Abstract
The p53 family is known as a family of transcription factors with functions in tumor suppression and development. Whereas the central DNA-binding domain is highly conserved among the three family members p53, p63 and p73, the C-terminal domains (CTDs) are diverse and subject to alternative splicing and post-translational modification. Here we demonstrate that the CTDs strongly influence DNA binding and transcriptional activity: while p53 and the p73 isoform p73γ have basic CTDs and form weak sequence-specific protein–DNA complexes, the major p73 isoforms have neutral CTDs and bind DNA strongly. A basic CTD has been previously shown to enable sliding along the DNA backbone and to facilitate the search for binding sites in the complex genome. Our experiments, however, reveal that a basic CTD also reduces protein–DNA complex stability, intranuclear mobility, promoter occupancy in vivo, target gene activation and induction of cell cycle arrest or apoptosis. A basic CTD therefore provides both positive and negative regulatory functions presumably to enable rapid switching of protein activity in response to stress. The different DNA-binding characteristics of the p53 family members could therefore reflect their predominant role in the cellular stress response (p53) or developmental processes (p73).
INTRODUCTION
The tumor suppressor p53 is known as ‘the guardian of the genome’ owing to its ability to integrate many signals that control life or death (1–3). Activated by various types of cellular stress, including DNA damage and oncogenic stress, p53 initiates transcriptional programs that ultimately arrest proliferation and prevent the generation of genetically altered cells. The spectrum of p53-based cell fate decisions ranges from a transient cell-cycle arrest enabling damage repair to an irreversible block of proliferation through the induction of senescence, differentiation or apoptosis (1). This central role of p53 in cell fate control is underlined by its frequent inactivation in human cancer with an estimated 50% of malignancies harboring a mutated p53 locus (2).
The p53 protein contains the classical features of a sequence-specific transcriptional activator, including an aminoterminal transactivation domain, a sequence-specific DNA binding ‘core’ domain in the center of the protein, and a tetramerization domain C-terminal to the core. In addition, the extreme C-terminus of the p53 protein contains an arginine- and lysine-rich basic domain which was shown to function as an autonomous sequence-nonspecific nucleic acid binding region (4,5). Initial studies suggested that the C-terminal domain (CTD) is a negative autoregulator of sequence-specific DNA binding as truncation of the entire CTD in p53Δ30, replacement of the CTD by alternative splicing, or blocking of the CTD by antibodies and post-translational modifications resulted in increased binding to short oligonucleotides containing p53 binding sites (6–10). Based on these findings p53 was postulated to exist in a latent state and to undergo a modification-induced allosteric conformational shift that enables the core domain to bind to p53 response elements.
This allosteric model, however, was challenged by a number of recent findings: NMR analysis demonstrated that full-length and CTD-deleted mutants of p53 have essentially similar structures (11). Furthermore, although acetylation appears to increase p53 binding to short double-stranded DNA fragments, it does not affect the ability of p53 to bind to its specific response element in the context of chromatin or long DNA molecules (12). Similarly, quantitative chromatin immunoprecipitation failed to demonstrate allosteric regulation as p53 target promoters do not exhibit significant enrichment in p53 binding after genotoxic stress (13).
Instead, some recent studies have worked out a positive regulatory role of the CTD (14,15): McKinney et al. have demonstrated the involvement of the CTD in the ability of p53 to diffuse linearly on DNA, a concept supported by previous time-lapse atomic force microscopy studies (14,16). Based on these findings, a sliding (or facilitated search) model has been postulated proposing that p53 binds to the genomic DNA in a sequence-nonspecific manner via its CTD. It slides along the DNA searching for p53 response elements which are ultimately bound in a sequence-specific manner via the core domain. The CTD would thus facilitate the search for specific targets in the context of a complex genome. Consistently, the CTD is required for efficient promoter activation in vivo early after protein induction when p53 protein levels are limiting (14,15). At later time points or at higher expression levels the favorable role of the CTD became negligible (17,18).
In 1997 the cloning of p73 as a second p53 family member was reported and rapidly followed by the discovery of p63 as the third member (19–22). The overall protein architecture is highly conserved among the three family members and the highest degree of sequence homology is seen within the DNA binding core domain. p63 and p73 share ∼65% amino acid identity with the core domain of p53, and even higher identity with each other (1). Despite this structural homology the proteins have fundamentally different functions in vivo as indicated by the analysis of knockout mice (23–26): whereas tumor susceptibility is the primary phenotype of the p53 knockout (26,27), p63 and p73 knockouts are both characterized by severe developmental abnormalities and only moderately enhanced tumor incidence (28).
All three genes express a multitude of differently spliced isoforms—a feature that was thought to be unique for p63 and p73 but has recently been shown to also be true for p53 (29). Most of the alternative splicing occurs at the 3′ end, involves exons 10 to 13, and generates transcripts encoding protein isoforms with different CTDs as depicted in Figure 1 for p73. Whereas the full-length protein appears to be the predominant p53 isoform, at least two differently spliced isoforms with different C-terminal sequences are commonly expressed in the case of p63 (p63α and p63γ) and p73 (p73α and p73β). A number of other splice variants (e.g. p73γ and p73δ) have been reported, however, their physiological functions still remain to be elucidated (30,31). From overexpression studies it is known that the various p73 isoforms have very different transactivation potential indicating crucial functions of the CTDs in transactivation control (30,32–36).
Figure 1.
C-terminal charge of p53 and p73 isoforms. Shown are the domain structure and C-terminal charge distribution of p53, the p73 isoforms p73α, β, γ and δ, the CTD-deleted p53Δ30 and the p53/p73 chimera p73α+30. C-terminal regions in p73β, γ and δ that differ from the sequence in p73α are shaded in dark grey. The percentage of conserved residues in the transactivation (TA), DNA binding core (DBD) and oligomerization (OD) domain of p53 and p73 is shown. The calculated isoelectric points (pI values) of the total protein, the C-terminus (CT) and the last 30 amino acids are shown in the table for each protein. In the case of p73, CT is defined as the sequence C-terminal to the conserved oligomerization domain.
Considering the role of the p53 CTD in linear diffusion we here investigated the function of the p73 CTD in comparison to p53. We show that the electrostatic charge of the CTDs of the various p53 family proteins differs significantly and correlates inversely with DNA binding in vitro and in vivo. p53 and p73γ which both have a basic, positively charged CTD form weak sequence-specific protein–DNA complexes. In contrast, most p73 isoforms (α, β and δ) have neutral C-terminal sequences and bind DNA strongly. Importantly, the DNA-binding properties observed in vitro directly correlate with promoter binding and target gene activation in vivo. Adding the basic p53 CTD to the p73α C-terminus inhibits p73 DNA binding in vitro and reduces the transcriptional activity in vivo. Together with the previously proposed facilitated search model our results indicate that the basic p53 CTD represents an autonomous, transposable domain that provides both positive and negative regulatory functions presumably to enable rapid switching of protein activity in response to stress. The major p73 isoforms p73α and p73β lack this functional domain consistent with their predominant role in developmental control. CTD diversity within the p53 family therefore provides an additional layer of complexity useful to fine-tune cell fate decisions that are regulated by the p53 family.
MATERIALS AND METHODS
Cell lines and cell culture
H1299, Saos-2 and 293 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Sigma) supplemented with 10% fetal bovine serum (Sigma), 1% penicillin/streptomycin (Sigma) and 0.4% amphotericin B (Sigma). Transfections were performed using Escort V reagent (Sigma) according to the manufacturer's instructions. Adenoviruses were produced using the AdEasy system (37) and amplified on 293 cells.
Plasmids
Expression vectors for HA-tagged p53, p73α, p73β, p73γ and p73δ have been described (30,38). cDNAs for p53, p53Δ30 (amino acids 1–363 of human p53), p73α and p73α+30 (amino acids 364–393 of p53 fused to the C-terminus of p73α) were generated by PCR using pCMV-p53wt (39) and pcDNA3-HA-p73α (30) as templates. The primers used are as follows: p53-F: 5′-CAC CAT GGA GGA GCC GCA GTC A-3′, p53-R: 5′-TCA GTC TGA GTC AGG CCC TTC-3′, p53Δ30-R: 5′-TCA CCT GCT CCC CCC TGG CTC-3′, p73α-F: 5′-CAC CAT GGC CCA GTC CAC CGC C-3′, p73α-R: 5′-TCA GTG GAT CTC GGC CTC CGT-3′, p73α+30-A: 5′-CGA GAT CCA CGC TCA CTC CAG CCA CCT GAA G-3′, p73α+30-B: 5′-TGG AGT GAG CGT GGA TCT CGG CCT CCG TGA A-3′. pENTR-p53/p53Δ30/p73α/p73α+30 plasmids were generated by topo-ligation of the gel-purified PCR products into pENTR/D-Topo (Invitrogen). Expression vectors for in vitro translation were generated by LR-recombination of pENTR vectors with pEXP1/DEST vectors leading to the expression of His-tagged proteins using the Gateway system (Invitrogen).
pAdTrack-HA-p53/p53Δ30/p73α/p73α+30 vectors were generated by in vitro recombination of Gateway-adapted pAdTrack-CMV-HA with respective pENTR vectors using the Gateway system (Invitrogen). pAdGFP-HA-p53/p53Δ30/p73α/p73α+30 were generated by bacterial recombination using the AdEasy system as described (37).
To generate YFP fusion proteins of p53, p53Δ30, p73α and p73α+30, the coding sequence of EYFP (Clontech) was cloned into XbaI and PacI sites of pUC19-SfiI/T7 (kindly provided by Rob Chapman). In addition, the respective p53/p73 cDNA were cloned into XhoI and XbaI sites and an HA-Flag-tag into AgeI and SpeI sites of pUC19-SfiI/T7. The complete resulting coding sequence was excised with SfiI and cloned into the episomal doxycycline-inducible expression vector pRTS-1 (40).
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed in 20 µl mixtures containing 20 mM HEPES (pH 7.8), 0.5 mM EDTA (pH 8.0), 6 mM MgCl2, 60 mM KCl, 1 mM DTT, 120 ng sheared salmon sperm DNA, 0.1 pmol of 32P end-labeled oligonucleotide, and 1–5 µl of the in vitro translated protein (TNT Quick Coupled Transcription/Translation Set; Promega). For supershift analysis 0.2 µg α-HA (Y-11, Santa Cruz) or α-His (Qiagen) antibody were added. The p53 CTD was blocked by addition of 0.2 µg α-p53 (PAb421, Calbiochem) antibody. For competition experiments a 200-fold molar excess of self-competitor (oligonucleotide with the same sequence as the labeled probe without the 32P labels) or unspecific competitor (unlabeled oligonucleotide with a scrambled sequence) were used. After 30 min incubation at room temperature, reaction mixtures were subjected to electrophoresis on a 3.5% native polyacrylamide gel (37.5:1 acrylamide/bisacrylamide) containing 0.5× Tris–borate–EDTA buffer at 150 V for 120 min at room temperature. DNA protein complexes were visualized by phosphorimaging (FLA-3000, Fujifilm). For quantification, phosphor-stimulated luminescence (PSL) values were background corrected and normalized to the sum of the bound and unbound oligonucleotide signals.
The following double-stranded oligonucleotides (p53-binding sites underlined) were used for EMSAs: p53 consensus sequence 5′-GGG TAG ACA TGC CTA GAC ATG CCT AAG CTC CC-3′, p53 consensus scrambled 5′-GGG CCA GCT AGC AGG CAG CAT CAG TAC TTC CC-3′, p21-5′ site (28 bp) 5′-TCA GGA ACA TGT CCC AAC ATG TTG AGC T-3′, p21-5′ site (66 bp) 5′-AGC ATT GGC TTT CTG GCC ATC AGG AAC ATG TCC CAA CAT GTT GAG CTC TGG CAT AGA AGA GGC GCT-3′.
Protein analysis
The isoelectric points of the total protein, the C-terminal region (CT) and the last 30 amino acids were calculated with VectorNTI software (Invitrogen). Cells were lysed in RIPA buffer (50 mM Tris–Cl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and total protein concentration was quantified by Bradford assay. Cell lysates or in vitro translated protein samples were separated by SDS–PAGE and transferred to nitrocellulose membranes (Amersham). After blocking with 10% nonfat dry milk, membranes were probed with antibodies specific for the His-tag (Qiagen), HA-tag (Y-11, SantaCruz) or β-actin (AC15, Abcam) and visualized using appropriate horseradish-peroxidase-conjugated secondary antibodies according to the enhanced chemiluminescence protocol.
Fluorescence recovery after photobleaching (FRAP)
Cells were seeded onto gelatine-coated coverslips, transfected with Tet-inducible expression vectors for p53, p53Δ30, p73α or p73α+30 as indicated and treated with 1 µg/ml doxycycline. Twenty-four hours post transfection, coverslips were transferred to a live-cell imaging chamber with DMEM without phenol red (PAN) supplemented with 10% fetal bovine serum. Photobleaching experiments were performed on a Leica TCS SP5 equipped with a 63/1.4 NA oil immersion objective. YFP was excited using the 514 nm laser line of an argon ion laser and imaged with a spectral detector with a bandpass set at 518–608 nm. A YFP fluorescence image of the whole cell was captured before and after the FRAP experiment to determine the location of the nucleus and to determine the photobleaching and recovery levels, including observations of immobile fractions. The FRAP protocol was performed similarly to the method of Hinow et al. (41). Briefly, a 41.01 by 2.56 µm strip selected from the cellular image that sliced out a central region through the nucleus was imaged 20 times before bleaching with 1% laser power of the 514 nm laser line. This strip was photobleached by a series of bleach pulses for 1 s using all of the argon ion laser lines combined at maximum (100%) power. Recovery of fluorescence was measured for 20 s with settings as before bleach in intervals of 100, 250 and 500 ms for 2, 3 and 15 s intervals, respectively.
Mean fluorescence intensity values of the nuclear part were analyzed for each time point with either the Leica SP5 software (LASAF ‘Frap Wizard’, Leica) or with ImageJ software (NIH). Background intensities (of each time point) were subtracted from each image before analysis. To allow direct comparison of individual FRAP curves, raw data were normalized to the mean prebleach intensity of each cell measurement. The recovery curves were then analyzed in Matlab (MathWorks, USA) with modified routines from Hinow et al. (41) to determine the parameters of the extended binding or free diffusion model. The percentage of immobile fraction is calculated by comparing prebleach intensity with recovered intensities. For the best average representation of the FRAP data, the recovery curves were averaged from at least 16 cells from several measurements on different days, depicting, the mean fluorescence intensity ± SE of the normalized values. Each of the individual curves was also tested for the extended binding model whose fitting to the model could be rejected by goodness of fit parameters suggested by Hinow et al. (41).
Chromatin immunoprecipitation
The ChIP assay was essentially performed as described (42). Briefly, H1299 cells infected with adenoviruses AdGFP, AdGFP-HA-p53 or AdGFP-HA-p53Δ30 were fixed with formaldehyde (final concentration 1% v/v) in the cell culture medium for 10 min at room temperature. Glycine was added to a final concentration of 0.125 M to stop cross-linking. Fixed cells were washed twice with ice cold PBS, scraped and pelleted by centrifugation. Cells were resuspended in SDS lysis buffer (50 mM Tris–HCl pH 8.1, 10 mM EDTA, 1% v/v SDS) including protease inhibitors and sonificated to yield chromatin fragments of approximately 500–1000 bp as assessed by agarose gel electrophoresis. The cell lysates were diluted with ChIP dilution buffer (167 mM NaCl, 16.7 mM Tris–HCl pH 8.1, 1.2 mM EDTA, 1.1% v/v Triton X-100, 0.01% v/v SDS), precleared by incubation with blocked protein G agarose (Upstate) and incubated with 1 µg of anti-HA (Y-11, SantaCruz) or anti-largeT (Pab 101, Santa Cruz) antibodies overnight at 4°C. Samples of total chromatin were taken at this point as input controls for the PCR reactions. Blocked protein G agarose was added and DNA–protein complexes were washed with low salt (150 mM NaCl, 20 mM Tris–HCl pH 8.1, 2 mM EDTA, 1% v/v Triton X-100, 0.1% v/v SDS), high salt (500 mM NaCl, 20 mM Tris–HCl pH 8.1, 2 mM EDTA, 1% v/v Triton X-100, 0.1% v/v SDS), LiCl immune complex wash buffer (20 mM Tris–HCl pH 8.1, 2 mM EDTA, 1% v/v deoxycholic acid, 1% v/v IGEPAL-CA630, 250 mM LiCl) and twice with TE buffer and finally eluted with two incubations in elution buffer (1% v/v SDS, 0.1 M NaHCO3) at room temperature for 15 min each. Cross-links were reversed by incubation at 65°C for 5 h followed by RNase A (37°C, 30 min) and proteinase K (45°C, 2 h) treatment. Samples were extracted twice with phenol-chloroform and ethanol precipitated overnight in the presence of 20 µg glycogen as carrier. Resuspended DNA fragments were used for quantitative PCR with primers for the GAPDH (5′-GTA TTC CCC CAG GTT TAC AT-3′, 5′-TTC TGT CTT CCA CTC ACT CC-3′) and the 5′ p53-binding site in the p21 promoter (5′-GTG GCT CTG ATT GGC TTT CTG-3′, 5′-CTG AAA ACA GGC AGC CCA AG-3′). ChIP data from triplicate measurements were expressed as promoter occupancy in percent of input DNA.
RNA isolation, reverse transcription PCR
RNA was isolated using the RNeasy Kit (Qiagen) and quantified with RiboGreen (Molecular Probes). Total RNA (1 µg) was reverse transcribed using Omniscript RT (Qiagen) with random hexamers or the QuantiTect Reverse Transcription Kit (Qiagen). Specific cDNAs were quantified by qPCR performed in triplicate reactions on an ABI Prism 7000 Sequence Detection System (Applied Biosystems) using the SYBR Green JumpStart Taq ReadyMix (Sigma) or the QuantiTect SYBR Green PCR Kit (Qiagen) in combination with QuantiTect Primer Assays for GDF15, JAG2, FDXR, PUMA/BBC3 and S100A2 (Qiagen). Primer sequences for p21: 5′-TGG AGA CTC TCA GGG TCG AAA-3′, 5′-GGC GTT TGG AGT GGT AGA AAT C-3′, GAPDH: 5′-AAT GGA AAT CCC ATC ACC ATC T-3′, 5′-CGC CCC ACT TGA TTT TGG-3′. Amplification specificity was verified by melting curve analysis and agarose gel electrophoresis of the PCR products. Expression data were normalized to GAPDH and the GFP-infected control sample using the ΔΔCt method.
Microarrays
H1299 cells were harvested 24 h after infection with adenovirus expressing GFP together with HA-tagged p53/p53Δ30/p73α/p73α+30 or GFP only to control for effects of adenoviral infection. Total RNA was extracted as described above. Equal amounts of RNA were pooled from three independent infections. The samples were analyzed for gene expression using Affymetrix GeneChip Human Genome U133A 2.0 Arrays and further analyzed with GeneSpring 7.0 (Silicon Genetics) to extract a list of 191 genes that were changed >3-fold in any p53/p73 expressing sample compared to the GFP control. The complete set of microarray data has been deposited in NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and is accessible through GEO Series accession number GSE8660.
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=fdojhkemwaiskvc&acc=GSE8660
Flow cytometry
H1299 or Saos-2 cells were collected 24 h after infection, fixed in 70% ethanol and stained for DNA content with propidium iodide (Sigma). Cell cycle profiles were measured directly on a FACScalibur with Cellquest software (Becton Dickinson). Cell cycle profiles were analyzed with ModFit LT software (Becton Dickinson).
Statistical analysis
All numerical data are reported as the mean ± standard deviation (SD) or standard error (SE) as indicated. Statistical significance was calculated using the two-sided Student's t-test.
RESULTS
C-terminal diversity of p53 and p73 isoforms influences sequence-specific DNA binding
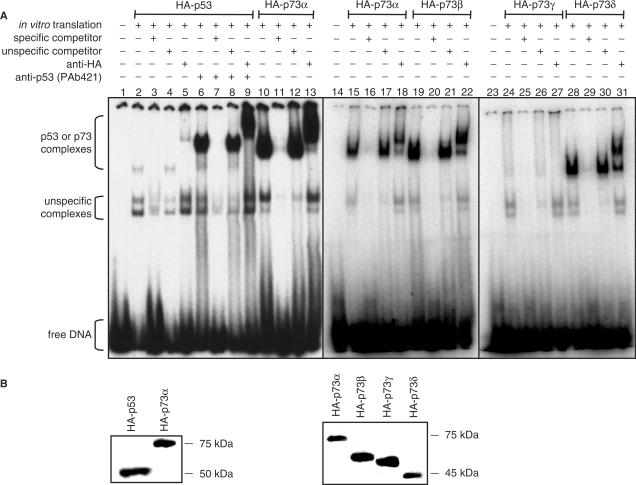
Although p73 shares ∼65% amino acid identity with the DNA binding domain of p53 and binds to the p53 consensus DNA sequence (43), the two proteins exhibit fundamentally different biological functions. To determine whether differences are already present on the level of DNA binding we performed electrophoretic mobility shift assays (EMSAs) with in vitro translated proteins and a 32P-labeled 28-bp double-stranded oligonucleotide containing the p53 consensus binding sequence. Consistent with previous studies (44–46) we observed only weak p53 complexes (Figure 2A, lanes 2–5). In contrast, sequence-specific p73α complexes were inherently strong (Figure 2A, lanes 10–13), despite equal amounts of p53 and p73α (Figure 2B, left panel). The specificity of these protein–DNA complexes was confirmed by efficient competition with the same but not a scrambled unlabeled oligonucleotide and by supershift of the complex upon addition of a specific antibody. Importantly, p53 complex intensity was increased substantially and reached the intensity of the p73 complexes following pre-incubation with the antibody PAb421 (Figure 2A, lanes 6–9). Since this antibody binds to a C-terminal epitope and blocks the p53 CTD, functional differences of the p53 and p73α CTDs presumably account for the different complex intensities.
Figure 2.
C-terminal diversity of p53 and p73 isoforms influences sequence-specific DNA binding. (A) EMSA showing binding of in vitro translated p53 and p73α, β, γ and δ to a double-stranded, 32P labeled oligonucleotide containing a consensus p53-binding site (5 nM). A 200-fold molar excess of the same oligonucleotide without 32P label was used as a specific competitor, a scrambled oligonucleotide as an unspecific competitor. 200 ng anti-HA antibody was added for supershift analysis as indicated. 200 ng anti-p53 (PAb421) was added to block the p53 CTD. (B) Western Blot demonstrating equal amounts of all in vitro translated proteins. Immunodetection was performed with an anti-HA antibody.
We therefore further examined other p73 isoforms that differ in their CTDs. p73β and p73δ are C-terminally truncated versions of p73α, whereas p73γ is characterized by a unique C-terminal stretch of 76 amino acids (Figure 1). Similar to p73α, the smaller p73 isoforms p73β and p73δ formed strong sequence-specific DNA complexes (Figure 2A, lanes 19–22, 28–31), whereas p73γ, although equally expressed (Figure 2B, right panel), formed only weak complexes similar to p53 (Figure 2A, lanes 24–27). Together, these data demonstrate that the various p53 family proteins differ substantially with respect to sequence-specific DNA binding in vitro and suggest that sequence-specific DNA binding is strongly influenced by functionally different CTDs.
Sliding properties explain differences in sequence-specific DNA binding
The inhibitory effect of the p53 CTD on sequence-specific DNA binding has repeatedly been observed in classical EMSA experiments (10,14,44–47) and has recently been explained by the ability of the C-terminus to promote linear diffusion (sliding) along the DNA via sequence-nonspecific DNA interactions (14,15). In this model, p53 can either dissociate from its specific binding site where it is tightly bound in a sequence-specific manner via its core domain or it can slide off the ends of a DNA fragment via weak nonspecific interactions mediated by the C-terminus. In the context of short oligonucleotides, linear diffusion is dominant and results in p53 rapidly sliding off the ends, which destabilizes the protein–DNA complexes. Increasing the length of the oligonucleotide or blocking the oligonucleotide ends with streptavidin prevents sliding off, eliminates this mode of complex dissociation and enhances overall complex stability (14,15). To investigate the role of linear diffusion for the observed differences in p53/p73 DNA binding we analyzed the effects of longer oligonucleotides on complex stability.
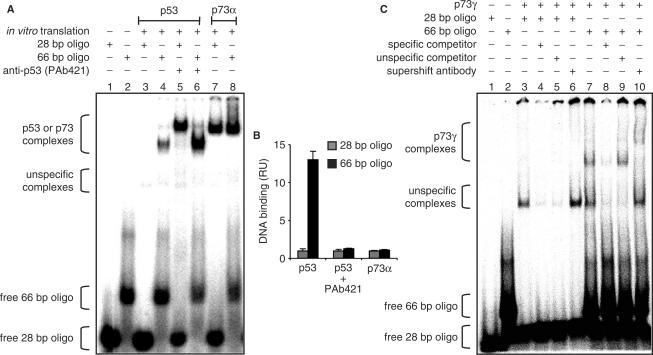
First, we performed EMSAs with 28 and 66 bp DNA fragments containing the 5′-binding site of the p21 promoter, a common target of p53 and p73. As expected, p53 complex intensity increased with the 66 bp DNA (Figure 3A, lanes 3 and 4). In the presence of PAb421, p53 complex intensity was even stronger but no longer dependent on oligonucleotide length, indicating that the negative impact of sliding on sequence-specific DNA binding is partially abrogated in the context of the longer DNA and completely disabled by blocking p53's CTD (Figure 3A, lanes 5 and 6, Figure 3B). Of note, the p53–DNA complex with the 66 bp oligonucleotide migrated faster than the 28 bp complex which might be due to bending of the longer DNA or a different complex stoichiometry (Figure 3A, lanes 5 and 6). Nevertheless, the p73α complex was completely independent of oligonucleotide length, providing evidence that DNA binding of p73α is not inhibited by sliding (Figure 3A lanes 7 and 8, Figure 3B). In contrast, DNA binding of the p73 isoform p73γ was stimulated by longer DNA (Figure 3C), suggesting that the p73γ C-terminus has sliding properties similar to p53. Similarly, complex intensities of p53 and p73γ, but not p73α, were enhanced when the DNA ends were blocked by streptavidin barriers (data not shown). Together these experiments indicate that p53 and p73γ but not p73α/β/δ are capable of sliding and that this accounts for the observed differences in sequence-specific DNA binding.
Figure 3.
Enhanced binding of p53 and p73γ but not p73α to longer DNA oligonucleotides. (A) EMSA showing binding of in vitro translated p53 and p73α to double-stranded 32P labeled oligonucleotides containing 28 bp or 66 bp from the 5′ p53-binding site of the human p21 promoter. The p53 CTD was blocked by addition of 200 ng anti-p53 (PAb421) antibody. Protein–DNA complexes were visualized and quantified by phosphorimaging. (B) Graphical analysis of the phosphorimaging data depicted in (A). Binding was normalized to the 28 bp oligonucleotide. Data represent the mean ± SD of three independent EMSA experiments. RU, relative units. (C) DNA binding of in vitro translated p73γ as described in (A). A 200-fold excess of the same oligonucleotide without 32P label was used as a specific competitor, a scrambled sequence as an unspecific competitor.
Sliding properties of p53 family proteins correlate with differences in C-terminal charge
Knowing that the p53 C-terminus is a highly positively charged, basic domain and that linear diffusion is largely based on electrostatic interactions between a positively charged protein domain and the negatively charged DNA backbone (48), we calculated the isoelectric points (pI) of the various p53 and p73 C-termini (Figure 1). In contrast to the p53 CTD, which has a basic isoelectric point (pI 10.0), the C-termini of p73α, p73β and p73δ have a neutral pI between 5.52 and 7.92. The isoelectric point of p73γ is much higher than that of the other p73 isoforms in respect to both the complete C-terminus (CT, pI 11.76) and the last 30 amino-acids (pI 8.77) resulting in a positively charged C-terminus. The calculated C-terminal charges therefore directly correlate with the observed DNA-binding properties and explain the different sliding abilities.
C-terminal charge influences sequence-specific DNA binding in vitro
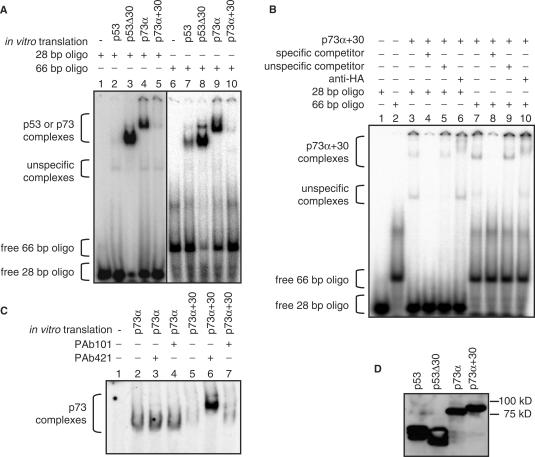
To test the impact of sliding on sequence-specific DNA-binding avoiding confounding effects of different protein backbones, we generated a chimeric construct p73α+30 containing the positively charged C-terminal 30 amino acids of p53 fused to the C-terminus of p73α. A deletion mutant of p53 lacking the C-terminal domain (p53Δ30) had been previously described and was used as a control (14). Deletion of p53's CTD strongly enhanced DNA binding whereas fusion of this domain to p73α reduced the DNA binding of p73α (Figure 4A) although respective protein levels were pairwise equal (Figure 4D). These effects were observed in the context of long 66 bp oligonucleotides (Figure 4A, right panel) and even more pronounced with short 28 bp oligonucleotides (Figure 4A, left panel). Similar to p53, DNA binding of p73α+30 was stimulated approximately 5-fold by long 66 bp compared to 28 bp oligonucleotides (Figure 4B). The DNA-binding activity of p73α+30 was restored to p73α levels upon pre-incubation with PAb421, ruling out that the fusion-caused misfolding of the p73 core DNA-binding domain (Figure 4C). These data demonstrate that the positively charged p53 CTD is an autonomous and transposable domain that confers sliding properties and negatively affects the sequence-specific DNA-binding properties of not only p53 but also when fused experimentally to p73.
Figure 4.
The p53 CTD is an autonomous transposable negative regulatory domain. (A, B) EMSA of the indicated in vitro translated proteins to 32P labeled oligonucleotides of the indicated size containing the 5′ p53-binding site of the p21 promoter. A 200-fold molar excess of the same oligonucleotide without 32P label was used as a specific competitor, a scrambled oligonucleotide as an unspecific competitor. A 200 ng anti-HA antibody was added for supershift analysis as indicated. (C) EMSA of p73 and p73 + 30 binding to the 32P labeled p53 consensus oligonucleotide. The p53 CTD was blocked by addition of 200 ng PAb421. PAb101 directed against the SV40 large T antigen was used as a control. (D) Western blot demonstrating pairwise equal amounts of protein.
p53 CTD limits intranuclear mobility in vivo
Sequence-nonspecific interactions with chromatin and linear diffusion along the DNA backbone are expected to significantly reduce the freedom in its motion. Hinow et al. have recently shown by fluorescence recovery after photobleaching (FRAP) experiments that the intranuclear mobility of p53 largely follows a sequence-nonspecific-binding model which requires a binding region of the protein apart from the DNA binding core domain (41). We therefore used FRAP to determine whether a charged CTD influences the mobility of p53 and p73 in the living cell (Figure 5). For this p53, p53Δ30, p73α and p73α+30 were fused to the yellow fluorescent protein (YFP) and transiently expressed in p53-null H1299 cells. All fusion proteins were expressed to equal levels and localized predominantly to the nucleus (Figure 5A). Photobleaching was performed on a 2.56 µm wide strip for 1 s. Sequential scanned images following photobleaching were utilized to determine the kinetics of fluorescence recovery (Figure 5B). Freely diffusing YFP and the immobile histone fusion protein H2B-YFP were also analyzed for comparison showing a fast, nearly nondetectable bleach and recovery for free YFP and a clearly deep bleach without recovery for the histone. Bleaching depths for p53 and p73α were comparable and both recovered to similar extent. Under equal bleaching conditions, the bleaching was less and recovery was higher for p53Δ30 compared to p53. In contrast, the fusion of the p53 CTD to the p73α C-terminus increased the bleaching and lowered the recovery level. A reduced bleaching depth in comparison to p53 as seen for p53Δ30 indicates a higher mobility similar to the free YFP and results from rapid recovery during the bleaching period before the first detected recovery data point. A high bleaching depth as seen for p73α+30 in comparison to p73α is typical for immobile proteins like the chromatin component histone H2B.
Figure 5.
Influence of p53 CTD on intranuclear mobility. (A) Fluorescence micrographs of H1299 cells transfected to express YFP fusion proteins of p53, p53Δ30, p73 and p73 + 30. Scale bar, 10 µm. (B) Fluorescence recovery after photobleaching (FRAP). H1299 cells were transfected with YFP fusion proteins. Shown are fluorescence recovery curves calculated from sequential scanned images recorded after photobleaching of a 2.56 µm wide strip for 1 s. Examples of the bleached areas are shown in (A). Fluorescence intensity before photobleaching was set to 100%. Each data point represents the mean ± SE of n cells. Recovery curves of immobile histone H2B and highly mobile free YFP are shown for comparison. (C) Representative FRAP data from individual cells fitted to the free-diffusion model (red curves) and the extended reaction-diffusion model (blue curves) according to Hinow et al. (41). P value, confidence level of improvement by the extended model.
As a further test, we applied the reaction-diffusion model suggested by Hinow et al. (41) which includes binding and release rates of the diffusing species to an immobile structure on the individual recovery curves for p53, p53Δ30, p73α and p73α+30 to find first that the YFP fusion of p53 showed similar behavior as reported for the GFP fusion to p53 (41). Second, in the case of p53Δ30 and p73α the fit to the reaction-diffusion model (Figure 5C, blue curves) provided no further improvement compared to a one-parameter model involving only free diffusion (Figure 5C, red curves), indicating no contribution of nonspecific DNA binding to intranuclear mobility (Figure 5C). In summary, the proteins containing the basic CTD were more similar to the immobile histone H2B, whereas the proteins lacking a basic CTD were more similar to free YFP. Thus we conclude that the charged p53 CTD mediates sequence-nonspecific interactions with genomic DNA in vivo.
p53 CTD inhibits p21 promoter binding in vivo
Considering that the ability to slide inhibits sequence-specific DNA binding in vitro and intranuclear mobility in vivo, we analyzed by chromatin immunoprecipitation (ChIP) whether a similar negative impact can be observed on gene expression in vivo. It has been previously shown that the p53 CTD functions as a positive regulator of DNA binding in vivo when p53 is exogenously expressed in p53-null cells under conditions where the amount of p53 protein is limiting (14,15). Under these conditions scanning of the genome by linear diffusion is expected to enhance the identification of p53-binding sites in the complex genome. However, in p53-wild-type cells, p53 is known to be present on target promoters like the p21 promoter under both stressed and unstressed conditions so that the identification of binding sites in the genome should not be the rate-limiting step in the stress response (13,49). To investigate if the p53 CTD influences promoter binding under these conditions, we expressed p53 (or p53Δ30) in H1299 cells by means of adenoviral infection and analyzed promoter binding 24 h later when constantly high steady-state levels of p53 were obtained.
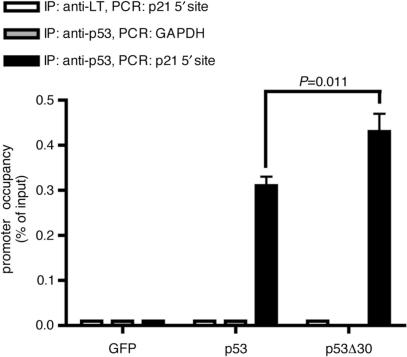
ChIP analyses revealed enrichment of p21CDKN1A promoter DNA by both p53 and p53Δ30 (Figure 6). p73α and p73α+30 did not significantly associate with the p21 promoter (data not shown) consistent with the lack of p21 transactivation under these conditions (Figure 7C). Importantly, promoter binding was significantly higher for p53Δ30 than for p53 (P = 0.011). The specificity of these results was confirmed by the lack of binding to the promoter of the non-target gene GAPDH and by the absence of a p21 promoter signal when using a nonspecific antibody (Figure 6). We conclude that the positively charged p53 CTD has a statistically significant negative regulatory impact on sequence-specific DNA binding not only in vitro but also in vivo when the amount of p53 protein is not limiting.
Figure 6.
Deletion of the p53 CTD enhances in vivo binding to the p21 promoter. ChIP (chromatin immunoprecipitation assay) comparing in vivo binding of p53 and p53Δ30 to the 5′ p53-binding site in the p21 promoter. Binding to a region within the GAPDH promoter is shown as a negative control. Binding to the p21 promoter of a control immunoprecipitation using an antibody directed against large T antigen (LT) is shown as an additional negative control. ChIP data were quantitated by qPCR and normalized to their respective inputs for p21 and GAPDH. The graph shows the mean and SD of promoter occupancy in percent of input DNA (n=3). p21 promoter occupancy of p53 and p53Δ30 were significantly different (P=0.011). Protein levels of p53 and p53Δ30 are shown in Figure 7B.
Figure 7.
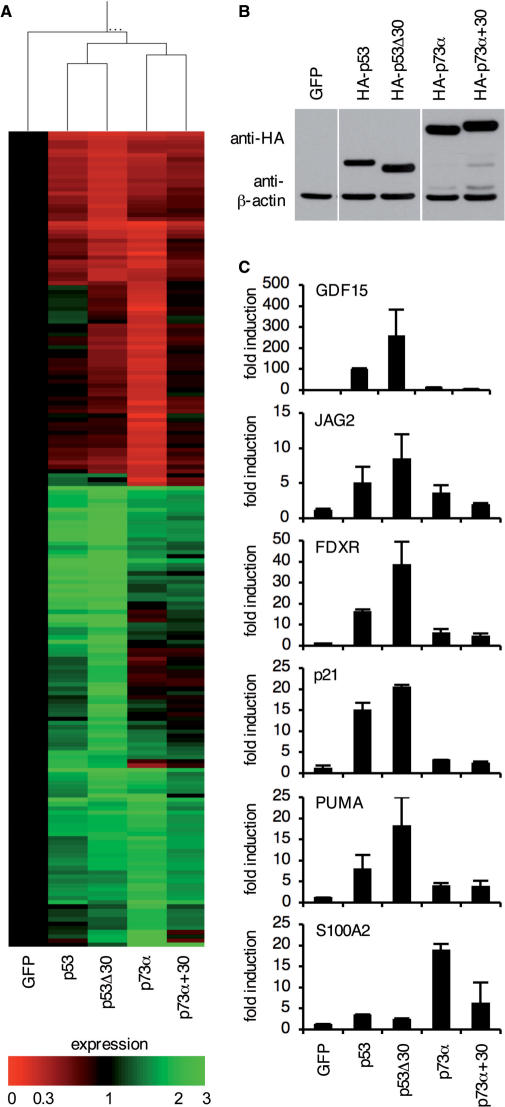
The p53 CTD reduces transactivation of target genes. (A) Gene expression profiles of H1299 cells infected with adenoviruses expressing GFP alone or in combination with p53, p53Δ30, p73α or p73α+30. Samples from three independent infections were pooled for microarray analysis using Affymetrix Human Genome U133A 2.0 Arrays. Shown is a heatmap of the 191 genes that were changed by more than 3-fold in at least one of the four p53/p73 samples compared to the GFP control. (B) Western blot demonstrating pairwise equal expression of p53 and p73 proteins following infection of H1299 cells with the indicated adenoviruses. β-actin expression is shown as a loading control. (C) Quantitative RT-PCR of the samples in (A) for expression of p53 target genes (GFP15, p21, JAG2, PUMA/BBC3, FDXR) and the p73 target gene S100A2. Shown is the mean fold induction ± SD of three replicate measurements.
p53 CTD reduces transcriptional activity of p53 and p73
As DNA binding of a transcription factor is a prerequisite for transactivation, we next examined differences in genome-wide gene expression profiles using unbiased oligonucleotide microarrays. For this we isolated mRNA of H1299 cells 24 h after infection with adenoviruses expressing pairwise equal amounts of p53 and p53Δ30 or p73α and p73α+30, respectively (Figure 7). mRNA from three independent experiments was pooled to increase the accuracy of the microarray results. 191 genes were changed by more than 3-fold in at least one of the four p53/p73-samples compared to the AdGFP-infected control (Figure 7A). Among these genes were several known p53/p73 target genes involved in cell cycle regulation (p21CDKN1A), differentiation and development (S100A2, GDF15, JAG2) and apoptosis (FDXR, PUMA/BBC3). The heat map depicts that the target gene spectrum is mainly determined by the protein backbone, i.e. despite significant overlap condition clustering clearly distinguished the p53-driven expression profiles from the p73 samples (Figure 7A). Deletion or addition of the p53 CTD did not shift the target gene spectrum but rather changed the level of target gene activation as was confirmed by quantitative RT-PCR (Figure 7C). p53Δ30-transactivated p53 target genes stronger than wildtype p53 whereas p73α-transactivated p73 target genes better than p73α+30 (Figure 7C). Thus the absence of a positively charged C-terminus in p53Δ30 and p73α not only enhanced promoter binding in vitro and in vivo but also resulted in elevated gene transcription providing further support for a negative regulatory role of a positively charged CTD.
C-terminal domain influences cell fate control
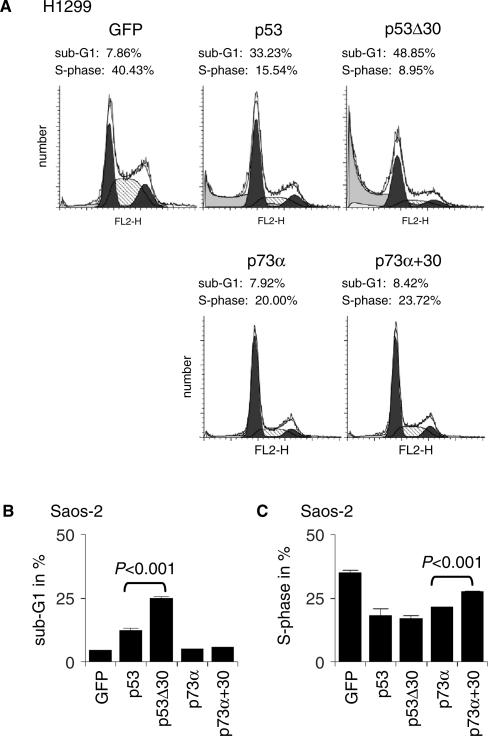
To determine whether the observed differences in DNA binding and transactivation influence cell fate decisions, we analyzed the induction of cell-cycle arrest and apoptosis. Infection of H1299 cells with adenoviruses expressing p53 or p53Δ30 resulted in strong induction of apoptosis compared to the GFP control cells (Figure 8A). As previously seen on the level of target gene expression, the effect of p53Δ30 on apoptosis induction was clearly enhanced compared to wild-type p53. p73α and p73α+30 did not increase the apoptosis rate in H1299 cells, but reduced the percentage of cells in S-phase similar to p53. Again p73α was more effective than p73α+30. The negative impact of the charged p53 CTD on the ability of p53 and p73 to regulate cell cycle progression and apoptosis was further confirmed in Saos-2 cells as a second p53-null cell line (Figure 8B and C). We conclude that a positively charged CTD reduces the anti-proliferative activity of p53 family members. The implications of these findings are discussed below.
Figure 8.
The p53 CTD inhibits the p53/p73 response. (A) H1299 cells were infected with adenoviruses expressing GFP alone or in combination with p53, p53Δ30, p73α or p73α+30. Protein levels obtained by adenoviral infection are shown in Figure 7B. Shown are the cell cycle profiles measured 24 h after infection. (B, C) Quantitative analysis of (B) apoptosis (sub-G1) and (C) S-phase 24 h after Saos-2 cells were infected and analyzed as described in (A). The observed differences are statistically significant as determined by two-sided Student's t-test.
DISCUSSION
In this study we show that the CTDs of p53 and various p73 isoforms differ in their electrostatic charge and therefore in their ability to interact with DNA in a sequence-nonspecific manner. Although the p53 CTD has long been known to be an independent DNA-binding module (5,17), our results demonstrate that a functionally equivalent domain is missing in the major p73 isoforms (with the exception of p73γ). In the case of p53, interactions of the CTD with DNA compromise the stability of sequence-specific complexes with short oligonucleotides which is elegantly explained by the recently proposed p53 sliding model (5,17). In the context of short DNA, CTD-mediated linear diffusion results in rapid sliding off the DNA ends and thus dissociation of the protein–DNA complex. Consistently, all experimental measures that interfere with this process such as blocking the DNA ends with streptavidin (14), ligating the DNA into circles (50), increasing the length of the DNA (12) or blocking the CTD with antibodies (10) have been shown to increase complex stability. As p73α, β and δ have no positively charged domain in their CTD they cannot engage in sequence-nonspecific electrostatic interactions with the negatively charged DNA backbone. Thus, their binding is not affected by any of the measures described above. Direct dissociation is rare because of strong sequence-specific protein–DNA interactions resulting in very stable p73–DNA complexes. In fact, p73α, β and δ formed equally strong complexes as p53 proteins in which the CTD had been blocked (Figure 2A) or deleted (Figure 4A). This high p73–DNA complex intensity has previously been observed in one of the first EMSAs to be reported for p73 (51). In contrast, no significant difference in DNA-binding strength was observed by Lokshin et al. (52). This might be explained by the use of short oligonucleotides as competitor DNA or recombinant proteins expressed in insect cells which show different post-translational modifications compared to p53/p73 proteins produced by in vitro translation using mammalian cell lysates (53).
In a living cell a second sequence-nonspecific DNA-binding domain would be expected to enhance chromatin association and therefore to reduce the freedom in its motion. In fact, Hinow et al. demonstrated that the intranuclear mobility of p53 is independent of its sequence-specific DNA-binding properties and largely determined by nonspecific binding to genomic DNA (41). Consistent with the CTD harboring the sequence-nonspecific DNA-binding function, we observed a change in intranuclear mobility when the CTD was altered (Figure 5). Deletion of the CTD increased the intranuclear mobility of p53, whereas addition of the p53 CTD to the p73α C-terminus reduced p73 mobility. This change in intranuclear mobility cannot simply be explained by the change in size which is only 30 amino acids and negligible compared to the total size of the protein as recently shown by small angle X-ray scattering (54). The intranuclear mobility therefore inversely correlates with the predicted degree of chromatin association indicating that protein–DNA interactions as determined in vitro are similarly observed in the living cell.
As expected from the negative regulatory role of the p53 CTD in DNA-binding experiments, our study demonstrates that the charged p53 CTD has a negative impact on p21 promoter binding and gene transcription on a genome-wide scale (Figures 6 and 7). These findings are in contrast to recent reports which demonstrated CTD-mediated sliding to enhance promoter binding and gene transcription (14,17). A possible explanation for this discrepancy could be based on the different time points and expression levels that were investigated. In contrast to focusing on the initial steps of promoter localization occurring when p53 is expressed at limited amounts in a p53-null cell, we analyzed transcription 24 h after the onset of p53 expression when p53 levels were constant and no longer limiting. We chose this late time point because endogenous p53 is bound to its target promoters in unstressed cells (13,49), so that the locating of p53-binding sites in the genome might not be the rate-limiting step of a p53-driven transcriptional stress response. It can be envisioned that instead of promoting the search for p53 response elements, sliding enhances the dissociation of promoter-bound p53 under steady-state conditions similar as observed in EMSAs and thus reduces gene transcription (Figure 9). Following initial steps where sliding is beneficial, sliding would therefore need to be turned off once a response element has been identified to stabilize the promoter-bound transactivation complex. This could be accomplished in vivo by kinases or acetylases that are known to reduce the C-terminal charge in response to stress signals (55).
Figure 9.
Model. Transcription factor binding sites in the genome are not only located and occupied but also left either directly via dissociation or indirectly via sliding. In the case of p53, p73γ and p73α+30 a charged CTD allows sequence-nonspecific interactions with the DNA, enables sliding and thus destabilizes the sequence-specific protein–DNA complex. In the case of p73α, p73β, p73δ and p53Δ30 these destablizing sequence-nonspecific interactions are absent so that stable protein–DNA complexes are formed. Blocking the DNA binding function of a basic CTD by antibodies (PAb421) in vitro or charge-neutralizing post-translational modifications (PTM) in vivo stabilizes promoter-bound complexes by preventing sliding off the binding site. Sequence-nonspecific interactions therefore not only help to identify response elements (RE) in the genome but also enable transcription factors to leave these sites and therefore render these DNA-binding activities a target for regulation.
Another important aspect revealed by our study is that the target gene spectrum is solely determined by the identity of the DNA-binding domain. Despite overlapping target genes, the p53 target gene profile can be distinguished clearly from the p73-initiated transcriptional response (Figure 7). Although molecules without a charged CTD show increased transcriptional activity, swapping CTDs does not switch the target gene spectrum. p21 is predominantly activated by p53 and p53Δ30, whereas S100A2 is only activated significantly by p73α and p73α+30. This is consistent with the model, that the CTD enhances dissociation of the protein–DNA complex by enabling sliding off without directly influencing the molecular interactions between the DNA-binding core domain and the response element that determine sequence specificity.
In contrast to p73α, β and δ, p73γ is characterized by a basic CTD that allows for sequence-nonspecific DNA binding. With respect to its DNA-binding properties p73γ is therefore the p73 isoform most similar to p53 and indeed was more similar to p53 than to p73α in all DNA binding assays. Unfortunately, very little is known about the expression pattern and physiological role of p73γ possibly due to the absence of validated antibodies that are specific for this isoform (30). Based on our results we would predict p73γ to be a latent protein similar to p53 that requires activation for full function. In line with this, it has been shown that p73γ cannot bind to p53 response elements in vitro unless acetylated by PCAF (56). Considering the different phenotype of p53 and p73 loss in mice, it is tempting to speculate that p73, which is involved primarily in developmental control, does not require rapid activity switching and therefore comes as a transcriptionally regulated protein with constitutive DNA-binding activity, whereas p53 and p73γ are endowed with a second regulatory DNA-binding domain that keeps the proteins functionally latent but primed for rapid activation in response to stress.
ACKNOWLEDGEMENTS
This work was funded by grants from the Deutsche Forschungsgemeinschaft (Transregio TR17 ‘Ras-dependent pathways in human cancer’, Forschungszentrum FZ82). A.R. is supported by the Interdisciplinary Center for Clinical Research (IZKF), University of Würzburg. We are grateful to Gerry Melino, Bert Vogelstein and Georg Bornkamm for providing plasmids, and Geoffrey Lambright and Mike Friedrich for their assistance with FRAP experiments. Funding to pay the Open Access publication charges for this article was provided by the Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stiewe T. The p53 family in differentiation and tumorigenesis. Nat. Rev. Cancer. 2007;7:165–168. doi: 10.1038/nrc2072. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Lane DP. p53 in health and disease. Nat. Rev. Mol. Cell. Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 4.Kim E, Deppert W. The versatile interactions of p53 with DNA: when flexibility serves specificity. Cell Death Differ. 2006;13:885–889. doi: 10.1038/sj.cdd.4401909. [DOI] [PubMed] [Google Scholar]
- 5.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- 7.Takenaka I, Morin F, Seizinger BR, Kley N. Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J. Biol. Chem. 1995;270:5405–5411. doi: 10.1074/jbc.270.10.5405. [DOI] [PubMed] [Google Scholar]
- 8.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 10.Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 11.Ayed A, Mulder FA, Yi GS, Lu Y, Kay LE, Arrowsmith CH. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- 12.Espinosa JM, Emerson BM. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- 13.Kaeser MD, Iggo RD. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl Acad. Sci. USA. 2002;99:95–100. doi: 10.1073/pnas.012283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKinney K, Mattia M, Gottifredi V, Prives C. p53 linear diffusion along DNA requires its C terminus. Mol. Cell. 2004;16:413–424. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Lagowski JP, Vanderbeek GE, Kulesz-Martin MF. Facilitated search for specific genomic targets by p53 C-terminal basic DNA binding domain. Cancer Biol. Ther. 2004;3:1102–1108. doi: 10.4161/cbt.3.11.1189. [DOI] [PubMed] [Google Scholar]
- 16.Jiao Y, Cherny DI, Heim G, Jovin TM, Schaffer TE. Dynamic interactions of p53 with DNA in solution by time-lapse atomic force microscopy. J. Mol. Biol. 2001;314:233–243. doi: 10.1006/jmbi.2001.5129. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Kulesz-Martin MF. Sliding into home: facilitated p53 search for targets by the basic DNA binding domain. Cell Death Differ. 2006;13:881–884. doi: 10.1038/sj.cdd.4401905. [DOI] [PubMed] [Google Scholar]
- 18.Crook T, Marston NJ, Sara EA, Vousden KH. Transcriptional activation by p53 correlates with suppression of growth but not transformation. Cell. 1994;79:817–827. doi: 10.1016/0092-8674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 19.Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, et al. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat. Med. 1998;4:839–843. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- 20.Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- 21.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 22.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 23.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 24.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 25.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 26.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery C.A., Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 27.Harvey M, McArthur MJ, Montgomery C.A., Jr, Butel JS, Bradley A, Donehower LA. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat. Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 28.Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J. Exp. Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Laurenzi VD, Catani MV, Terrinoni A, Corazzari M, Melino G, Costanzo A, Levrero M, Knight RA. Additional complexity in p73: induction by mitogens in lymphoid cells and identification of two new splicing variants epsilon and zeta. Cell Death Differ. 1999;6:389–390. doi: 10.1038/sj.cdd.4400521. [DOI] [PubMed] [Google Scholar]
- 32.Zhu J, Jiang J, Zhou W, Chen X. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 1998;58:5061–5065. [PubMed] [Google Scholar]
- 33.Yu J, Zhang L, Hwang PM, Rago C, Kinzler KW, Vogelstein B. Identification and classification of p53-regulated genes. Proc. Natl Acad. Sci. USA. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozaki T, Naka M, Takada N, Tada M, Sakiyama S, Nakagawara A. Deletion of the COOH-terminal region of p73alpha enhances both its transactivation function and DNA-binding activity but inhibits induction of apoptosis in mammalian cells. Cancer Res. 1999;59:5902–5907. [PubMed] [Google Scholar]
- 35.Ueda Y, Hijikata M, Takagi S, Chiba T, Shimotohno K. Transcriptional activities of p73 splicing variants are regulated by inter-variant association. Biochem. J. 2001;356:859–866. doi: 10.1042/0264-6021:3560859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee CW, La Thangue NB. Promoter specificity and stability control of the p53-related protein p73. Oncogene. 1999;18:4171–4181. doi: 10.1038/sj.onc.1202793. [DOI] [PubMed] [Google Scholar]
- 37.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl Acad. Sci. USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stiewe T, Theseling CC, Putzer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J. Biol. Chem. 2002;277:14177–14185. doi: 10.1074/jbc.M200480200. [DOI] [PubMed] [Google Scholar]
- 39.Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 40.Bornkamm GW, Berens C, Kuklik-Roos C, Bechet JM, Laux G, Bachl J, Korndoerfer M, Schlee M, Holzel M, Malamoussi A, et al. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 2005;33:e137. doi: 10.1093/nar/gni137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinow P, Rogers CE, Barbieri CE, Pietenpol JA, Kenworthy AK, DiBenedetto E. The DNA binding activity of p53 displays reaction-diffusion kinetics. Biophys. J. 2006;91:330–342. doi: 10.1529/biophysj.105.078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cam H, Griesmann H, Beitzinger M, Hofmann L, Beinoraviciute-Kellner R, Sauer M, Huttinger-Kirchhof N, Oswald C, Friedl P, Gattenlohner S, et al. p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer Cell. 2006;10:281–293. doi: 10.1016/j.ccr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 43.Jost CA, Marin MC, Kaelin W.G., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 44.Hupp TR, Lane DP. Allosteric activation of latent p53 tetramers. Curr. Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 45.Hupp TR, Lane DP. Regulation of the cryptic sequence-specific DNA-binding function of p53 by protein kinases. Cold Spring Harb. Symp. Quant. Biol. 1994;59:195–206. doi: 10.1101/sqb.1994.059.01.024. [DOI] [PubMed] [Google Scholar]
- 46.Delphin C, Cahen P, Lawrence JJ, Baudier J. Characterization of baculovirus recombinant wild-type p53. Dimerization of p53 is required for high-affinity DNA binding and cysteine oxidation inhibits p53 DNA binding. Eur. J. Biochem. 1994;223:683–692. doi: 10.1111/j.1432-1033.1994.tb19041.x. [DOI] [PubMed] [Google Scholar]
- 47.Jayaraman L, Moorthy NC, Murthy KG, Manley JL, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 49.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol. Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 50.McKinney K, Prives C. Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol. Cell. Biol. 2002;22:6797–6808. doi: 10.1128/MCB.22.19.6797-6808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marin MC, Jost CA, Irwin MS, DeCaprio JA, Caput D, Kaelin W.G., Jr Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol. Cell. Biol. 1998;18:6316–6324. doi: 10.1128/mcb.18.11.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lokshin M, Li Y, Gaiddon C, Prives C. p53 and p73 display common and distinct requirements for sequence specific binding to DNA. Nucleic Acids Res. 2007;35:340–352. doi: 10.1093/nar/gkl1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warnock LJ, Raines SA, Mee TR, Milner J. Role of phosphorylation in p53 acetylation and PAb421 epitope recognition in baculoviral and mammalian expressed proteins. FEBS J. 2005;272:1669–1675. doi: 10.1111/j.1742-4658.2005.04589.x. [DOI] [PubMed] [Google Scholar]
- 54.Tidow H, Melero R, Mylonas E, Freund SM, Grossmann JG, Carazo JM, Svergun DI, Valle M, Fersht AR. Quaternary structures of tumor suppressor p53 and a specific p53-DNA complex. Proc. Natl Acad. Sci. USA. 2007;104:12324–12329. doi: 10.1073/pnas.0705069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 56.Momii Y, Izumi H, Shiota M, Onitsuka T, Abe T, Kobayashi H, Miyamoto N, Uchiumi T, Kohno K. p73gamma transactivates the p21 promoter through preferential interaction with the p300/CBP-associated factor in human prostate cancer cells. Oncol. Rep. 2007;18:411–416. [PubMed] [Google Scholar]