Abstract
A novel family of transcription factors responsible for regulation of various aspects of NAD synthesis in a broad range of bacteria was identified by comparative genomics approach. Regulators of this family (here termed NrtR for Nudix-related transcriptional regulators), currently annotated as ADP-ribose pyrophosphatases from the Nudix family, are composed of an N-terminal Nudix-like effector domain and a C-terminal DNA-binding HTH-like domain. NrtR regulons were reconstructed in diverse bacterial genomes by identification and comparative analysis of NrtR-binding sites upstream of genes involved in NAD biosynthetic pathways. The candidate NrtR-binding DNA motifs showed significant variability between microbial lineages, although the common consensus sequence could be traced for most of them. Bioinformatics predictions were experimentally validated by gel mobility shift assays for two NrtR family representatives. ADP-ribose, the product of glycohydrolytic cleavage of NAD, was found to suppress the in vitro binding of NrtR proteins to their DNA target sites. In addition to a major role in the direct regulation of NAD homeostasis, some members of NrtR family appear to have been recruited for the regulation of other metabolic pathways, including sugar pentoses utilization and biogenesis of phosphoribosyl pyrophosphate. This work and the accompanying study of NiaR regulon demonstrate significant variability of regulatory strategies for control of NAD metabolic pathway in bacteria.
INTRODUCTION
NAD cofactor, in addition to its role in innumerable redox reactions, is utilized in many metabolic and regulatory processes as a consumable co-substrate (1). Among NAD-consuming enzymes are histone/protein deacetylase (2), bacterial DNA ligase (3) and a variety of ADP-ribosyltransferases (4). Maintaining homeostasis of NAD cofactor pool via regulation of biosynthetic and recycling pathways in a variety of growth conditions appears to be of paramount importance. Whereas most biochemical pathways related to NAD metabolism were studied in detail [for reviews, see (5–7)], our current knowledge of respective regulatory mechanisms is rather limited. Thus, prior to this study, only two types of bacterial transcriptional regulators related to NAD metabolism have been identified in a limited set of bacterial species (see subsequently). This prompted us to search for new candidate transcriptional factors and regulons associated with NAD metabolism in other bacteria using the comparative genomics approach [as recently reviewed in (8)].
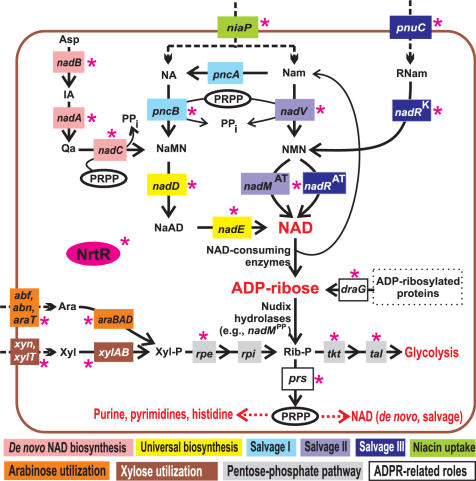
A schematic representation of the key pathways of NAD biogenesis in bacteria, including de novo biosynthesis from aspartate and various salvage pathways from the exogenous precursors—nicotinamide (Nam), nicotinic acid (NA) and ribosyl nicotinamide (RNam)—is provided in Figure 1 and described in more details in the accompanying paper (9). Different combinations of these metabolic routes result in a substantial diversity of the NAD biosynthetic machinery in various species. Using a subsystem-based approach to comparative genome analysis implemented in the SEED genomic platform (10), multiple versions of NAD metabolism were mapped in hundreds of completely sequenced bacterial genomes [as captured in the ‘NAD regulation’ subsystem at http://theseed.uchicago.edu/FIG/subsys.cgi and briefly overviewed in (11)].
Figure 1.
Overview of NAD biosynthesis and salvage pathways and a link with other metabolic pathways via ADP-ribose. NrtR-controlled steps are indicated by a red asterisk. Metabolic enzymes and uptake transporters are shown by solid and dashed lines, respectively and colored by a metabolic pathway. De novo NAD biosynthesis pathway utilizes L-aspartate oxydase (the product of nadB gene), quinolinate synthase (nadA) and quinolinate phosphoribosyltransferase (nadC). Universal NaMN to NAD pathway utilizes nicotinate mononucleotide adenylyltransferase (nadD), and NAD synthetase (nadE). In Nam salvage pathways, NaMN is synthesized from NA and Nam precursors that are taken up by niacin transporter (niaP). Salvage I pathway involves nicotinamide deaminase (pncA), and nicotinate phosphoribosyltransferase (pncB). Salvage II pathway uses nicotinamide phosphoribosyltransferase (nadV), and nicotinamide mononucleotide adenylyltransferase (nadMAT) (31,41). In the third salvage pathway, NAD is synthesized from the exogenous RNam precursor delivered by the RNam transporter (pnuC) via consecutive reactions catalyzed by two separate domains of NadR, nicotinamide mononucleotide adenylyltransferase (nadRAT), and nicotinamide riboside kinase (nadRK)(47,48). Endogenous Nam and ADP-ribose are generated by enzymes hydrolyzing the N-glycosidic bond of NAD. Enzymes linked to NAD metabolism via ADP-ribose are ribose phosphate pyrophosphokinase (prs); ribose phosphate isomerase (rpi), ribulose phosphate epimerase (rpe), transketolase (tkt), transaldolase (tal), as well as xylose (xylAB), and arabinose (araBAD) utilization enzymes. Asp, aspartate; Trp, tryptophan; IA, iminoaspartate; Qa, quinolinic acid; NaMN, nicotinate mononucleotide; NaAD, nicotinate adenine dinucleotide; NA, nicotinic acid; Nam, nicotinamide; RNam, ribosyl nicotinamide; NMN, nicotinamide mononucleotide; ADPR, ADP-ribose; PRPP, phosphoribosyl pyrophosphate; Rib-P, ribose-5-phosphate; Xyl-P, xylulose-5-phosphate; Ara, l-arabinose; Xyl, d-xylose.
The first transcriptional regulatory function for NAD synthesis was originally linked to the nadR (nadI) gene of Salmonella and Escherichia coli (12–14) prior to identification of the two mentioned enzymatic activities of this multifunctional protein. The repressor function of NadR (hence the name) is provided by an N-terminal helix-turn-helix (HTH) domain, which is present only in enterobacterial members of the NadR family. The NadR dimer in complex with the NAD co-repressor binds to a palindromic 18-bp operator with consensus sequence TGTTTA-N6-TAAACA in the promoter region of genes involved in de novo NAD biosynthesis and salvage pathways (15,16). NadR provides an interesting example of a new transcriptional regulator emerging via fusion of a DNA-binding domain with a metabolic enzyme. In contrast to other known examples of this evolutionary scenario [e.g. members of the ROK family (17)], the enzymatic domains of NadR remain functionally active. A recent comparative genomic analysis of HTH-containing members of NadR family and corresponding regulons confirmed that their occurrence is restricted to a compact phylogenetic group of Enterobacteria (18).
The second, structurally and mechanistically distinct transcriptional regulator of NAD synthesis was recently discovered and characterized in Bacillus subtilis (19) and studied in more details in the accompanying paper (9). The B. subtilis niacin-responsive DNA-binding regulator YrxA (tentatively re-named to NiaR) represses transcription of the de novo biosynthesis operon nadABC and the niacin transporter niaP (formerly yceI) by direct binding to operator sites in the promoter regions of target genes (9,19). The comparative genomic reconstruction of orthologous NiaR regulons confirmed conservation of this regulon in bacteria from the Bacillus/Clostridium group and in the deep-branched group of Thermotogales (9). However, the mechanism of transcriptional regulation of the NAD metabolism in many other bacterial lineages still remained unknown.
In this study, we expanded the use of comparative genomic techniques toward a prediction of a novel transcription regulator of NAD metabolism and de novo reconstruction of respective regulons in many diverse bacterial lineages beyond those containing NadR or NiaR regulators. The analysis of conserved NAD biosynthetic operons led to a tentative identification of a previously uncharacterized family of Nudix-related transcriptional regulators (termed here NrtR) that are composed of an N-terminal domain homologous to ADPR pyrophosphatase of the Nudix family and a C-terminal HTH-like domain. In the proposed mechanism of transcriptional regulation, the Nudix domain is responsible for a specific binding of an effector molecule, interfering with the ability of HTH-domain to bind to operator sites in the promoter regions of regulated genes. NrtR-binding sites were predicted in a variety of bacterial genomes allowing in silico reconstruction of NrtR regulons that primarily include genes involved in NAD metabolism. Two selected representatives of the NrtR family from diverse bacteria, Synechocystis sp. (Slr1690) and Shewanella oneidensis (SO1979), were experimentally characterized using gel-shift mobility assays. This analysis validated NrtR cognate binding sites deduced for these species and showed that ADP-ribose (ADPR), one of the intermediates of NAD degradation, can operate as an effector molecule weakening the NrtR–DNA complex. The detailed bioinformatic analysis revealed a number of remarkable features of the NrtR family, such as (i) conservation over a large phylogenetic distance, along with (ii) significant variations in DNA-binding motifs between species and (iii) functional diversity of regulated biochemical processes that, in addition to NAD metabolic pathways, include utilization of sugar pentoses and biogenesis of phosphoribosyl pyrophosphate (PRPP), a central precursor for the synthesis of nucleotides and amino acids.
MATERIALS AND METHODS
Bioinformatics tools and resources
Functional annotations of genes involved in NAD metabolism and related pathways in ∼400 bacterial genomes were from the collection of metabolic subsystems in The SEED comparative genomic database (http://theseed.uchicago.edu/) (10). Complete and partial bacterial genomes were downloaded from GenBank (20). Preliminary sequence data were also obtained from the web sites of The Institute for Genomic Research (http://www.tigr.org), the Welcome Trust Sanger Institute (http://www.sanger.ac.uk) and the DOE Joint Genome Institute (http://jgi.doe.gov). Multiple sequence alignments of protein sequences were produced by the Clustal series of program (21). The PHYLIP package was used for construction of maximum likelihood phylogenetic tree for the NrtR protein family including bootstrapping with 100 replicates and drawing of a consensus tree (22), The Protein Families database (Pfam) (http://www.sanger.ac.uk/Software/Pfam/) and the Clusters of Orthologous Groups (COG) database (23) were used to identify conserved functional domains. Three-dimensional protein structures were obtained from the Protein Data Bank (www.rcsb.org/pdb/). Structural similarity searches were performed using the SSM server (http://www.ebi.ac.uk/msd-srv/ssm/ssmstart.html) (24). Functional coupling of genes via clustering on the chromosome were analyzed using The SEED tools.
To identify candidate regulatory motifs, we started from sets of potentially coregulated genes (using functional and genome context considerations). An iterative motif detection procedure implemented in the program SignalX was used to identify common regulatory DNA motifs in a set of upstream gene fragments and to construct the motif recognition profiles as previously described (25). For each group of NrtR proteins on the phylogenetic tree, we used a separate training set of the upstream regions of candidate target genes to construct the NrtR binding site profile. The constructed group-specific recognition rules were used to scan a subset of genomes that encode an NrtR regulator from the corresponding group. Positional nucleotide weights in the recognition profile and Z-scores of candidate sites were calculated as the sum of the respective positional nucleotide weights [as previously described in (26)]. Genome scanning for additional candidate sites recognized by specific regulatory motifs was performed using GenomeExplorer software (27). The threshold for the site search was defined as the lowest score observed in the training set. Sequence logos for the derived group-specific DNA binding motifs were drawn using WebLogo package v.2.6 (http://weblogo.berkeley.edu/) (28).
Cloning and expression of Synechocystis sp. and Shewanella oneidensis nrtR genes
NrtR genes were amplified from genomic DNA of Synechocystis sp. strain PCC 6803 and S. oneidensis MR-1 with the set of primers shown in Supplementary Data, Table S3 and designed to incorporate a BamHI or NdeI restriction site at the 5′ end and a Bpu1102I or a BamHI site at the 3′ end of each gene, respectively. Polymerase chain reaction (PCR) conditions were as follows: 5 min at 94°C; 1 min at 94°C, 1 min at 58°C, 3 min at 72°C for 25 cycles. Reactions were performed in the presence of 0.3 pmol/μl of each primer, 1.5 mM MgCl2, 0.2 nmol/μl deoxynucleoside triphosphates, and 2 units of 0.04 U/μl Taq polymerase (Finnzymes, Espoo, Finland). PCR fragments were purified, digested with the respective enzymes, and inserted into the corresponding restriction sites of the expression vector pET15b. Escherichia coli TOP10 F′ was used for plasmid propagation, and the nucleotide sequences of the inserts were confirmed by DNA sequencing. For protein expression, the constructs were used to transform E. coli BL21 (DE3). Cells were grown at 37°C in LB medium supplemented with ampicillin (100 μg/ml). After reaching an OD600 of 0.3, cultures were shifted at 25°C and expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside at an OD600 of 0.6. After 12 h induction, cells were harvested by centrifugation at 5000g for 10 min and stored at −20°C.
Purification of recombinant NrtR
Cell pellets were resuspended in one-twentieth of the original culture volume of 10 mM potassium phosphate buffer, pH 7.5, 0.3 M NaCl, 7 mM β-mercaptoethanol (buffer A), containing 1 mM phenylmethanesulfonylfluoride (PMSF) and 0.02 mg/ml each of leupeptin, antipain, chymostatin and pepstatin. After disruption by French Press at 1000 p.s.i., suspensions were centrifuged at 40 000g for 20 min. Supernatants were diluted 10-fold with buffer A containing 1 mM PMSF and subjected to Ni+2-chelating chromatography. For syNrtR purification, the cell free extract deriving from 40 ml culture was loaded onto 500 μl Ni-NTA agarose (Qiagen) column, equilibrated with buffer A. After washing with 100 mM imidazole in buffer A, elution was performed with 350 mM imidazole in the same buffer. For soNrtR purification, the cell-free extract deriving from 40 ml culture was applied to a 1-ml HisTrap HP column (Amersham Biosciences), equilibrated with buffer A. The column was washed with 30 mM imidazole in buffer A, and elution was performed with an imidazole gradient from 30 mM to 130 mM in buffer A. Fractions containing the target proteins were collected, concentrated by ultrafiltration using YM-10 membranes (Centricon, Amicon, Bedford, MA) and subjected to gel filtration chromatography on a Superose 12 HR 10/30 column (Amersham Biosciences). For elution, a buffer containing 50 mM Tris/HCl, pH 8.0, 0.3 M NaCl and 1 mM dithiothreitol (DTT) was used. Samples were analyzed by Tricine Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) (29). Protein concentration was determined as described by Bradford (30), using bovine serum albumin as the standard.
Assay of enzymatic activity
The HPLC-based assay described in (31) was used to measure the Nudix hydrolase activity of both NrtR proteins. The reaction mixture contained 50 mM HEPES, pH 8.0, 5 mM MgCl2 or 5 mM MnCl2, 1 mM DTT and 1 mM nucleoside diphosphate derivatives as the substrate, and varying amounts of pure recombinant proteins. After incubation at 37°C for 30 min, the reaction was stopped with HClO4.
Electrophoretic mobility shift assay
Regions containing the predicted regulatory sites were amplified by PCR from genomic DNA by using the primer pairs shown in Supplementary Table S3. One of the primers was 5′-biotinylated by Sigma-Aldrich Corp. (St Louis, MO, USA). By using Synechocystis sp. DNA as the template, primers nadMVfw and nadMVrev amplified a 55-bp fragment in the upstream region of the operon sequence, primers nadAfw and nadArev generated a 71-bp fragment consisting of 68 bp of upstream and 3 bp of 5′ nadA sequence, primers nadEfw and nadErev produced a 60-bp fragment including 3 bp of 3′ nrtR sequence, all of the intergenic region between nrtR and nadE (49 bp), and 8 bp of 5′nadE sequence. By using the S. oneidensis DNA as the template, primers prsfw and prsrev generated a 144-bp fragment including the two predicted DNA sites in the intergenic region between nrtR and prs sequences. PCR products were purified by using the High Pure PCR Product Purification kit (Roche, Basel, Switzerland) and spectrophotometrically quantitated.
For the elecrophoretic mobility shift assay, the biotin-labeled DNA (1 nM) was incubated with the indicated amount of purified NrtR in 20 μl of binding buffer containing 10 mM Tris, pH 7.5, 50 mM KCl, 1 mM DTT, 2.5% glycerol, 5 mM MgCl2, 0.05% NP-40 and 0.50 mg/ml of bovine serum albumin. After 20 min incubation at 25°C, the reaction mixtures were electrophoresed on a 5% native polyacrylamide gel in 0.5 × Tris–borate–EDTA for 60 min at 120 V, at 4°C. The gel was electrophoretically transferred (30 min, at 380 mA) onto a nylon membrane (Pierce, Rockford, Ill.) and fixed by UV cross-linking. Biotin-labeled DNA was detected with the LightShift Chemiluminescent EMSA kit (Pierce, Rockford, Ill.), as recommended by the manufacturer. Specificity of the NrtR–DNA interaction was established by including a 200-fold molar excess of non-biotinylated target DNA (specific competitor) or 1 μg poly(dI-dC) (non-specific competitor) in binding reaction mixtures. The effect of potential effectors on NrtR-DNA binding was tested by pre-incubation of the proteins with NAD metabolites at the indicated concentrations for 10 min at 25°C before the biotinylated DNA was added.
RESULTS
We began this study by performing a bioinformatics survey of candidate genes for transcriptional regulators of NAD metabolism in those bacterial species that do not contain orthologs of known regulators, NadR or NiaR. We considered a conserved clustering on the chromosome with known genes of NAD biogenesis as primary evidence for implication of such candidates (32). Further prioritization of candidate genes was performed based on their domain structure analysis for a presence of putative DNA-binding motifs as well as on additional evidence of functional coupling, such as occurrence profiles and presence of shared regulatory sites (8,33), as metabolic transcriptional regulators are often auto regulated.
Reconstruction of NAD metabolic and regulatory networks in ∼400 bacterial species with completely sequenced genomes was performed using a subsystems-based approach (10) implemented in the SEED genomic platform (see ‘NAD regulation’ subsystem at http://theseed.uchicago.edu/FIG/subsys.cgi). A genome context analysis evidenced a strong tendency of genes involved in NAD biogenesis (including regulatory gene niaR) to form conserved operon-like clusters (Supplementary Data, Table S1). Among other genes with similar chromosomal clustering patterns, of particular interest was a family of Nudix hydrolase homologs. Members of this family were considered primary candidates for the NAD regulatory role and annotated as possible NrtR based on the following key observations:
presence of a C-terminal domain with winged HTH-fold characteristic of many prokaryotic transcription factors (34);
homology of the NrtR N-terminal domain to members of the Nudix hydrolase family related to NAD metabolism, such as ADPR pyrophosphatase involved in NAD recycling;
lack of conservation within the Nudix hydrolase signature sequence suggesting that many NrtR members could have lost a catalytic activity, while potentially retaining an ability to specifically bind relevant metabolites (e.g. ADPR).
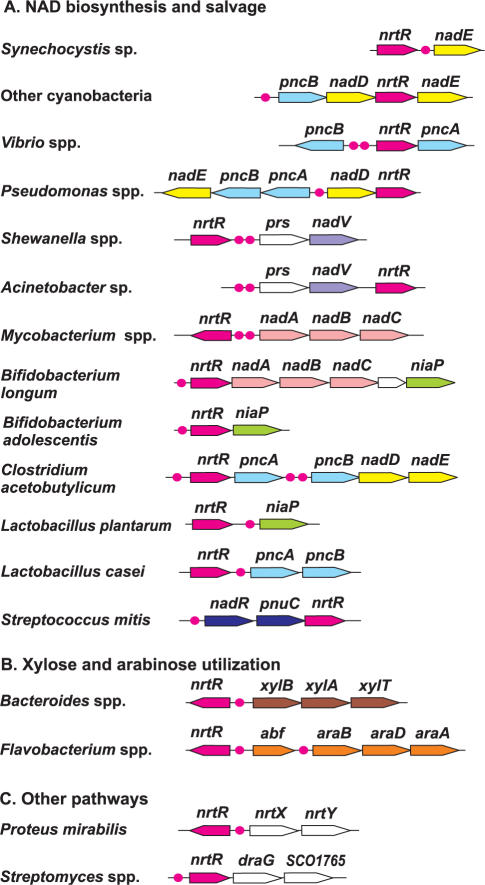
Chromosomal co-localization of nrtR genes
In most cases (42 out of 62 analyzed) nrtR genes are positionally linked to various NAD metabolism genes (Supplementary Table S1). The tendency of nrtR genes to cluster on the chromosome with NAD biosynthesis genes is illustrated in Figure 2. For instance, nrtR genes occur in clusters with nadABC in Actinobacteria; nadD-nadE and pncB in Cyanobacteria; prs-nadV in γ-proteobacteria; nadR-pnuC in Streptococcus and niaP in Bifidobacterium.
Figure 2.
Genomic organization of nrtR-containing loci involved in NAD metabolism (A), pentose utilization (B) and other pathways (C). Genes encoding the predicted NrtR regulator are shown by red arrows; the color code and the abbreviations for other genes correspond to those used in Figure 1. Red circles indicate the predicted NrtR-binding sites.
In Streptomyces species, the nrtR gene occurs next to a gene coding for a putative ADP-ribosyl-glycohydrolase (draG), which catalyzes the removal of the ADPR group covalently linked to target proteins (Figure 1). Protein ADP-ribosylation was proven to be an indispensable regulatory mechanism in Streptomyces species (35) even though its role remains to be elucidated.
Additional types of conserved chromosomal clusters were observed pointing to possible functional coupling of NrtR with other metabolic pathways. For example nine cases of nrtR chromosomal clustering with genes involved in utilization of pentoses (l-arabinose and d-xylose) were detected in some γ-proteobacteria and in Bacteroidetes. Notably, the pentose utilization pathways have connections with NAD metabolism via the shared intermediate ribose 5-phosphate (Rib-P). In fact, glycohydrolitic degradation of NAD generates ADPR, which is further converted to Rib-P by Nudix hydrolases. Rib-P can be recycled to generate NAD, via PRPP formation (Figure 1). Rib-P is also produced by the pentoses utilization routes, after they merged to the pentose phosphate pathway. The result is a combination of pentoses utilization, NAD degradation and recycling in a compact subnetwork (Figure 1).
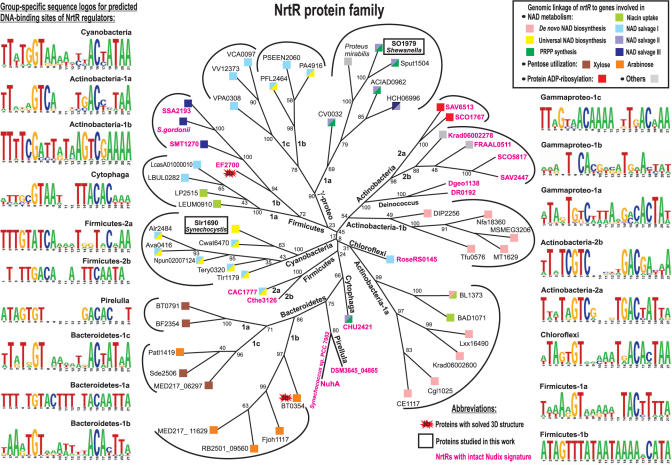
Phylogenetic distribution and domain composition of NrtR proteins
We constructed the maximum likelihood phylogenetic tree for 62 NrtR protein family representatives selected from diverse bacterial species (Figure 3). The distribution of NrtR orthologs largely coincides with known taxonomic groups with several exceptions, e.g. in the case of two proteins from γ-proteobacteria, Pseudoalteromonas atlantica and Saccharophagus degradans, whose clustering with the Bacteroidetes group is a likely result of lateral gene transfer. NrtR proteins from Actinobacteria and Firmicutes are split on the tree into two separate groups, which may reflect the actual functional divergence, e.g. by the set of co-regulated genes or by the consensus DNA-binding motif (see next section). Moreover, some of these species contain two NrtR paralogs. For instance, two NrtR paralogs in Streptomyces species (groups 2a and 2b) likely resulted from a recent duplication event. In another species of Actinobacteria, Kineococcus radiodurans, the situation is different: two NrtR paralogs are located on the most divergent branches of the tree (groups 1a and 2b).
Figure 3.
Maximum likelihood phylogenetic tree and DNA recognition motifs for the NrtR family of transcriptional regulators. NrtR proteins recognizing the same DNA motif are grouped (the group names are given), and the corresponding motif sequence logos are shown on the left and right sides. Species content of the NrtR groups, as well as the content of corresponding regulons and the NrtR-regulated pathways are summarized in Table 1. Genome context of nrtR genes is shown by squares with colors corresponding to the color code of functional roles in Figure 1. NrtR proteins possessing intact Nudix signature are in red. Proteins from B. thetaiotaomicron and E. faecalis with solved 3D structures are highlighted. Proteins studied in this work are boxed. The numbers indicate the number of bootstrap replications, out of 100, that support each node on the tree.
A common feature of the NrtR family is the invariant presence of the N-terminal Nudix domain (PF00293 or COG1051) fused with a characteristic C-terminal domain (PB002540), which is similar to C-terminal part of proteins from uncharacterized COG4111 family. However, COG4111 and NrtR proteins have extremely divergent N-terminal domains (see Discussion section for further details on COG4111). The schematic representation of the NrtR domain arrangement compared with that of some of the known members of the Nudix hydrolase family (where the Nudix domain is combined with other domains) and the COG4111 protein family is shown in Supplementary Fig. S1. A multiple alignment of selected NrtR proteins, including two proteins with known 3D structure, is provided in Supplementary Fig. S2. The Nudix domain is typical of a family of hydrolases found in nearly all known species in all three domains of life. Typical substrates of Nudix hydrolases are nucleoside diphosphates with large variation of residues (x) attached to the phosphate moiety (hence the name, nudix). These enzymes hydrolyze a pyrophosphate bond in a wide range of organic pyrophosphates, including nucleoside di- and triphosphates, dinucleoside polyphosphates and nucleotide sugars (such as ADPR), with varying degrees of substrate specificity. The number of Nudix-like genes in prokaryotic genomes is a subject of significant variation reaching up to 30 copies, depending on metabolic complexity and adaptability of species [reviewed in (36)]. Among various Nudix hydrolases, those with established substrate preference for ADPR (i.e. ADPR pyrophosphatases, catalyzing ADPR hydrolysis to AMP and Rib-P) show the most significant similarity with the Nudix domain of NrtR proteins.
A detailed analysis of conserved sequence motifs within the NrtR family indicated that, in contrast to functional Nudix hydrolases, many members of this family could have lost a catalytic activity. This conjecture is based on the apparent lack of conservation within a signature sequence, GX5EX7REUXEEXGU (where U is a hydrophobic residue and X is any residue), which is strictly conserved in all active Nudix hydrolases identified so far (37). In most members of the NrtR family, at least one or more of the conserved signature residues are replaced in a more or less random fashion (Supplementary Fig. S2). Previously published results of biochemical characterization of two divergent members of the NrtR family from cyanobacteria, NuhA from Synechococcus sp. PCC 7002 and Slr1690 from Synechocystis sp., provided additional insights for interpretation of functional motifs in this family. Whereas the NuhA protein with an intact Nudix signature displayed a high ADPR pyrophosphatase activity (38), the hydrolytic activity of its homolog Slr1690 with a severely perturbed signature was barely detectable (kcat ∼ 1.4 × 10−4s−1) (39). Restoring the canonical Nudix signature by a directed mutagenesis of the slr1690 gene led to a 600-fold increase of the catalytic rate (39). These facts suggest that the presence of an intact Nudix signature in NrtR proteins correlate with their catalytic activity.
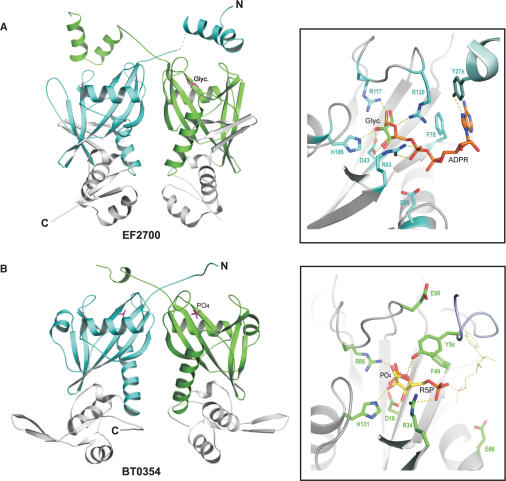
Structural analysis of NrtR proteins
A comparative structural analysis of C-terminal domains in NrtR proteins provided the key evidence for their role in the regulation of transcription. This analysis was enabled by the availability of 3D structures determined at the Midwest Center for Structural Genomics (http://www.mcsg.anl.gov/) for the two NrtR family members from Bacteroides thetaiotaomicron (BT0354, PDB code 2FB1) and Enterococcus faecalis (EF2700, PDB code 2FML) (Figure 4). It is important to note that while the results of similarity searches for NrtR C-terminal domain based solely on sequence comparison were rather inconclusive, a structure-based search by the SSM server (24) revealed a substantial similarity with winged helix-turn-helix (wHTH) domains. A three-stranded wHTH fold (α1-β1-α2-α3-β2-β3) of the C-terminal domain (Supplementary Fig. S2) is typical for DNA-binding domains present in many families of prokaryotic transcription factors (34).
Figure 4.
Crystal structures and ligand-binding sites of E. faecalis EF2700 (A) and B. thetaiotaomicron BT0354 (B). Structures of EF2700 (PDB accession number 2FML) and BT0354 (2FB1) were solved at the Midwest Center for Structural Genomics. The C-terminal wHTH domains are shown in grey, and the N-terminal Nudix domains of each monomer are shown in cyan and green, respectively. The co-crystallized glycerol and phosphate molecules are indicated. In the insets are the close-ups of the Nudix domain active sites with a modeled ADPR molecule in EF2700 and a Rib-P molecule in BT0354, respectively, based on the superposition with the Synechocystis sp. NadM–ADPR complex structure. In the binding pocket of BT0354, the Rib-P moiety (R5P) of ADPR is shown as sticks while the rest of the molecule is show as thin lines. Protein residues that are predicted to interact with the bound ligand are also shown as sticks. Dotted lines represent potential hydrogen bonds. Residues Y27B and Y5B of EF2700 and BT0354, respectively, come from the second subunit of the dimer.
The 3D structures of BT0354 and EF2700 show that both NrtR proteins form dimers with clear domain swapping. The Nudix domain of both proteins is very similar to the ADPR pyrophosphatase domain of the bifunctional enzyme NadM from Synechocystis sp., whose structure has been recently solved in complex with ADPR (Huang, N. and Zhang, H., unpublished data). In particular, root mean square deviations of 1.4 Å and 1.5 Å have been calculated for 115 and 108 superimposed Cα atoms between NadM and EF2700 and NadM and BT0354, respectively. The residues demonstrated to be directly involved in ADPR binding in the NadM structure are well conserved in EF2700 (Figure 4A). Most of the residues interacting with ADPR are also conserved in the BT0354 active site; however, only the ribose-phosphate moiety of ADPR fits well into the binding pocket (Figure 4B). These structural considerations allowed us to suggest ADPR and Rib-P primary candidates as possible NrtR effector molecules.
Prediction of NrtR-binding sites and reconstruction of NrtR regulons in bacterial genomes
To identify possible DNA motifs specifically recognized by NrtR in various taxonomic groups, we created training sets by combining the upstream regions of NAD metabolic genes from complete bacterial genomes containing nrtR genes. Based on the phylogenetic analysis, the NrtR family was divided into several taxonomic groups, and some of them were further split to distinct branches (Figure 3). They were used to compile group-specific training sets. For nrtR genes that occur in conserved chromosomal clusters with genes other than those involved in NAD metabolism, the respective training sets included upstream regions of these gene clusters (e.g. nrtR-draG locus in Streptomyces and ara or xyl loci in Bacteroidetes).
By applying the motif-detection program SignalX with the inverted repeat option (40), we have identified candidate NrtR recognition sites conserved in each of the compiled training sets and constructed the corresponding NrtR profiles and sequence logos (Figure 3). Although the derived motifs are substantially different in consensus sequence and information content (depending on the number and diversity of species within groups and the number of candidate target genes), most of them share a 21-bp palindrome symmetry and a conserved core with consensus GT-N7-AC. The most divergent NrtR consensus sequences were detected for two groups of the γ-proteobacteria (Vibrio and Pseudomonas) and one group of the Firmicutes (Streptococcus). For the Deinococcus and Enterococcus species, we were unable to identify NrtR recognition profiles and regulons.
The constructed NrtR-binding site recognition profiles were used to detect new candidate members of the NrtR regulons in the genomes containing nrtR genes. Table 1 gives a list of genes and operons predicted to be under control of NrtR. The detailed information about the sequence, position, and score of each predicted NrtR site, as well as the genomic identification numbers of downstream genes, are provided in Supplementary Table S2. The key features of the reconstructed NrtR regulons are outlined in details by taxonomic groups in Supplementary Text S1.
Table 1.
Operon structure for nrtR genes and predicted NrtR sites in bacteria
| Organism | NrtR operon/regulon structurea | NrtR-regulated pathwayb | NrtR DNA-binding sequence logo profilec |
|---|---|---|---|
| Synechocystis sp. PCC 6803 | nrtR-#nadE; #nadM-nadV; #nadA | NAD(d,u,s2) | Cyanobacteria |
| Nostoc, Anabaena spp; Crocosphaera watsonii; Thermosynechococcus elongatus | #pncB-nadD-nrtR-nadE; #nadA | NAD(d,u,s1) | Cyanobacteria |
| Trichodesmium erythraeum | #pncB-nadD-nrtR | NAD(u,s1) | Cyanobacteria |
| Corynebacterium glutamicum, C.efficiens | #nrtR-nadA-nadC-sufS | NAD(d) | Actinobacteria-1a |
| Kineococcus radiotolerans, Leifsonia xyli | #nrtR-nadA-nadB-nadC-sufS | NAD(d) | Actinobacteria-1a |
| Bifidobacterium longum | #nrtR-nadA-nadB-nadC-sufS-niaP | NAD(d,t) | Actinobacteria-1a |
| Bifidobacterium adolescentis | #nrtR-niaP; #pncB | NAD(t,s1) | Actinobacteria-1a |
| Corynebacterium diphtheriae | nrtR<#>nadA-nadB-nadC | NAD(d) | Actinobacteria-1b |
| Mycobacterium species, Nocardia farcinica | nrtR<#>nadA-nadB-nadC | NAD(d) | Actinobacteria-1b |
| Thermobifida fusca | nrtR<#>nadA | NAD(d) | Actinobacteria-1b |
| Streptomyces species - (1) | #nrtR1-draG-SCO1765 | ADPR | Actinobacteria-2a |
| Streptomyces species - (2) | #nrtR2; #nrtX-nrtY | ? | Actinobacteria-2b |
| Kineococcus radiotolerans - (2), Frankia alni | nrtR2<#>nrtX-nrtY | ? | Actinobacteria-2b |
| Lactobacillus casei, L.delbrueckii | nrtR-#pncA-pncB | NAD(s1) | Firmicutes-1a |
| Lactobacillus plantarum, L.mesenteroides | nrtR-#niaP | NAD(t) | Firmicutes-1a |
| Streptococcus mitis, S.gordonii, S.sanguinis | #nadR-pnuC-nrtR | NAD(s3) | Firmicutes-1b |
| Clostridium acetobutylicum | #nrtR-pncA-#pncB-nadD-nadE | NAD(u,s1) | Firmicutes-2a |
| Clostridium thermocellum | #nrtR; #pncA-pncB | NAD(s1) | Firmicutes-2b |
| Shewanella oneidensis, S.putrefaciens | nrtR-#prs-nadV | NAD(s2,p) | Gammaproteo-1a |
| Acinetobacter sp. ADP1 | #prs-nadV-nrtR | NAD(s2,p) | Gammaproteo-1a |
| Proteus mirabilis | nrtR<#>nrtX-nrtY | ? | Gammaproteo-1a |
| Hahella chejuensis | nrtR<#>pnuC-nadR-#nadM-pncB; #nrtX-nrtY | NAD(s1,s2,s3) | Gammaproteo-1a |
| Chromobacterium violaceum | nrtR-#prs-nadV | NAD(s2,p) | Gammaproteo-1a |
| Pseudomonas aeruginosa, P.fluorescens | nrtR-nadD<#>pncA-pncB-nadE | NAD(u,s1) | Gammaproteo-1b |
| Pseudomonas entomophila | nrtR<#>pncA-pncB | NAD(s1) | Gammaproteo-1b |
| Vibrio cholerae | nrtR<#>pncB | NAD(s1) | Gammaproteo-1c |
| Vibrio parahaemolyticus, V.vulnificus | pncA-nrtR<#>pncB | NAD(s1) | Gammaproteo-1c |
| Cytophaga hutchinsonii | nrtR-#prs-nadV; #nadE | NAD(u,s2,p) | Cytophaga |
| Roseiflexus sp. RS-1 | #nrtR-pncA; #pncB; #nadC-nadA-nadB | NAD(b,s1) | Chloroflexi |
| Blastopirellula marina | #pncB-nrtR-pncA; #nadE | NAD(u,s1) | Pirellula |
| Bacteroides fragilis, B. thetaiotaomicron - (1) | #nrtR-xylB-xylA-xylT | Xyl | Bacteroidetes-1a |
| B. thetaiotaomicron - (2) | #galM-araT-nrtR2-araD-araB-araA | Ara | Bacteroidetes-1b |
| Robiginitalea biformata | nrtR<#>abf-araB-araD-araA-araT | Ara | Bacteroidetes-1b |
| Flavobacterium sp. MED217 - (2) | nrtR2<#>abf-abn-xyn-xyl; #araB-araD-araT-araA | Xyl, Ara, PP | Bacteroidetes-1b |
| Flavobacterium johnsoniae UW101 - (2) | nrtR2<#>abf-#araB-araD-araA; # other genes | Ara | Bacteroidetes-1b |
| Saccharophagus degradans | nrtR<#>xylB-xylA; # other genes | Xyl, PP | Bacteroidetes-1c |
| Pseudoalteromonas atlantica | nrtR-#abf | Ara | Bacteroidetes-1c |
| Flavobacterium sp. MED217 - (1) | nrtR1<#>xylB-xylA | Xyl | Bacteroidetes-1c |
aGenes forming one putative operon (with spacer <100 bp) are separated by dashes. Different loci are separated by semicolons. The direction of transcription in divergons (two oppositely directed transcriptional units) is shown by angel brackets. Predicted NrtR-binding sites are denoted by ‘#’. Functions of NrtR-regulated genes are described in the legend to Figure 1. SufS is a homologue of IscS cysteine desulfurase involved in the in vivo maturation of Fe-S clusters, possibly necessary to assemble NadAB complex. Other predicted NrtR-regulated genes of unknown function are denoted nrtX and nrtY.
bBiochemical pathways predicted to be regulated by NrtR are abbreviated according to Figure 1. Functional roles involved in NAD synthesis: d, de novo biosynthesis; u, universal NAD synthesis; s1, s2 and s3 stand for salvage pathways I, II, and III; p, PRPP synthesis; t, niacin transport. Other metabolic pathways are pentose-phosphate pathway (PP), xylose (Xyl), arabinose (Ara) utilization and reversible protein ADP-ribosylation (ADPR).
cGenomes are grouped according to the predicted DNA-binding profiles (see the phylogenetic tree of NrtR proteins on Figure 3), and in most cases, the NrtR groups coincide with the taxonomic groups of organisms.
Functional gene content of the reconstructed NrtR regulons varies significantly between different taxonomic groups of bacteria (Table 1 and Figure 3). The NrtR-regulated pathways include the universal NAD biosynthesis and salvage I pathways in Cyanobacteria and γ-proteobacteria, Pirellula and Chloroflexi; the de novo NAD biosynthesis pathway in Actinobacteria; niacin uptake and salvage I and III pathways in Lactobacillales (Firmicutes); and the pentose utilization pathways in Bacteroidetes (see color square code in Figure 3). Taxonomic distribution of NrtR regulators is complementing to the distribution of two other transcriptional regulators of NAD metabolism, NadR (18) and NiaR [see the accompanying paper (9)] with the exception of some species of Firmicutes, where both NrtR and NiaR appear to regulate non-overlapping aspects of NAD metabolism (Supplementary Table S1). For instance, in Clostridium actobutylicum, NrtR regulates the nicotinamide salvage pathway (pncAB) and the universal NAD synthesis (nadDE), whereas NiaR controls the de novo NAD synthesis (nadBCA). The Lactobacillales species represent another example of the large variability in the content of NAD regulons. In Lactobacillus casei, NiaR and NrtR regulate the niaP and pncAB genes, respectively, whereas in L. plantarum they control the pncB and niaP genes, respectively (9).
The position of the candidate NrtR-binding sites in the regulatory gene regions, either overlapping the predicted promoter elements or lying between the promoter and the translation start site of the downstream gene, strongly suggests that these regulators might act as repressors of transcription (Supplementary Figure S3). They are expected to bind to target genes via the wHTH C-terminal domain and a postulated interaction of the Nudix N-terminal domain with an effector molecule is anticipated to weaken the NrtR–DNA complex, leading to derepression of target genes.
While many aspects of the proposed mechanism are yet to be investigated, in this study we chose to perform an experimental validation of selected NrtR family members in order to provide minimal required support for the suggested novel functional annotations. For these validation experiments we have chosen two representatives of the NrtR family, Slr1690 from Synechocystis sp. and SO1979 from S. oneidensis. The candidate NrtR-binding sites in Synechocystis sp. precede the nadA, nadE and nadM-nadV genes, whereas the predicted NrtR regulon in S. oneidensis contains a single target operon, prs-nadV (Table 1). The choice of these two species was dictated by the following considerations: (i) unambiguous association of the chosen NrtR groups with NAD metabolism; (ii) a divergent nature of these groups at the level of taxonomic placement of respective organisms as well as at the level of protein sequences (position on the NrtR tree and consensus DNA motifs); (iii) availability of biochemical data for the Slr1690 protein (39) from the best studied model cyanobacterium Synechocystis sp., including the detailed analysis of the NAD biosynthetic machinery (41) and (iv) S. oneidensis being an important model organism with a rapidly growing body of physiological and genomic data (42).
Experimental characterization of two NrtR family representatives
To experimentally test the ability of NrtR to specifically bind to the predicted DNA sites and to assess possible effectors, slr1690 from the Synechocystis sp. (further referred to as syNrtR) and SO1979 from S. oneidensis (soNrtR), were cloned and overexpressed in E. coli. Both His6-tagged recombinant proteins were purified to homogeneity by Ni+2-chelating chromatography, followed by gel filtration as described in the Materials and Methods section. SDS-PAGE of pure syNrtR and soNrtR proteins revealed molecular masses of about 32 and 29 kDa, respectively, in agreement with the expected size (Supplementary Figure S4).
As expected, no appreciable ADPR pyrophosphatase activity could be detected for syNrtR and soNrtR proteins in the presence of Mg2+ or Mn2+. This is consistent with observed alterations in their Nudix signatures (Supplementary Figure S2) as well as with the previous report of extremely low enzymatic activity of Slr1690 protein, several orders of magnitude lower than that measured for other ADPR pyrophosphatases in Synechocystis sp. (39). Likewise, both NrtR proteins were unable to hydrolyze other nucleoside diphosphate compounds, including (2′)phospho-ADP-ribose (pADPR), NAD, flavine adenine dinucleotide, diadenosine polyphosphates (ApnA, n = 2, 4) and GDP-mannose.
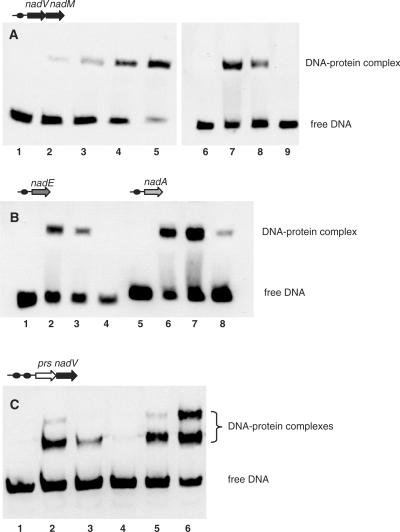
We used electophoretic mobility shift assay (EMSA) to test the specific DNA-binding of the purified syNrtR and soNrtR proteins to their predicted operator sites derived from the upstream regions of nadA and nadE genes, the nadM-nadV operon from Synechocystis sp. and the prs-nadV operon from S. oneidensis. A substantial shift of the DNA band was observed in all cases upon incubation of the target DNA fragments with respective proteins (Figure 5). A typical protein concentration dependence of DNA-binding is illustrated in Figure 5A showing the increasing intensity of the shifted DNA band (corresponding to a predicted nadM-nadV target site) in the presence of increasing amounts of the syNrtR protein. The band shift was suppressed in the presence of 200-fold excess unlabeled DNA fragments but not in the presence of negative control DNA, poly(dC/dI), confirming a specific nature of the NrtR–DNA interactions (Figure 5A). A similar specific binding in the presence of competing DNA was observed between syNrtR and its cognate DNA-sites from the upstream regions of nadE and nadA genes (Figure 5B). Two distinctly shifted protein–DNA complexes were observed in the case of soNrtR interaction with the prs-nadV DNA target, confirming a functional competence of both predicted tandem NrtR-binding sites (Figure 5C).
Figure 5.
EMSA demonstrating specific NrtR binding to DNA. DNA fragments used in the assays are defined by their genomic positions and are shown as dark circles in the top of each panel. (A) EMSA with nadM-nadV DNA fragment (0.7 ng) in the absence (lane 1) and in the presence (lanes 2–5) of increasing syNrtR protein concentrations (0.25, 0.5, 1.0 and 5.0 nM, respectively). The specificity of interaction of syNrtR (2 nM) with DNA fragment (lane 7) was tested by competition with 1 μg polydC/dI (lane 8) and 140 ng unlabeled DNA fragment (lane 9). Lane 6 contains 0.7 ng of the biotylinated DNA fragment only. (B) EMSA with nadE and nadA DNA fragments in the absence (lanes 1 and 5) and in the presence of syNrtR (2 nM) (lanes 2 and 6). The specificity of interaction was tested with polydC/dI (lanes 3 and 7) and unlabeled DNA fragment (lanes 4 and 8). (C) EMSA with prs-nadV DNA fragment in the absence (lane 1) and in the presence of 2 nM soNrtR (lanes 2 and 5) and 5 nM soNrtR (lane 6). Competition assays were performed with 2 nM soNrtR in the presence of polydC/dI (lane 3) and unlabeled DNA (lane 4).
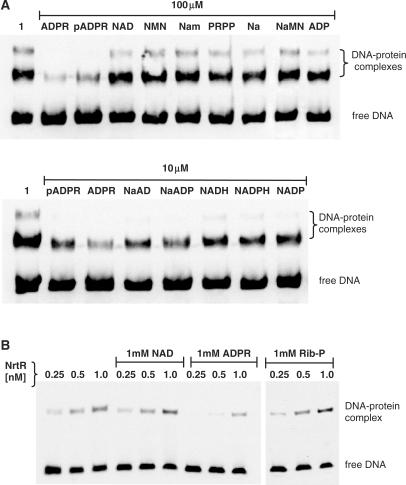
Of the tested intermediary metabolites associated with NAD biosynthetic, salvage, and recycling pathways (Figure 1), Nam, NA, quinolinic acid, NMN, NaMN, NAD, PRPP, Rib-P, ADP and AMP at 100 μM concentration did not interfere with complex formation between soNrtR and the prs-nadV tandem DNA-site. This is illustrated for some of these compounds by the results of EMSA analysis (Figure 6A). In the same experiment, 100 μM of ADPR and pADPR significantly suppressed soNrtR-DNA binding (Figure 6A), and a similar effect was observed with 100 μM of nicotinate adenine dinucleotide (NaAD), nicotinate adenine dinucleotide phosphate (NaADP), NADH, NADPH and NADP (data not shown). As shown in the lower panel of Figure 6A, these metabolites exerted a similar effect on soNrtR-DNA binding complex formation even at 10 μM, with ADPR being the most effective of all. ADPR was also shown to suppress specific DNA-binding of syNrtR-DNA, as illustrated in Figure 6B for the nadM-nadV target, whereas NAD and Rib-P had no effect under the same conditions.
Figure 6.
Effect of NAD metabolites on NrtR–DNA binding. (A) Electrophoretic mobility of prs-nadV DNA fragment incubated with purified soNrtR (2 nM) in the absence (lane 1) and in the presence of 100 and 10 μM of the indicated compounds. (B) Electrophoretic mobility of nadM-nadV DNA fragment incubated with increasing concentrations of purified syNrtR in the absence and in the presence of 1 mM NAD, 1 mM ADPR and 1 mM Rib-P.
DISCUSSION
Identification of a novel family of transcriptional factors for the NAD metabolism allowed us to fill a substantial gap in the knowledge of transcriptional regulation of the key metabolic pathways in bacteria. Prior to this study, transcriptional regulation of NAD biosynthesis was elucidated only in the classic model systems, E. coli/Salmonella and B. subtilis (15,19). Of the two known families of transcriptional regulators of NAD biosynthesis, NadR appears to be operational within only a limited group of Enterobacteria (18), whereas the distribution of the NiaR (YrxA) family is mostly restricted to species from the Bacillus/Clostridium and Thermotogales group (9). This limited knowledge is in marked contrast with a comprehensive understanding of NAD biosynthetic machinery that was reliably reconstructed by comparative genomic analysis in nearly all bacteria with completely sequenced genomes (11).
In this study, we used a combination of comparative genomic techniques including analysis of genome context and regulatory DNA motifs (8) to predict the novel regulator NrtR and reconstruct respective regulons in several divergent groups of bacteria. The NrtR protein family of transcriptional regulators described here is the first known example of a functional fusion between a Nudix domain and a DNA-binding domain. The comparative analysis of predicted NrtR-binding motifs revealed their significant variability between taxonomic groups that were largely in agreement with a phylogenetic tree of the NrtR protein family (Figure 3). The derived motifs for 12 NrtR groups have a relatively conserved core (TG-N7-CA), whereas the other six groups appear to have adopted dissimilar NrtR-binding sites. This degree of variability of NrtR DNA-binding motifs is higher than what is usually observed for most other broadly conserved bacterial transcription factors such as BirA, ArgR and NrdR (8,43–45).
The predicted composition of NrtR regulons in different bacterial genomes appears to be the subject of substantial variations that to some extent reflect various combinations of pathways involved in NAD biosynthesis (Figure 1 and Table 1). NrtR regulation appears to play a central role in the transcriptional control of all aspects of NAD biosynthesis, salvage and recycling in Cyanobacteria [for a comparative genomic reconstruction of NAD metabolism in this bacterial lineage, see (41)]. In contrast, the NrtR regulon in S. oneidensis includes only one target operon, prs-nadV. Although the functional association between these two genes is straightforward (Figure 1), an inferred co-regulation of the PRPP–synthesizing enzyme Prs with a rather local Nam salvage pathway via NadV is quite unexpected. However, unlike most bacteria having a single copy of the prs gene, S. oneidensis and several other species with prs-nadV operons contain two prs paralogs, only one of which is predicted to be regulated by NrtR. A functional significance of having separate machinery for PRPP production committed to Nam to NMN conversion is yet to be elucidated.
We chose representatives of the two NrtR groups discussed earlier to experimentally assess their specific DNA-binding properties and to test possible small molecule effectors. Both recombinant purified proteins from Synechocystis sp. and from S. oneidensis were shown to form a complex with their cognate target DNA sequences (Figure 5). Among various tested metabolites associated with NAD metabolism, ADPR was the most effective in suppressing in vitro binding of both regulators to their target sequences (Figure 6). At the same time, enzymatic characterization of both proteins revealed no appreciable catalytic (ADPR pyrophosphatase) activity in agreement with their substantially perturbed Nudix signature sequences as well as with the previous report about the Synechocystis sp. protein (39). Although the in vitro EMSA data alone were insufficient for unambiguous identification of the actual physiological NrtR effector molecule(s), ADPR appears to be the most likely candidate for this role. As discussed earlier, this effector specificity could be anticipated from the sequence and structure similarity between the NrtR Nudix domain and the ADPR pyrophosphatase domain of the bifunctional enzyme NadM (Figure 4). Notwithstanding these caveats, the obtained experimental data are generally supportive of a role of NrtR–effector interactions in the proposed mechanism of de-repression of NrtR-regulated genes. They also allowed us to exclude common NAD precursors (including NA, the known NiaR co-repressor), as well as NAD (the known effector of the NadR regulator) from the list of possible NrtR effectors.
The proposed physiological role of ADPR as an anti-repressor of NAD synthesis is based on the assumption that the cell may interpret the accumulation of ADPR as a signal to replenish the NAD cofactor pool. This assumption is reasonable as the only source of ADPR in the cell is consumption of NAD by direct enzymatic hydrolysis. Binding of ADPR to an NrtR Nudix domain would promote dissociation of NrtR–DNA complexes, leading to de-repression of transcription of NAD biosynthetic genes. ADPR itself may contribute to NAD production by providing a Rib-P precursor for PRPP synthesis upon its Nudix-mediated hydrolytic cleavage. This provides an additional rationale for the observed regulatory connections between the NAD biosynthetic machinery and prs gene. Intriguingly, some members of the NrtR family that have an intact Nudix signature may directly contribute to ADPR hydrolysis in addition to their role in regulation of transcription.
The most remarkable aspect of the already-mentioned functional plasticity of the NrtR family is its apparent adaptation to the regulation of metabolic pathways that are not immediately related to NAD metabolism. Among the examples analyzed in this study are pentoses utilization, and the pentose phosphate pathway in Bacteroidetes and in two species of γ-proteobacteria (Table 1). Indeed, these pathways share the intermediate Rib-P with NAD degradation and recycling (Figure 1). It is tempting to speculate that in these species NrtR regulons may utilize Rib-P (rather than ADPR) as an effector molecule, as also suggested by ligand modeling in the active site of NrtR from B. thetaiotaomicron (Figure 4). Further experiments are required to test this hypothesis.
The data analyzed in this study allowed us to suggest a likely scenario for the evolution of the NrtR family that includes a fusion of an ADPR-preferring Nudix hydrolase with a DNA-binding domain, giving rise to a transcriptional regulator. An ancestral version of NrtR regulator could retain ADPR pyrophosphatase activity. Later in speciation, NrtR proteins could lose the hydrolase activity that was redundant in the presence of other active Nudix hydrolase(s). Regardless of the actual effector specificity, which is yet to be explored across the NrtR family, this evolutionary scenario is another example of a novel transcription factor emerging via a fusion between the enzymatic and DNA-binding domains. This appears to be quite an efficient strategy, as may be deduced from a large and growing number of transcription factors ‘designed’ using similar principles. Among other prominent examples in bacteria are: biotin repressor BirA, a fusion of functional biotin-ligase and DNA-binding HTH domain (43); xylose repressor XylR and other regulators of the ROK family composed of a sugar kinase-like domain fused with HTH domain (17); transcriptional repressor PutA that also carries out the enzymatic steps in proline catabolism (46); and NadR, a NAD salvage enzyme and the transcriptional regulator of NAD synthesis in Enterobacteria (15,47,48).
In this work and the accompanying study (9), we characterized the NrtR and NiaR families of transcriptional factors implicated in the control of NAD homeostasis. Although these findings substantially expanded our knowledge in this important but relatively unexplored area, mechanism of transcriptional regulation of NAD metabolism remains unknown in many other bacterial lineages including α-, β-, δ- and ε-proteobacteria. Notably, another family of putative transcriptional regulators of NAD metabolism (COG4111, here named NadQ) was identified in 15 species of α-proteobacteria and in some pathogenic β-proteobacteria (Supplementary Table S1). Members of this family share similar C-terminal HTH domains with NrtR proteins but have highly divergent N-terminal domains (as illustrated in Supplementary Figure S1). NadQ encoding genes also have a strong tendency to cluster on the chromosome with genes involved in NAD metabolism. The genomic identification of NadQ-binding sites and functional characterization of NadQ regulon are currently underway. Altogether these findings demonstrate the significant variability of regulatory strategies for control of NAD metabolic pathways in bacteria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We want to thank Dr Giulio Magni and Dr Silverio Ruggieri (Università Politecnica delle Marche, Ancona, Italy) for discussions and advice in the field of NAD metabolic biochemistry, Dr Mikhail Gelfand (IITP, Moscow, Russia) for useful discussions and comments during the preparation of this article and Dr Andrei Mironov (Moscow State University, Russia) for software for the analysis of regulons. We are grateful to the Midwest Center for Structural Genomics team for sharing the results of structural analysis that were instrumental for our functional prediction of the novel family of transcriptional regulators. We thank Ross Overbeek, Veronika Vonstein, Gordon Push and other members of The SEED development team at Fellowship for Interpretation of Genomes (FIG) for their help with the use of The SEED genomic resource. This work was partially supported by grants from Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) ‘Characterization of key enzymes of NAD(P) biosynthesis in bacteria and their regulation:essential steps to find targets of new drugs’ (PRIN 2005 to N.R.); National Institute of Health (NIH) ‘Genomics of Coenzyme Metabolism in Bacterial Pathogens’ (1-R01-AI066244-01A2 to A.O.) and Department of Energy (DOE) ‘Integrated Genome-Based Studies of Shewanella Ecophysiology’ (DE-FG02-07ER64384 to A.O.). Funding to pay the Open Access publication charges for this article was provided by National Institute of Health research grant 1-R01-AI066244-01A2.
Conflict of interest statement. None declared.
REFERENCES
- 1.Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S. Enzymology of NAD+ homeostasis in man. Cell Mol. Life Sci. 2004;61:19–34. doi: 10.1007/s00018-003-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J. Biol. Chem. 2004;279:40122–40129. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson A, Day J, Bowater R. Bacterial DNA ligases. Mol. Microbiol. 2001;40:1241–1248. doi: 10.1046/j.1365-2958.2001.02479.x. [DOI] [PubMed] [Google Scholar]
- 4.Domenighini M, Rappuoli R. Three conserved consensus sequences identify the NAD-binding site of ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Mol. Microbiol. 1996;21:667–674. doi: 10.1046/j.1365-2958.1996.321396.x. [DOI] [PubMed] [Google Scholar]
- 5.Begley TP, Kinsland C, Mehl RA, Osterman A, Dorrestein P. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam. Horm. 2001;61:103–119. doi: 10.1016/s0083-6729(01)61003-3. [DOI] [PubMed] [Google Scholar]
- 6.Magni G, Amici A, Emanuelli M, Raffaelli N, Ruggieri S. Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 1999;73:135–182. doi: 10.1002/9780470123195.ch5. xi. [DOI] [PubMed] [Google Scholar]
- 7.Gerlach G, Reidl J. NAD+ utilization in Pasteurellaceae: simplification of a complex pathway. J. Bacteriol. 2006;188:6719–6727. doi: 10.1128/JB.00432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodionov DA. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 2007;107:3467–3497. doi: 10.1021/cr068309+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodionov DA, Li X, Rodionova IA, Yang C, Sorci L, Dervyn E, Martynowski D, Zhang H, Gelfand MS, Osterman AL. Nucleic Acids Res. 2008. Transcriptional regulation of NAD metabolism in bacteria: Genomic reconstruction of NiaR (YrxA) regulon. X, XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osterman AL, Begley TP. A subsystems-based approach to the identification of drug targets in bacterial pathogens. Prog. Drug Res. 2007;64:131, 133–170. doi: 10.1007/978-3-7643-7567-6_6. [DOI] [PubMed] [Google Scholar]
- 12.Holley EA, Spector MP, Foster JW. Regulation of NAD biosynthesis in Salmonella typhimurium: expression of nad-lac gene fusions and identification of a nad regulatory locus. J. Gen. Microbiol. 1985;131:2759–2770. doi: 10.1099/00221287-131-10-2759. [DOI] [PubMed] [Google Scholar]
- 13.Tritz GJ, Chandler JL. Recognition of a gene involved in the regulation of nicotinamide adenine dinucleotide biosynthesis. J. Bacteriol. 1973;114:128–136. doi: 10.1128/jb.114.1.128-136.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu N, Roth JR. The nadI region of Salmonella typhimurium encodes a bifunctional regulatory protein. J. Bacteriol. 1991;173:1302–1310. doi: 10.1128/jb.173.3.1302-1310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grose JH, Bergthorsson U, Roth JR. Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica. J. Bacteriol. 2005;187:2774–2782. doi: 10.1128/JB.187.8.2774-2782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penfound T, Foster JW. NAD-dependent DNA-binding activity of the bifunctional NadR regulator of Salmonella typhimurium. J. Bacteriol. 1999;181:648–655. doi: 10.1128/jb.181.2.648-655.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Titgemeyer F, Reizer J, Reizer A, Saier M.H., Jr. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology. 1994;140(Pt 9):2349–2354. doi: 10.1099/13500872-140-9-2349. [DOI] [PubMed] [Google Scholar]
- 18.Gerasimova AV, Gelfand MS. Evolution of the NadR regulon in Enterobacteriaceae. J. Bioinform. Comput. Biol. 2005;3:1007–1019. doi: 10.1142/s0219720005001387. [DOI] [PubMed] [Google Scholar]
- 19.Rossolillo P, Marinoni I, Galli E, Colosimo A, Albertini AM. YrxA is the transcriptional regulator that represses de novo NAD biosynthesis in Bacillus subtilis. J. Bacteriol. 2005;187:7155–7160. doi: 10.1128/JB.187.20.7155-7160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 23.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta. Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 25.Gelfand MS, Novichkov PS, Novichkova ES, Mironov AA. Comparative analysis of regulatory patterns in bacterial genomes. Brief Bioinform. 2000;1:357–371. doi: 10.1093/bib/1.4.357. [DOI] [PubMed] [Google Scholar]
- 26.Mironov AA, Koonin EV, Roytberg MA, Gelfand MS. Computer analysis of transcription regulatory patterns in completely sequenced bacterial genomes. Nucleic Acids Res. 1999;27:2981–2989. doi: 10.1093/nar/27.14.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mironov AA, Vinokurova NP, Gel'fand MS. [Software for analyzing bacterial genomes] Mol. Biol. 2000;34:253–262. [PubMed] [Google Scholar]
- 28.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 30.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Raffaelli N, Lorenzi T, Amici A, Emanuelli M, Ruggieri S, Magni G. Synechocystis sp. slr0787 protein is a novel bifunctional enzyme endowed with both nicotinamide mononucleotide adenylyltransferase and ‘Nudix’ hydrolase activities. FEBS Lett. 1999;444:222–226. doi: 10.1016/s0014-5793(99)00068-x. [DOI] [PubMed] [Google Scholar]
- 32.Huynen M, Snel B, Lathe W., 3rd, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osterman A, Overbeek R. Missing genes in metabolic pathways: a comparative genomics approach. Curr. Opin. Chem. Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 34.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Shima J, Penyige A, Ochi K. Changes in patterns of ADP-ribosylated proteins during differentiation of Streptomyces coelicolor A3(2) and its development mutants. J. Bacteriol. 1996;178:3785–3790. doi: 10.1128/jb.178.13.3785-3790.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLennan AG. The Nudix hydrolase superfamily. Cell Mol. Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bessman MJ, Frick DN, O’Handley SF. The MutT proteins or ‘Nudix’ hydrolases, a family of versatile, widely distributed, ‘housecleaning’ enzymes. J. Biol. Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 38.Okuda K, Nishiyama Y, Morita EH, Hayashi H. Identification and characterization of NuhA, a novel Nudix hydrolase specific for ADP-ribose in the cyanobacterium Synechococcus sp. PCC 7002. Biochim. Biophys. Acta. 2004;1699:245–252. doi: 10.1016/j.bbapap.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Okuda K, Hayashi H, Nishiyama Y. Systematic characterization of the ADP-ribose pyrophosphatase family in the Cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2005;187:4984–4991. doi: 10.1128/JB.187.14.4984-4991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gelfand MS, Koonin EV, Mironov AA. Prediction of transcription regulatory sites in Archaea by a comparative genomic approach. Nucleic Acids Res. 2000;28:695–705. doi: 10.1093/nar/28.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerdes SY, Kurnasov OV, Shatalin K, Polanuyer B, Sloutsky R, Vonstein V, Overbeek R, Osterman AL. Comparative genomics of NAD biosynthesis in cyanobacteria. J. Bacteriol. 2006;188:3012–3023. doi: 10.1128/JB.188.8.3012-3023.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gralnick JA, Hau HH. Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 2007;61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 43.Rodionov DA, Mironov AA, Gelfand MS. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makarova KS, Mironov AA, Gelfand MS. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2001;2:Research0013. doi: 10.1186/gb-2001-2-4-research0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodionov DA, Gelfand MS. Identification of a bacterial regulatory system for ribonucleotide reductases by phylogenetic profiling. Trends Genet. 2005;21:385–389. doi: 10.1016/j.tig.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 46.Ostrovsky de Spicer P, Maloy S. PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc. Natl Acad. Sci. USA. 1993;90:4295–4298. doi: 10.1073/pnas.90.9.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurnasov OV, Polanuyer BM, Ananta S, Sloutsky R, Tam A, Gerdes SY, Osterman AL. Ribosylnicotinamide kinase domain of NadR protein: identification and implications in NAD biosynthesis. J. Bacteriol. 2002;184:6906–6917. doi: 10.1128/JB.184.24.6906-6917.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raffaelli N, Lorenzi T, Mariani PL, Emanuelli M, Amici A, Ruggieri S, Magni G. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 1999;181:5509–5511. doi: 10.1128/jb.181.17.5509-5511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]