT, p = 2.8 × 10−4), and 4.5 + 0.23 (1.3-fold WT, p = 5.8 × 10−3) for the V48D and L125Q mutants, respectively. In contrast, the H215 mutant did not affect nonsense suppression (3.76 + 0.14%, 1.1-fold WT, p = 0.1) consistent with its wild-type phenotype with respect to paromomycin sensitivity.
The V48D and L125Q mutants promote strong defects in 60S biogenesis and polysome profiles
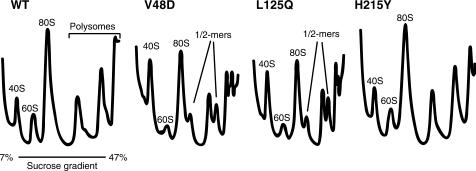
Since loss of the Killer phenotype has also been associated with 60S biogenesis defects (38), S30 fractions of cycloheximide-arrested cell lysates were sedimented through linear 7–47% sucrose gradients ribosomes to examine the effects of the mutants on this parameter. A254 tracings of the fractionated gradients are shown in Figure 4. Notably, the profile of the ‘wild-type’ strain showed slight defects typically associated with 60S subunit defects. Specifically, these were (i) decreased 60S peak heights relative to 40S peak heights, and (ii) the presence of small ‘shoulders’ on the right sides of the 80S and polysome peaks indicative of half-mer formation. Shoulders and half-mer peaks indicate a population of mRNAs with an extra 40S subunit compared to the associated primary peak. The extra mass is due to a 40S subunit stalled at the start codon either due to a shortage of 60S subunits, or a subunit-joining defect. We believe that this is an artifact of expression of only one isoform of L2, consistent with recent findings of specialized roles for different ribosomal protein isoforms in yeast (41). The polysome profile of the H215Y mutant was not significantly different from those of cells expressing wild-type L2. In contrast, the 60S and half-mer defects were strongly accentuated in the polysome profiles of cells expressing the V48D and L125Q mutations.
Figure 4.
The V48D and L125Q mutants promote strong 60S biogenesis and subunit-joining defects. Cytoplasmic extracts from isogenic strains were loaded onto 7–47% sucrose gradients, centrifuged in an SW41 rotor at 40 000 r.p.m. for 180 min at 4°C, fractionated and analyzed by continuous monitoring of A254 (48). The locations of 40S, 60S, 80S, polysome fractions and half-mers are labeled. The presence of half-mers is indicative of subunit-joining defects.
Ribosome biochemistry
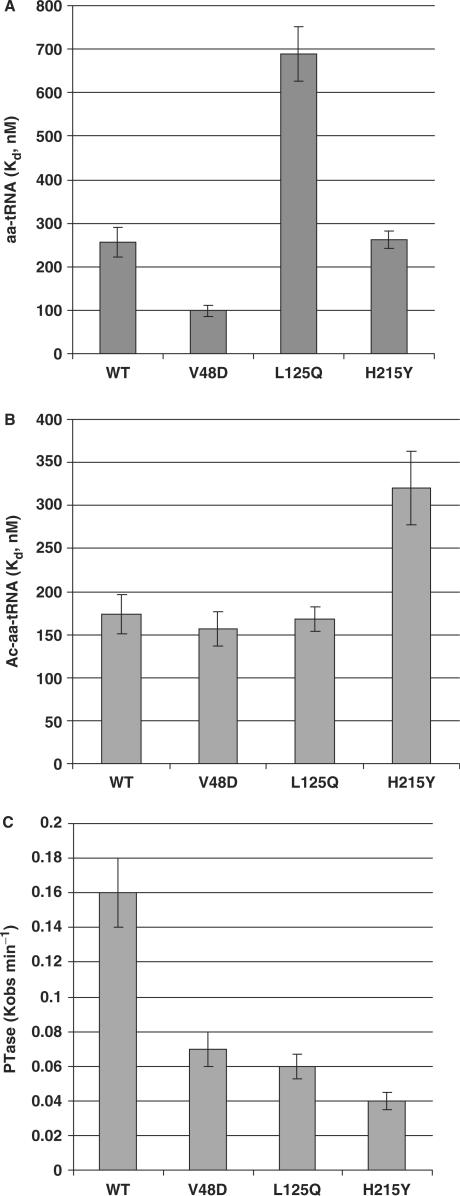
Anisomycin and sparsomycin are peptidyltransferase inhibitors that target and affect interactions of aa–tRNA and peptidyl–tRNAs with the ribosomal A- and P-sites, respectively (42). In contrast, paromomycin targets the decoding center in the small subunit (43), which can affect the overall interaction of aa–tRNA with the ribosome. Thus, changes in the interactions between ribosomes and tRNAs at the biochemical level can manifest themselves at the biological level as changes in cellular drug sensitivity. To determine the effects of the mutants on ribosome-tRNA interactions, filter-binding assays were performed under equilibrium conditions to determine dissociation constants of aa–tRNA to the A-sites and of Ac–aa–tRNA to the P-sites of wild-type and mutant ribosomes. The results of these experiments are shown in Figure 5. Binding of aa–tRNA to the A-site was significantly affected by the V48D and L125Q mutants (Figure 5A). Specifically, while the Kd for aa–tRNA in wild-type ribosomes was 256.6 + 35.1 nM, Kd values were ∼0.4-fold and ∼2.7-fold (98.5 + 12.3 nM and 688.6 + 62.8 nM) of wild-type for V48D and L125Q ribosomes, respectively. Notably, these mutants displayed changes in phenotypes in response to the A-site specific drugs anisomycin and paromomycin. The H215Y mutant, which did not promote A-site drug-specific defects, did not significantly affect ribosome–aa–tRNA interactions (Kd = 262.2 + 19.1 nM). In contrast, this mutant was resistant to the P-site specific inhibitor sparsomycin, consistent with its ability to promote a >1.8-fold increase in Kd for Ac–aa–tRNA (320.4 + 42.8 nM) as compared to wild-type ribosomes (173.6 + 22.2 nM) (Figure 5B). The V48D and L125Q mutants did not affect this parameter (156.6 + 19.4 nM and 167.7 + 14.1 nM, respectively). All of the mutants negatively impacted ribosomal peptidyltransferase activity (Figure 5C). Specifically, Kobs of wild-type ribosomes was 0.16 + 0.2 min−1, while the values for the V48D, L125Q and H215Y mutants were 0.07 + 0.01 min−1, 0.6 + 0.01 min−1 and 0.04 + 0.01 min−1, respectively. These findings are consistent with previous observations that decreased rates of peptidyltransfer can stimulate −1 PRF (44).
Figure 5.
The L2 mutants affect tRNA binding and peptidyltransferase activity. (A) Dissociation constants generated by analysis of single-site-binding isotherms of eEF-1A stimulated binding of [14C]Phe-tRNA to ribosomal A-sites.(B) Dissociation constants generated by analysis of single-site-binding isotherms of Ac-[14C]Phe-tRNA to ribosomal P-sites. (C) Characterization of peptidyltransferase activities of wild-type and mutant ribosomes generated by analyses of first order time plots of Ac-[14C]Phe-puromycin formation. Error bars denote mean and standard deviation.
The L2 mutants affect rRNA structure in the vicinity of the peptidyltransferase center
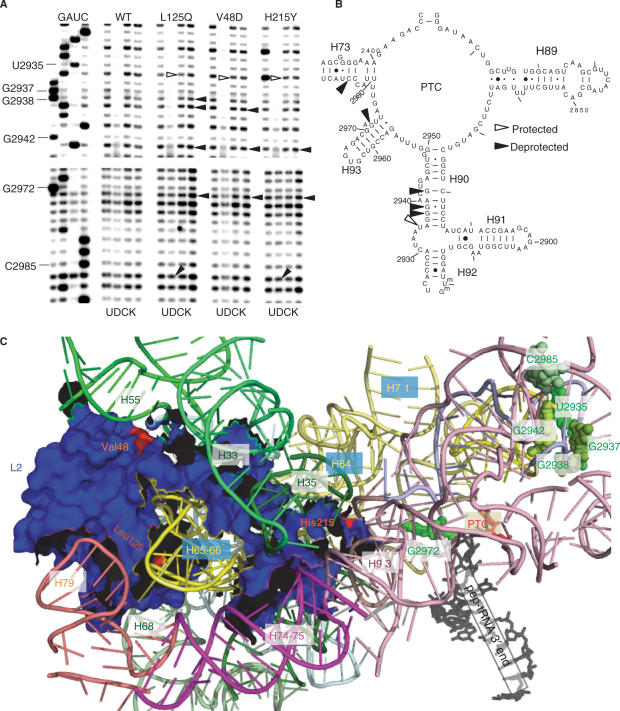
Given the effects of the mutants on ribosome biochemistry, chemical protection methods were used to probe for changes in rRNA structure in the peptidyltransferase center. Ribosomes purified from wild-type and mutant cells were probed for structural changes using DMS, kethoxal and CMCT. rRNAs were extracted, and modified bases were identified by primer extension using reverse transcriptase to detect methylation at the N1 position of adenosines and N3 position of cytidines (DMS), at the N1 and N2 positions of guanosines (kethoxal), and at the N3 position of uridines and the N1 position of guanosines (CMCT) (1). The primers used (see Materials and Methods section) were designed to probe rRNA bases in the PTC that interact with peptidyl- and aa–tRNAs, (from 5′ to 3′ Helix 89–Helix 73). Changes in chemical protection patterns are observed as changes in band intensities 1 nt 5′ of the modified base due to the inability of reverse transcriptase to utilize modified bases.
The results of these experiments are shown in Figure 6A and mapped onto a 2-dimensional representation of yeast 25S rRNA in Figure 6B. In Helix 90, all three of the mutants promoted hyperprotection of U2935 (E. coli A2566) and deprotection of G2942 (E. coli G2574). Also in this region, G2938 (E. coli G2570) was deprotected by both the V48D and L125Q mutants, while it 5′ neighbor G2937 (E. coli G2560) was only deprotected in L125Q ribosomes. In Helix 93, all three mutants promoted deprotection of G2972 (E. coli G2603), and the L125Q and H215Y mutants promoted slightly greater levels of deprotection of C2985 (E. coli C2616) in Helix 73. Mapping of these changes onto the 3-dimensional structure of yeast L2 as determined by cryo-EM and fitted into the H. marismortui 50S X-ray crystallographic structure (13) reveal that all of the other bases are clustered in the A-site region of the PTC, with the exception of G2972 which is located near the P-site (Figure 6C).
Figure 6.
Structure probing of wild-type and mutant ribosomes. (A) Autoradiograms of reverse transcriptase primer extension reactions spanning sequence in helices 90–73. Sequencing reactions (left sides of panels) are labeled corresponding to the rRNA sense strand. Below each panel, U stands for untreated, D is DMS, C denotes CMCT and K indicates kethoxal. Sources of ribosomes are indicated at top. Protected and deprotected bases are indicated by open and filled arrows, respectively. Chemical modification results in strong stops 1-nt 5′ of the base, and bases with altered chemical protection patterns are identified at left. (B) Localization of bases in the vicinity of the peptidyltransferase center (PTC) of yeast 25S rRNA whose chemical modification patterns were affected by the L2 mutants. (C) Mapping of the L2 mutants and rRNA protection data into the yeast ribosome structure generated by cryo-EM and fitted onto the H. marismortui large subunit crystal structure (13). L2, the PTC, the 3′ end of the peptidyl–tRNA and rRNA helices are labeled. The V48D and L125Q mutants map to the globular domain of L2 where they interact with helices 55 and 65/66, respectively. The H215Y mutant maps to the tip of the extended domain where it inserts into the major groove of helix 93. Nucleotides with altered chemical protection patterns are shown in shades of green. Base numbering follows the S. cerevisiae sequence shown in panel (B).
DISCUSSION
The goal of this study was to create a library of yeast ribosomal protein L2 mutants and identify structurally and functionally important amino acid residues. Screening for loss of the killer virus as a general indicator of defective ribosome function allowed high-throughput identification of randomly generated mutants that conferred translational defects. Most of the mutants so identified contained mutations of more than one amino acid residue. While it is possible that multiple mutations could have been caused by the random mutagenesis protocol, this is unlikely as primer concentrations and reaction conditions were adjusted to generate primarily single mutations. Although only strains containing single mutations were selected for further study, the fact that multiple amino acid changes were generally necessary to promote killer virus loss suggests that this protein, which is firmly integrated in the large subunit through contacts with multiple domains of 25S rRNA, is functionally and structurally robust. This hypothesis is supported by the observation that human and archael L2 proteins were functional in E. coli ribosomes (8). The observations that all of the single mutants impacted cell growth (Supplementary Figure S2), and that its expression as the sole form of the protein resulted in a subtle 60S subunit biogenesis defect as manifested by decreased 60S peak heights relative to 40S and small half-mer peaks, suggest that that both the structure and expression of L2 has been fine-tuned to promote optimal cell growth in yeast.
Although all three of the single mutants promoted decreased peptidyltransferase activities, thus stimulating −1 PRF and loss of the killer virus, differences in other genetic biochemical, and sturctural aspects allow them to be grouped into two classes: V48D and L125Q versus H215Y. V48D and L125Q are both located on the surface of the globular domain of L2, near the ‘neck’ region linking it to the extended domain, and immediately adjacent to two helices of 25S rRNA (Figure 6C). Both mutants change small aliphatic side chains which should minimally interact with nucleoside bases to larger, polar (acidic and amide) side chains that could cause changes in both the local stereochemical and biochemical interactions with the Helix 55 and Helix 65–66. These helices appear to anchor the globular domain of L2 into the large subunit, and it is reasonable to conjecture that altering these interactions could propagate outward, resulting in more severe functional and biological defects. This is supported by the decreased growth rates, strong 60S biogenesis, and subunit-joining defects associated with the V48D and L125Q mutants. A recent study in E. coli showing that Helix 66 mutants promoted similar subunit association defects, and that the architecture of H66 rather than of its base sequence was the most important parameter (45) provides a complementary supporting dataset. The V48D and L125Q mutants appeared to impact on A-site-specific functions as manifested by their altered sensitivities to paromomycin and anisomycin, enhanced suppression of nonsense-codons and specific changes in aa–tRNA binding. In E. coli, it is notable that the D83N mutant (analogous to D50 in yeast) did not significantly affect aa–tRNA binding but did promote an ∼28% decrease in affinity for Ac–aa–tRNA in the P-site (9). The finding of structural alterations of bases in the A-site (U2935, G2937, G2938, G2943 and C2985), and one base in the P-site (G2972), all located quite distantly from V48 and L125 is also intriguing. Mutants in the W-finger domain and the N-terminal extension of ribosomal protein L3 mutants had similar structural effects on the same region of Helix 90 (46, Meskauskas and Dinman, unpublished). These studies have suggested that L3 helps to coordinate the stepwise processes of translation elongation. It is tempting to speculate that the opposing effects of the L2 mutants on aa–tRNA binding, and their long-distance effects on rRNA structure may be indicative of a certain degree of flexibility in L2 in the neck region between the two domains which might similarly be involved in coordinating tRNA–ribosome interactions.
In contrast, the nature and location of the H215Y mutant distinguish it from the V48D and L125Q mutants. This amino acid is located at the tip of the internal extension domain, nestled in the major groove of Helix 93, proximal to the P-site of the PTC and the binding sites for anisomycin and sparsomycin (47). Unlike the other two mutants the H215Y strain was relatively healthy, and its functional affects, i.e. slight anisomycin resistance, strong sparsomycin resistance and decreased affinity for Ac–aa–tRNA, suggests that its effects are more directly targeted to the PTC. Notably, in E. coli, mutants of D288, H229 and H213 (analogous to yeast D207, H208 and H210), which map to the tip of the L2 extension, also inhibited peptidyltransfer and Ac–aa–tRNA binding (9). These observations recommend this region of L2 for more detailed reverse-genetics studies by targeting specific amino acids across a range of biochemical properties in order to generate a more detailed picture of how the structure of L2 contributes to ribosome function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Rasa Rakauskaitė, Karen Jack, Trey Belew and Michael Rhodin for their help and insightful comments. This work was supported by grants to A.M. from the American Heart Association (AHA 0630163N), and to J.D.D. from the National Institutes of Health (R01 GM058859). J.R.R. was partially supported by an NIH training grant (T32 AI051967-05). Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Inoue T, Sullivan FX, Cech TR. Intermolecular exon ligation of the rRNA precursor of Tetrahymena: oligonucleotides can function as 5′ exons. Cell. 1985;43:431–437. doi: 10.1016/0092-8674(85)90173-4. [DOI] [PubMed] [Google Scholar]
- 2.Kazemie M. Binding of aminoacyl-tRNA to reconstituted subparticles of Escherichia coli large ribosomal subunits. Eur. J. Biochem. 1976;67:373–378. doi: 10.1111/j.1432-1033.1976.tb10701.x. [DOI] [PubMed] [Google Scholar]
- 3.Schulze H, Nierhaus KH. Minimal set of ribosomal components for reconstitution of the peptidyltransferase activity. EMBO J. 1982;1:609–613. doi: 10.1002/j.1460-2075.1982.tb01216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khaitovich P, Mankin AS, Green R, Lancaster L, Noller HF. Characterization of functionally active subribosomal particles from Thermus aquaticus. Proc. Natl Acad. Sci. USA. 1999;96:85–90. doi: 10.1073/pnas.96.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahnestock SR. Evidence of the involvement of a 50S ribosomal protein in several active sites. Biochemistry. 1975;14:5321–5327. doi: 10.1021/bi00695a016. [DOI] [PubMed] [Google Scholar]
- 6.Sonenberg N, Wilchek M, Zamir A. Mapping of Escherichia coli ribosomal components involved in peptidyl transferase activity. Proc. Natl Acad. Sci. USA. 1973;70:1423–1426. doi: 10.1073/pnas.70.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harada N, Maemura K, Yamasaki N, Kimura M. Identification by site-directed mutagenesis of amino acid residues in ribosomal protein L2 that are essential for binding to 23S ribosomal RNA. Biochim. Biophys. Acta. 1998;1429:176–186. doi: 10.1016/s0167-4838(98)00230-1. [DOI] [PubMed] [Google Scholar]
- 8.Uhlein M, Weglohner W, Urlaub H, Wittmann-Liebold B. Functional implications of ribosomal protein L2 in protein biosynthesis as shown by in vivo replacement studies. Biochem. J. 1998;331:423–430. doi: 10.1042/bj3310423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diedrich G, Spahn CM, Stelzl U, Schafer MA, Wooten T, Bochkariov DE, Cooperman BS, Traut RR, Nierhaus KH. Ribosomal protein L2 is involved in the association of the ribosomal subunits, tRNA binding to A and P sites and peptidyl transfer. EMBO J. 2000;19:5241–5250. doi: 10.1093/emboj/19.19.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 11.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 12.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 13.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA- ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 14.Oen H, Pellegrini M, Cantor CR. Peptidyl-transferase inhibitors alter the covalent reaction of BrAcPhe-tRNA with the E. coli ribosome. FEBS Lett. 1974;45:218–222. doi: 10.1016/0014-5793(74)80848-3. [DOI] [PubMed] [Google Scholar]
- 15.Wower J, Hixson SS, Zimmermann RA. Photochemical cross-linking of yeast tRNA(Phe) containing 8-azidoadenosine at positions 73 and 76 to the Escherichia coli ribosome. Biochemistry. 1988;27:8114–8121. doi: 10.1021/bi00421a021. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa A, Nakashima T, Taniguchi M, Hosaka H, Kimura M, Tanaka I. The three-dimensional structure of the RNA-binding domain of ribosomal protein L2; a protein at the peptidyl transferase center of the ribosome. EMBO J. 1999;18:1459–1467. doi: 10.1093/emboj/18.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Gosalbez M, Vazquez D, Ballesta JP. Affinity labelling of yeast ribosomal peptidyl transferase. Mol. Gen. Genet. 1978;163:29–34. doi: 10.1007/BF00268961. [DOI] [PubMed] [Google Scholar]
- 18.Reyes R, Vazquez D, Ballesta JP. Structure of the yeast ribosomes. Proteins associated with the rRNA. Biochim. Biophys. Acta. 1978;521:229–234. doi: 10.1016/0005-2787(78)90265-4. [DOI] [PubMed] [Google Scholar]
- 19.Kudlicki W, Szyszka R, Palen E, Gasior E. Evidence for a highly specific protein kinase phosphorylating two strongly acidic proteins of yeast 60 S ribosomal subunit. Biochim. Biophys. Acta. 1980;633:376–385. doi: 10.1016/0304-4165(80)90196-8. [DOI] [PubMed] [Google Scholar]
- 20.Otaka E, Higo K, Itoh T. Yeast ribosomal proteins: VII. Cytoplasmic ribosomal proteins from Schizosaccharomyces pombe. Mol. Gen. Genet. 1983;191:519–524. doi: 10.1007/BF00425772. [DOI] [PubMed] [Google Scholar]
- 21.Lucioli A, Presutti C, Ciafre S, Caffarelli E, Fragapane P, Bozzoni I. Gene dosage alteration of L2 ribosomal protein genes in Saccharomyces cerevisiae: effects on ribosome synthesis. Mol. Cell. Biol. 1988;8:4792–4798. doi: 10.1128/mcb.8.11.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kudlicki W, Grankowski N, Gasior E. Ribosomal protein as substrate for a GTP-dependent protein kinase from yeast. Mol. Biol. Rep. 1976;3:121–129. doi: 10.1007/BF00423225. [DOI] [PubMed] [Google Scholar]
- 23.Dinman JD, Wickner RB. Ribosomal frameshifting efficiency and Gag/Gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- 25.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dinman JD, Wickner RB. Translational maintenance of frame: mutants of Saccharomyces cerevisiae with altered -1 ribosomal frameshifting efficiencies. Genetics. 1994;136:75–86. doi: 10.1093/genetics/136.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickner RB, Leibowitz MJ. Mak mutants of yeast: mapping and characterization. J. Bact. 1979;140:154–160. doi: 10.1128/jb.140.1.154-160.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 30.Dinman JD, Kinzy TG. Translational misreading: Mutations in translation elongation factor 1α differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA. 1997;3:870–881. [PMC free article] [PubMed] [Google Scholar]
- 31.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobs JL, Dinman JD. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32:e160–e170. doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sachs AB, Davis RW. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 34.Meskauskas A, Petrov AN, Dinman JD. Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis. Mol. Cell. Biol. 2005;25:10863–10874. doi: 10.1128/MCB.25.24.10863-10874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 36.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 37.DeLano WL. The PyMOL Molecular Graphics System. 2006. http://www.pymol.org.
- 38.Ohtake Y, Wickner RB. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol. Cell. Biol. 1995;15:2772–2781. doi: 10.1128/mcb.15.5.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dinman JD, Berry MJ. Regulation of termination and recoding. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2006. [Google Scholar]
- 40.Harger JW, Dinman JD. Evidence against a direct role for the Upf proteins in frameshfiting or nonsense codon readthrough. RNA. 2004;10:1721–1729. doi: 10.1261/rna.7120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pestka S. Inhibitors of protein synthesis. In: Weissbach H, Pestka S, editors. Molecular Mechanismns of Protein Biosynthesis. New York: Academic Press; 1977. pp. 467–553. [Google Scholar]
- 43.Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 44.Meskauskas A, Harger JW, Jacobs KLM, Dinman JD. Decreased peptidyltransferase activity correlates with increased programmed -1 ribosomal frameshifting and viral maintenance defects in the yeast Saccharomyces cerevisiae. RNA. 2003;9:982–992. doi: 10.1261/rna.2165803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kitahara K, Kajiura A, Sato NS, Suzuki T. Functional genetic selection of Helix 66 in Escherichia coli 23S rRNA identified the eukaryotic-binding sequence for ribosomal protein L2. Nucleic Acids Res. 2007;35:4018–4029. doi: 10.1093/nar/gkm356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meskauskas A, Dinman JD. Ribosomal protein L3: gatekeeper to the A site. Molecular Cell. 2007;25:877–888. doi: 10.1016/j.molcel.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- 48.Ohtake Y, Wickner RB. KRB1, a suppressor of mak7-1 (a mutant RPL4A), is RPL4B, a second ribosomal protein L4 gene, on a fragment of Saccharomyces chromosome XII. Genetics. 1995;140:129–137. doi: 10.1093/genetics/140.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]