Abstract
Crassulacean acid metabolism (CAM) is a specialized mode of photosynthesis that improves water use efficiency by shifting part or all of net atmospheric CO2 uptake to the night. Genetic dissection of regulatory and metabolic attributes of CAM has been limited by the difficulty of identifying a reliable phenotype for mutant screening. We developed a novel and simple colorimetric assay to measure leaf pH to screen fast neutron-mutagenized populations of common ice plant (Mesembryanthemum crystallinum), a facultative CAM species, to detect CAM-deficient mutants with limited nocturnal acidification. The isolated CAM-deficient mutants showed negligible net dark CO2 uptake compared with wild-type plants following the imposition of salinity stress. The mutants and wild-type plants accumulated nearly comparable levels of sodium in leaves, but the mutants grew more slowly than the wild-type plants. The mutants also had substantially reduced seed set and seed weight relative to wild type under salinity stress. Carbon-isotope ratios of seed collected from 4-month-old plants indicated that C3 photosynthesis made a greater contribution to seed production in mutants compared to wild type. The CAM-deficient mutants were deficient in leaf starch and lacked plastidic phosphoglucomutase, an enzyme critical for gluconeogenesis and starch formation, resulting in substrate limitation of nocturnal C4 acid formation. The restoration of nocturnal acidification by feeding detached leaves of salt-stressed mutants with glucose or sucrose supported this defect and served to illustrate the flexibility of CAM. The CAM-deficient mutants described here constitute important models for exploring regulatory features and metabolic consequences of CAM.
Crassulacean acid metabolism (CAM), one of three modes of photosynthetic assimilation of atmospheric CO2, has evolved multiple times in approximately 7% of vascular plant species. The high number of independent origins of CAM in over 30 taxonomically diverse plant families is indicative of convergent evolution (Crayn et al., 2004; Silvera et al., 2005) and, as such, serves as a useful model for probing how complex traits evolve in response to a changing environment. CAM is typically characterized by nocturnal CO2 uptake via the enzyme phosphoenolpyruvate (PEP) carboxylase (PEPC), which is activated via phosphorylation by a dedicated protein kinase, PEPC kinase (PPCK; Nimmo, 2000). The 3-C substrate for nocturnal carboxylation, PEP, is produced via the glycolytic breakdown of carbohydrates at night. The organic acids produced via the dark carboxylation of PEP are stored overnight in a large central vacuole (Epimashko et al., 2004) and are subsequently decarboxylated during the day to release CO2 for fixation by Rubisco behind closed stomata. CAM facilitates CO2 acquisition in water-limited environments and is typically found in species that inhabit semiarid regions (Cushman and Borland, 2002), including tropical epiphytes that must endure intermittent or seasonal water availability (Zotz, 2004). In addition, CAM can lead to greater carbon acquisition in aquatic environments where daytime CO2 availability can be limiting (Keeley, 1998).
In recent years, efforts to elucidate the metabolic, regulatory, and signaling elements that define CAM have focused largely on the facultative CAM halophyte common ice plant (Mesembryanthemum crystallinum). When grown under well-watered, nonstressed conditions, common ice plant performs C3 photosynthesis from the seedling stage through seed set (Winter and Holtum, 2007), but can switch to CAM if subjected to various treatments that reduce water availability, including high salinity (Winter and Holtum, 2005). Thus, C3 photosynthesis and CAM can be clearly discerned in plants of similar developmental status, allowing a comparison between molecular, biochemical, and physiological processes associated with C3 photosynthesis versus those governing CAM. The inducibility of CAM in common ice plant has made it a favorite model for studying the molecular genetics of CAM (Bohnert and Cushman, 2000; Cushman, 2001). More than 20 cDNA libraries exist from common ice plant from different tissues, such as meristems, roots, shoots, leaves, epidermal bladder cells, flowers, and seed capsules and different stress treatments (Bohnert et al., 2001), and extensive EST collections have been generated to facilitate gene discovery efforts (Kore-eda et al., 2004). Large-scale gene expression profiling using a custom oligonucleotide-based microarray containing probes for more than 8,400 genes derived from these EST collections have documented large-scale steady-state mRNA abundance changes following CAM induction by salinity treatment (Cushman et al., 2008). Many genes exhibited circadian patterns of abundance, suggesting that control by the circadian clock is important for synchronizing and regulating the metabolic and transport processes that underpin operation of the 24-h CAM cycle (Borland and Taybi, 2004; Boxall et al., 2005).
To date, genetic dissection of the regulatory and metabolic attributes of CAM in common ice plant has been limited by the difficulty of identifying a reliable phenotype for mutant screening. Here, we report the establishment of large-scale mutant collections of any CAM species and the development of a novel screening strategy for the isolation of CAM-deficient mutants in common ice plant. Fast neutron (Nf) mutagenesis was selected over chemical mutagenesis because mutants generated in this way have kilobase pair-size deletions or rearrangements, rather than point mutations as in chemically mutagenized plants (Bruggemann et al., 1996; Ceccini et al., 1998). We have designed a mutant screen using a simple pH indicator to rapidly identify salt-stressed plants that fail to perform nocturnal acidification, a diagnostic characteristic of CAM. A prerequisite of CAM in common ice plant is the ability to accumulate and degrade sufficient starch reserves during the day to supply via glycolysis sufficient PEP for nocturnal C4 carboxylation (Borland and Taybi, 2004). Thus, one possible type of CAM-deficient mutant phenotype that might be anticipated is a starch-deficient phenotype. A secondary, complementary screen, based on iodine staining, was used to detect defects in daytime starch accumulation in salt-stressed plants. The CAM-deficient mutants isolated here failed to express plastidic phosphoglucomutase (PGM), the enzyme that catalyzes the interconversion of Glc-6-P↔Glc-1-P and is critical to subsequent gluconeogenesis and starch formation. Validation of this genetic lesion was demonstrated by the restoration of the CAM phenotype by feeding detached leaves with Glc or Suc. We discuss how the availability of such CAM-deficient mutants could improve our understanding of the complex day/night metabolic and circadian regulatory processes that govern CAM.
RESULTS
Isolation of CAM-Deficient Mutants
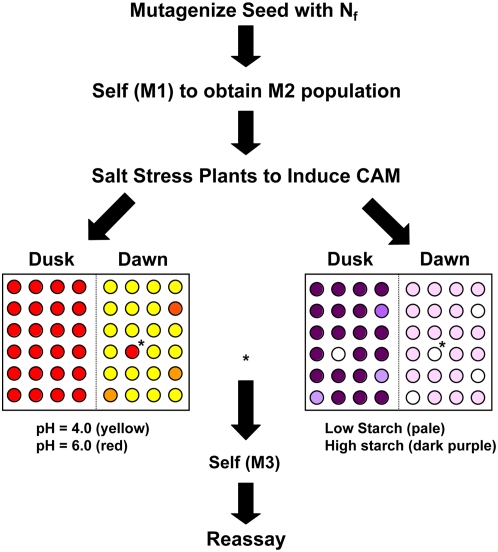
We investigated whether a simple visual screen using a colorimetric pH indicator dye could be used for high-throughput screening of ice plant mutants with a reduced ability to conduct nocturnal acidification, a key feature of CAM. Traditionally, nocturnal acidification is measured by the change in titratable acidity in leaf sample extracts collected at the end of the dark period (dawn) and at the end of the light period (dusk). However, this approach is too labor intensive for high-throughput screening. To simplify the screening process, leaf discs were sampled from plants at dawn and dusk in a microtiter plate assay containing the pH indicator, chlorophenol red (see Supplemental Fig. S1), rather than by measuring the absolute leaf pH of each individual plant independently (Fig. 1). To confirm putative CAM mutants, a secondary, complementary iodine staining assay (Caspar et al., 1985), which monitored directly changes in starch accumulation in leaves associated with CAM induction, was performed. Mutants with defects in either starch degradation or synthesis were identified by a lack of purple staining (Fig. 1).
Figure 1.
Screening strategy for CAM mutants of common ice plant. Mutagenized M2 plants were irrigated with 0.3 m NaCl for 7 to 14 d to induce CAM. Duplicate leaf discs were collected at dawn or at dusk in a 48-well microtiter plate. Leaf discs were assayed using the pH indicator chlorophenol red; wild-type plants performing CAM show nocturnal acidification, whereas CAM-deficient mutants (marked as *) do not. As a secondary screen, leaf discs were screened for starch content at dawn and dusk using a mixture of iodine and potassium iodide. Wild-type plants that accumulate normal levels of starch show intense iodine staining, whereas mutants deficient in starch accumulation show weak iodine staining. Plants failing one or both tests were scored as putative CAM mutants.
From an estimated total of 11,666 M2 plants (derived from 779 M1 plants irradiated with 50 Gy) and 5,954 M2 plants screened (derived from 292 M1 plants irradiated with 40 Gy), 167 candidates were selected during the primary screen on the basis of a failure to undergo nocturnal acidification following salinity stress treatment. Within these populations, 50 lines contained morphological aberrations (e.g. alterations in pigmentation or morphology of leaves and stems, etc.), including an epidermal bladder cell-less mutant with reduced salt tolerance (Agarie et al., 2007). The frequency of putative CAM mutants within the M2 generation irradiated at 40 and 50 Gy were 0.3% and 0.42%, respectively. In the subsequent M3 generation, the frequency of putative CAM-deficient mutants increased to 0.7%. Two lines were isolated and designated 351 and 371, which showed 18 and 22, respectively, of 24 plants assayed that were CAM deficient. These two mutant lines were subjected to further physiological and biochemical characterization.
Physiological Characterization of Putative CAM-Deficient Mutants
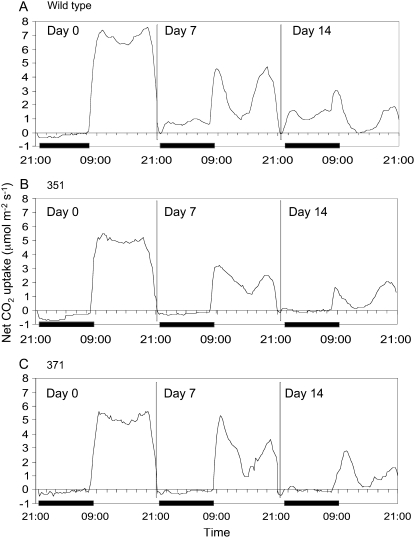
Patterns of net CO2 assimilation in leaves over a day/night cycle were monitored over a 2-week time course of salinity treatment and confirmed that M4 generation lines 351 and 371 were deficient in CAM (Fig. 2). Net dark CO2 uptake was apparent in wild-type plants after 7 d of salinity treatment and increased further by 14 d of salinity (Fig. 2). In contrast, there was negligible net dark CO2 uptake in either of the mutant lines after 14-d treatment with 0.3 m NaCl. Both mutant lines also showed lower rates of daytime net CO2 assimilation compared to wild type in well-watered conditions (day 0; Fig. 2), and this was also reflected in lower stomatal conductance and transpiration rates in the mutant lines (data not shown).
Figure 2.
Day/night net CO2 exchange profiles of wild type and two CAM-deficient lines of common ice plant grown under well-watered conditions (day 0) and after 7 and 14 d of treatment with 0.3 m NaCl. The solid bars on the x axes represent the periods of darkness. Each gas exchange profile is for a representative leaf from three replicate determinations.
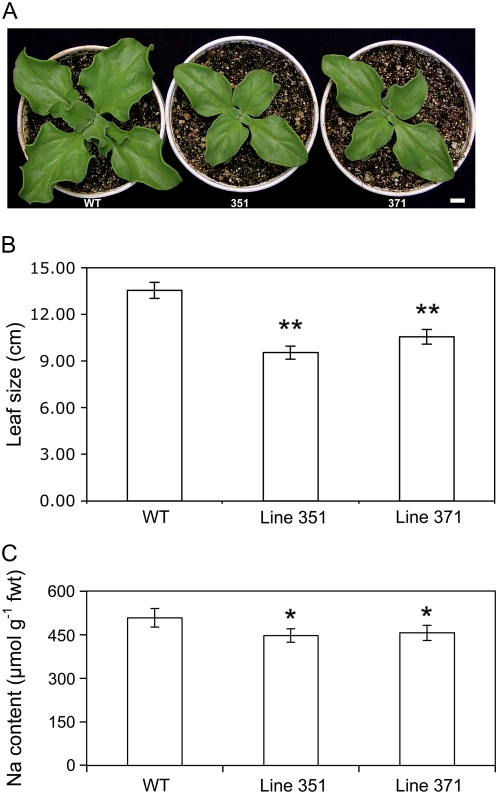
Growth rate measurements of wild type and M4 generation mutant lines under well-watered (unstressed) conditions up to 4 weeks of age showed that both mutant lines displayed significant retardation in growth (Fig. 3, A and B). To determine whether the lower transpiration and growth rates in the mutants, as compared to wild type, might be due to sodium toxicity, sodium accumulation was measured after 2 weeks of exposure to salinity. The mutant lines contained slightly less sodium than wild-type plants. Although the sodium content differences observed were statistically significant, the growth differences were unlikely to be due to sodium toxicity (Fig. 3C).
Figure 3.
Growth and sodium content of the leaves and shoots of wild type (WT) and two CAM-deficient lines of common ice plant. A, Representative plants (bar = 1 cm). B, Shoot growth of WT and two CAM-deficient lines (351 and 371) sampled after 28 d of growth under unstressed conditions. Each bar is the mean of 39 plants ± sd. Student's t test P value between WT and 351: P = 2.2 × 10−16** and between WT and 371: P = 1.6 × 10−10**. C, Sodium content of the leaves and shoots of WT and two CAM-deficient lines (351 and 371) sampled after 14-d treatment with 0.3 m NaCl. Each bar is the mean of four plants ± sd. Student's t test P value between WT and 351: P = 2.5 × 10−5* and between WT and 371: P = 1.9 × 10−4*.
Reproductive Output
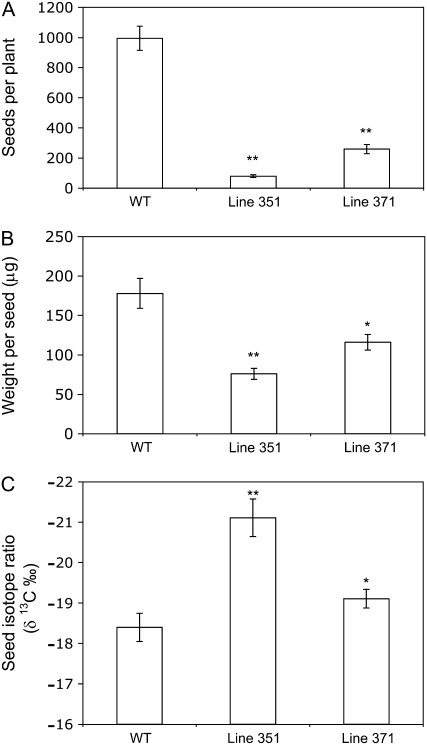
Wild-type plants and M4 generations of both mutant lines were taken through to seed. Both mutant lines produced substantially fewer and lighter seeds compared to wild type (Fig. 4, A and B). Line 351 showed the most significant reduction in reproductive output as compared to wild type, with seed production only 10% of that shown by wild type and average seed weight less than one-half of that noted in wild type. Carbon-isotope ratios of harvested seed from mutant line 351 were depleted in 13C by up to 3‰ (i.e. more C3-like) compared to wild-type seed, whereas seed from mutant line 371 was depleted in 13C by up to 1‰ compared to wild type (Fig. 4C).
Figure 4.
Reproductive capacity and carbon-isotope ratios in seed of wild type (WT) and two CAM-deficient lines of common ice plant treated with 0.3 m NaCl over a 4-month growth period. A, Number of seeds per plant. Each data point is the mean of 69 (351), 92 (371), or 131 (WT) plants ± se. Student's t test P value between WT and 351: P = 2.2 × 10−16** and between WT and 371: P = 2.2 × 10−16**. B, Individual seed weight. Each data point is the mean of 69 (351), 92 (371), or 131 (WT) individual plants ± se. Student's t test P value between WT and 351: P = 1.8 × 10−11** and between WT and 371: P = 1.2 × 10−4*. C, Carbon-isotope ratios of seeds. Each data point is the mean of 10 replicates ± se. Student's t test P value between WT and 351: P = 9.8 × 10−13** and between WT and 371: P = 1.2 × 10−3*.
Metabolite Levels and CAM Expression
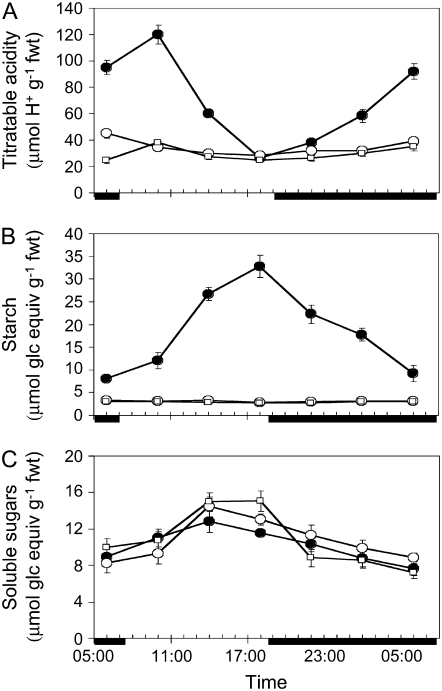
Following treatment with 0.3 m NaCl for 14 d, wild-type plants showed a more than 4.5-fold day/night change in the content of leaf titratable acids, whereas both mutant lines showed less than a 1.5-fold change in leaf titratable acids (Fig. 5A). Salt-stressed wild-type plants also showed more than a 4-fold day/night change in leaf starch content, whereas the leaves of both mutant lines showed very low levels of starch (3-fold lower than wild type at dawn) that failed to exhibit any diel fluxes (Fig. 5B). Control (unstressed) plants of both mutant lines showed similarly depleted levels of leaf starch relative to wild-type plants (data not shown). Assuming that two H+ are equivalent to one Mal and one Glc equivalent can generate two PEP, there was a close stoichiometry between starch degradation and titratable acid accumulation at night in the salt-stressed wild type. The day/night changes in leaf soluble sugar content were more pronounced and greater in absolute abundance during the afternoon period in both of the mutant lines compared to wild type (Fig. 5C). In both of the mutant lines, the small amounts of acid accumulated overnight could have been furnished by PEP produced from soluble sugar degradation using the same stoichiometric relationship as described above.
Figure 5.
Day/night changes in titratable acidity (A), starch (B), and soluble sugar (C) content in leaves sampled from wild type (black circles) and two CAM-deficient lines, 351 (white squares) and 371 (white circles), after exposure to 0.3 m NaCl for 14 d. Each point is the mean of four replicates ± se. Solid bars on the x axes represent the periods of darkness.
Restoration of CAM by Sugar Feeding
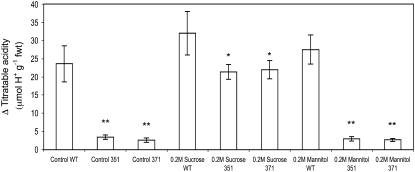
To test the hypothesis that the CAM deficiency in the mutants was due to a lack of substrate for nocturnal malate synthesis, detached salted leaves of line 351 were incubated with either 0.2 m Glc or Suc. Suc feeding restored the overnight accumulation of titratable acidity in detached leaves of mutant lines 351 and 371 (Fig. 6) to levels comparable to those found in salted wild-type plants (Fig. 5A). Feeding mutant leaves with 0.2 m Glc also resulted in a nearly identical restoration of titratable acidity in detached leaves of the mutant lines (data not shown). Such results imply that Suc was degraded to Fru and Glc, which were then processed to produce PEP for organic acid synthesis. Bulk analysis and HPLC analyses of the sugar-fed leaves confirmed that the mutant was capable of metabolizing exogenous Suc because there was an increase in total soluble sugars and Glc and Fru content in the leaf in parallel with the increase in Suc while starch content remained unchanged (E. Antony and T. Taybi, unpublished data). Detached leaves were also fed with 0.2 m mannitol to confirm that alterations in titratable acidity were not due to osmotic effects. Mannitol treatment had no significant impact on titratable acidity changes in either wild type or mutant lines (Fig. 6).
Figure 6.
Restoration of CAM-deficient phenotype by sugar feeding of detached leaves. Leaves of wild type (WT) and CAM-deficient lines 351 and 371 were detached from plants that had been treated with 0.3 m NaCl for 10 d and then placed in water (control) or fed with 0.2 m Suc or mannitol for the latter 8 h of the photoperiod. Measurements of the change (Δ) in leaf titratable acidity at the start and end of the subsequent dark period were determined and plotted as the difference between the two measurements. Each point is the mean of five replicates ± se. Student's t test P value for control between WT and 351: P = 5.4 × 10−4** and WT and 371: P = 3.3 × 10−4**. Student's t test P value for Suc feeding between WT and 351: P = 1.4 × 10−2* and WT and 371: P = 1.8 × 10−2*. Student's t test P value for mannitol feeding between WT and 351: P = 2.0 × 10−4** and WT and 371: P = 1.2 × 10−6**.
Plastidic PGM Enzyme Activity and mRNA Expression
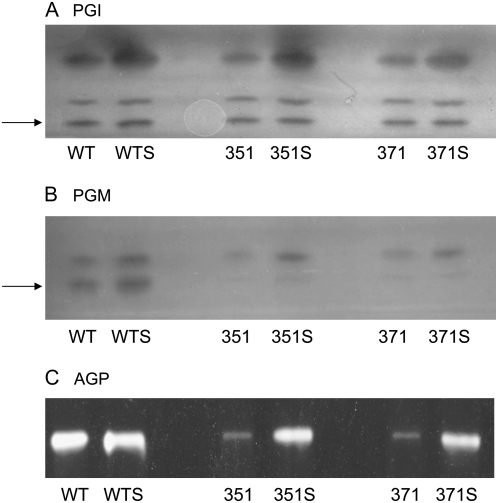
To test the hypothesis that a defect in starch biosynthesis and/or breakdown was responsible for the observed CAM-deficient phenotype, key gluconeogenic enzymes leading to starch biosynthesis were surveyed by in-gel activity staining assays. Phosphoglucoisomerase (PGI), which catalyzes the interconversion of Fru-6-P and Glc-6-P, is present as three distinct isozymes in the ice plant, one of which is localized to the plastid (Fig. 7A). All three isozymes of PGI showed similar activity in the wild type and mutant lines 351 and 371 under control conditions, whereas salinity elicited an increase in the activity of the highest Mr isozyme in all plants (Fig. 7A). Two isozymes of PGM, which catalyzes the interconversion of Glc-6-P↔Glc-1-P, were present in leaves of wild-type plants and the activity of both isozymes increased in response to salinity in the wild-type plants (Fig. 7B). In both mutant lines, salinity also elicited an increase in activity of the higher Mr cytosolic isoform of PGM, but both 351 and 371 were deficient in the lower Mr plastidic isoform of PGM (Fig. 7B). As an additional measure of the specificity of the genetic lesion associated with starch biosynthesis, the activity of ADP-Glc pyrophosphorylase (AGP) was also monitored. AGP catalyzes the first committed step in starch biosynthesis in plants: Glc-1-P + ATP→ADP-Glc + pyrophosphate (PPi; Lee et al., 2007). In noncereal plants, most AGP activity is localized to the plastid. Mutant lines 351 and 371 showed reduced AGP activity under control conditions relative to wild-type plants, but in salt-stressed plants the detectable activity of AGP was largely restored (Fig. 7C), presumably through a salt stress-induced increase in AGP expression. The salinity-induced increases in enzyme abundance for PGI and AGP are paralleled by increased steady-state transcript abundance (Cushman et al., 2008).
Figure 7.
In-gel activity stained gels of key glycolytic/gluconeogenic enzymes. A, PGI (10 μg protein/lane). B, PGM (10 μg protein/lane). C, AGP (50 μg protein/lane) in wild type (WT) and CAM-deficient lines 351 and 371 grown under control conditions or subjected to 0.3 m NaCl for 14 d (WTS, 351S, and 371S). Representative gels from triplicate assays are shown. Arrows indicate the position of the plastidic isozymes.
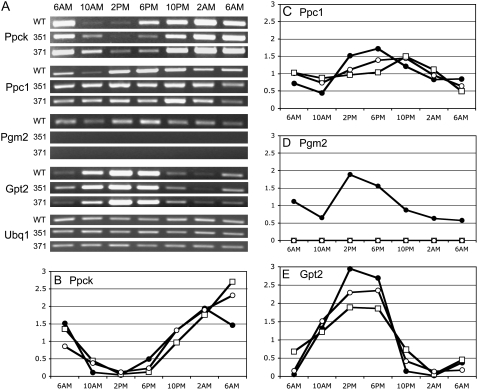
To verify that plastidic PGM expression was impaired in the CAM-deficient mutant lines, transcripts for a variety of genes important for CAM, including Ppck1 (Taybi et al., 2000), CAM-specific PEPC (Ppc1; Cushman et al., 1989), CAM-specific Glc-6-P/Pi translocator (Gpt2; Kore-eda et al., 2005), and plastidic Pgm2 were monitored by semiquantitative reverse transcription (RT)-PCR. The transcript abundances of all genes were similar in wild type and CAM-deficient mutant lines (Fig. 8), with the exception of plastidic PGM, which lacked any detectable mRNA in both mutant lines 351 and 371. Uniform transcript abundance of constitutively expressed Ubq1 indicated that the total RNA input was similar for all reactions (Fig. 8A). Quantitative analyses, however, revealed approximately 4-h shifts in the day/night patterns of transcript abundance of the CAM-related genes in the mutant lines in comparison to wild type (Fig. 8, C–E). This was particularly marked for both Ppc1 and Gpt2, which exhibited reduced transcript abundance during afternoon and dusk (2 pm and 6 pm) and elevated abundance during the night (10 pm and 2 am) in both 351 and 371 relative to wild type. Ppck1 also exhibited approximately a 4-h shift in relative transcript abundance over the course of the day/night period (Fig. 8). Such observations indicate a shift in circadian and/or metabolite-mediated control of transcription in the CAM-deficient lines.
Figure 8.
Relative steady-state mRNA abundance of key CAM-specific enzymes over a 24-h period in plants exposed to 0.3 m NaCl for 10 d. A, RT-PCR products from Ppck, Ppc1, plastidic Pgm2, Glc-6-P, Gpt2, and Ubq1 in wild type (WT) and two CAM-deficient lines, 351 and 371. Representative gels from duplicate assays are shown. B to E, Relative transcript abundance normalized to Ubq1 expression for Ppck1 (B), Ppc1 (C), Pgm2 (D), and Gpt2 (E) in WT (black circles) and two CAM-deficient lines, 351 (white squares) and 371 (white circles). Values plotted are the mean of two replicates.
DISCUSSION
Isolation and Characterization of CAM-Deficient Mutants
We have shown that a novel, yet simple, visual screen using a colorimetric pH indicator dye can be used for high-throughput screening and isolation of ice plant mutants with a reduced ability to conduct nocturnal acidification, a key feature of CAM (Fig. 5A). Two mutant lines, designated 351 and 371, were isolated using this screening protocol and were confirmed to be CAM deficient by their failure to conduct nocturnal net CO2 uptake after 2 weeks of treatment with 0.3 m NaCl (Fig. 2). In wild-type plants, the same salinity regime induced net dark CO2 uptake within 7 d. After 2 weeks of salinity stress, approximately 50% of net 24-h carbon gain occurred at night in wild-type plants. However, lines 351 and 371 could not be designated as CAM-null mutants because a slight, but significant, change in titratable acidity was measured over 24 h in plants from both mutant lines exposed to salinity for 2 weeks. Such nocturnal acidification can be attributed to the refixation of respiratory CO2 by PEPC (Griffiths, 1988). Under control conditions (day 0), the reduced net CO2 uptake in the mutant lines was similar to that observed in a starchless mutant of Arabidopsis (Arabidopsis thaliana) deficient in chloroplast PGM activity (Caspar et al., 1985) and reduced CO2 assimilation rates in a starchless mutant of Nicotiana sylvestris defective in plastidic PGM (Hanson, 1990) or in transgenic potato (Solanum tuberosum) in which plastidic PGM expression was suppressed by antisense inhibition (Lytovchenko et al., 2002). The inability to use triose-P/Glc-6-P for starch synthesis results in Pi limitation for ATP synthesis and ribulose-1,5-bisP regeneration and could account for the observed reductions in photosynthetic capacity (Sharkey et al., 1986; Lytovchenko et al., 2002).
CAM-Deficient Mutants Show Reduced Vegetative Growth and Reproductive Output
Both of the mutant lines isolated in this study showed a substantial reduction in leaf growth, seed number, and seed weight compared to the wild-type plants (Fig. 4). At least part of the reduced growth and reproductive output in mutant lines 351 and 371 can be attributed to starch deficiency. The slow-growth phenotype is similar to that observed for the starchless mutant of Arabidopsis deficient in chloroplast PGM activity also grown under a 12-h photoperiod (Caspar et al., 1985), but not for the starchless mutant of N. sylvestris defective in plastid PGM grown under a 16-h light photoperiod (Hanson and McHale, 1988). Floral initiation can be delayed and seed production reduced by up to 50% in starch-deficient mutants of Arabidopsis (Schulze et al., 1994; Yu et al., 2000). However, the 90% reduction in seed number from line 351 greatly exceeded that predicted just from starch deficiency alone and is comparable to the reduction in reproductive output reported when common ice plants were deprived of CO2 each night to minimize nocturnal carbon gain (Winter and Ziegler, 1992). The seeds produced by line 351 were more depleted in 13C (i.e. more C3-like) compared to seeds from line 371, suggesting that the latter conducted more nocturnal CO2 uptake during the reproductive phase. Presumably, differences occurred between the mutant lines in terms of carbohydrate allocation at the reproductive phase, as indicated by the faster growth rate of line 371, thereby resulting in higher CAM activity in this line, which produced twice as many, and significantly heavier, seeds than line 351. In summary, it would appear that the magnitude of CAM engaged under saline conditions influenced the reproductive success of common ice plant.
CAM-Deficient Mutants Are Deficient in Leaf Starch and Plastidic PGM
As a complementary screen, and to confirm the putative CAM defect indicated via the colorimetric pH screen, salt-stressed mutants were also tested for defects in day/night starch accumulation/depletion using a high-throughput iodine-staining protocol (Fig. 1). A diagnostic feature of the CAM cycle in common ice plant is reciprocal cycling of organic acids and starch, so a failure to accumulate starch over the day might be anticipated of a CAM-deficient mutant after the imposition of salt stress. Both of the isolated mutant lines showed this starch-deficient phenotype (Fig. 5B), but, because well-watered plants of both lines also had very low levels of leaf starch, it appears that starch deficiency may be the cause, rather than a consequence, of CAM deficiency. Interestingly, both CAM-deficient mutant lines also exhibited elevated soluble sugar accumulation (Fig. 5B). Starch deficiency and increased soluble sugar accumulation is typically observed in the leaves of Arabidopsis and N. sylvestris plastidic PGM mutants (Caspar et al., 1985; Hanson and McHale, 1988) or following the antisense inhibition of plastidic PGM expression in potato leaves (Lytovchenko et al., 2002) or tubers (Tauberger et al., 2000). Native activity gels (Fig. 7) and semiquantitative RT-PCR (Fig. 8) revealed that the CAM-deficient lines of common ice plant lacked expression of plastidic PGM, which can account for the observed starch deficiency and increased accumulation of soluble sugars.
Previous work has indicated that starch turnover and content are key regulatory factors for the expression of CAM in common ice plant (Borland and Dodd, 2002). Salinity elicits substantial increases in a range of enzyme activities that participate in glycolysis (Winter et al., 1982) and starch degradation in common ice plant (Paul et al., 1993), as well as their corresponding steady-state transcripts (Cushman et al., 2008), which together are thought to satisfy the demand for substrate for PEPC-mediated dark CO2 uptake as CAM is induced. Moreover, in leaves of common ice plant that were depleted in starch by 50% via exposure to CO2-free air for 24 h, subsequent net dark CO2 uptake in ambient air was reduced by 50% relative to controls, demonstrating the close stoichiometric relationship between starch degradation and malate accumulation (Dodd et al., 2003). Similar stoichiometric considerations suggested that the low levels of titratable acids that were accumulated overnight in leaves of the salt-stressed mutants could have been derived from PEP that was generated from the breakdown of soluble sugars. Consequently, sugar feeding of detached salted leaves of lines 351 and 371 restored nocturnal acid accumulation to levels that were comparable to those in wild-type plants (Fig. 6). Thus, the CAM deficiency in these mutants appears to be due to substrate limitation of dark carboxylation. Although the feeding of detached leaves with Glc or Suc was able to restore the CAM phenotype short term in detached leaves, it remains to be determined if restoration of carbohydrate alone is necessary and sufficient for the long-term support of CAM. Expression of various 3-C and 6-C carbon and carboxylic acid transport functions might be dependent upon the availability of sufficient and/or appropriate carbohydrate reserves. However, more importantly, the restoration of acid accumulation by sugar feeding demonstrates the flexibility of the CAM pathway in terms of utilizing different carbohydrate sources to generate substrate for nocturnal carboxylation. Such flexibility is mirrored by the diversity of soluble and insoluble storage carbohydrates utilized by different CAM species within and between different genera to provide 3-C substrate for nocturnal acidification (Christopher and Holtum, 1996). This observation also suggests that CAM has few, if any, unique biochemical or regulatory requirements for nocturnal carbohydrate mobilization. Thus, the convergent evolution of CAM appears to have been facilitated by exploiting existing biochemistry resulting from different evolutionary histories for the breakdown of starch and/or soluble sugars at night.
CAM Mutants Reveal Regulatory Features and Metabolic Consequences of CAM
In CAM plants, the expression, abundance, and activity of PEPC serve as a diagnostic marker for the potential capacity of nocturnal carboxylation and magnitude of CAM (Winter, 1982; Borland et al., 1998). In common ice plant, salinity is known to elicit an increase in the transcript abundance of Ppc1, which encodes a CAM-specific isoform of PEPC (Cushman et al., 1989), Ppck, the dedicated kinase that activates PEPC at night in CAM plants (Taybi et al., 2000), and a CAM-specific Gpt2, which is responsible for Glc-6-P export across the chloroplast envelope during nocturnal starch mobilization and/or daytime starch synthesis (Kore-eda et al., 2005). In this article, the wild type and CAM-deficient mutants of common ice plant showed comparable steady-state transcript abundances of Ppc1, Ppck, and Gpt2. Moreover, lines 351 and 371 showed comparable levels of PEPC protein to those in wild type as indicated by western blotting (data not shown). Such observations imply that the signaling elements responsible for engaging these key drivers of CAM expression were not perturbed significantly by the mutation and that the CAM deficiency in lines 351 and 371 was not attributable to a deficiency in the potential capacity for nocturnal carboxylation. However, the peak afternoon mRNA abundance of Ppc1 and Gpt2 was reduced in the CAM-deficient mutants, and there were shifts in the day/night patterns of transcript abundance of Ppc1, Ppck, and Gpt2 in the mutant lines relative to wild type. PPCK is known to be regulated at the level of transcript abundance (Taybi et al., 2000) and circadian oscillations in Ppck transcripts are known to be modulated by metabolites such as malate (Borland et al., 1999). The altered 24-h patterns of transcript abundance observed in the mutants may be explained by the dampened 24-h cycling of malate and/or carbohydrates in these plants compared to the wild type. Thus, the CAM-deficient mutants of common ice plant will provide further opportunities for determining which genes, known to show circadian patterns of transcript abundance, do so as a consequence of the metabolite cycling associated with CAM. Such large-scale mRNA expression studies can now be accomplished using a custom microarray developed recently for ice plant (Cushman et al., 2008).
In conclusion, as a consequence of Nf mutagenesis and high-throughput screening methods, a large collection of morphological and salt-sensitive (Agarie et al., 2007) mutants, as well as the CAM-deficient mutants described here, have been obtained for common ice plant. Such mutants will provide useful material for future investigations into developmental and salt tolerance studies. Whereas the CAM deficiency of the two mutant lines described here appears to be a corollary of starch deficiency, lines 351 and 371 provide a means for further elucidating which molecular and metabolic attributes of CAM are a consequence of the dynamic day/night changes in organic acid content and internal [CO2] and [O2] that typify this photosynthetic specialization.
MATERIALS AND METHODS
Establishment of Mutant Seed Banks
Common ice plant (Mesembryanthemum crystallinum) seeds were irradiated by Nf bombardment at a range of doses from 20 to 120 Gy using a cobalt 60 source at the reactor of the Atomic Energy Agency (courtesy of Dr. H. Brunner, Vienna). Wild-type Nf-irradiated seeds (M0) were surface sterilized and germinated in 150-mm disposable plastic petri dishes containing Murashige and Skoog agar (0.43% Murashige and Skoog salts, 1× B5 vitamins, 3% Suc, and 0.5% agar). The seedlings (M1) were grown in a growth chamber under 100 μmol m−2 s−1 of cool-white fluorescent light on a 12-h (26°C) light/12-h (18°C) dark photoperiod. After 1 month, the survival of the seedlings that grew to an expanded second leaf pair and beyond was scored as positive for survival. Based on survival rates, wild-type seeds that were irradiated with Nf doses of 40 and 50 Gy (55%–60% survival) were taken forward for mutant screening and were germinated on agar plates as described above. The plants were selfed to obtain an M2 population.
Plant Material
The M2 seeds were germinated on agar as described above and 1-week-old seedlings were transplanted into commercial soil mix (MetroMix 200; Scotts Sierra Horticultural Products) and irrigated daily with 0.5× Hoagland solution number 2 (Hoagland and Arnon, 1950). Plants were grown in a greenhouse under natural daylight supplemented with high-pressure sodium lamps providing a photon flux density of 350 μmol m−2 s−1 on a 12-h light (26°C)/12-h dark (18°C) cycle. Five-week-old plants were irrigated with 0.5× Hoagland solution containing 0.3 m NaCl for 14 d. This treatment was found to give uniform and adequate stress to fully induce CAM. Sampling of plants at younger developmental stages or under shorter durations of stress treatment often resulted in false positives. To increase the number of plants that could be screened, plants were grown in 24-well flats with a relatively small rooting volume (0.2 L). This format allowed each flat of plants to be sampled at both dawn and dusk using a single 48-well microtiter plate. If a putative mutant were detected, the arrayed format allowed the corresponding plant to be easily identified. This also reduced the size of the plants and also minimized the plant growth space needed for mutant screens.
CAM Mutant Screen
Leaf pH assays were conducted on 24 individuals from each M2 line before and after the imposition of salinity by punching out a 0.23-cm2 leaf disc with a paper punch from the fourth or fifth leaves both at the end of the dark period (dawn) and at the end of the light period (dusk), and immersing the leaf discs in a solution of 0.025% (w/v) chlorophenol red (Sigma-Aldrich; indicator grade no. 199524), a colorimetric pH indicator dye. Chlorophenol red has a visual transition interval from pH = 4.8 (yellow) to pH = 6.4 (red), which matches the pH difference observed between morning and evening measurements made in CAM-performing ice plant (see Supplemental Fig. S1). The goal was to identify individual mutants amid a population of wild-type plants by simply visualizing the color differences between leaf samples in a microtiter plate assay, rather than by measuring the absolute leaf pH of each individual plant independently. After 4 h, the plates were examined for color development. Plants that failed to undergo nocturnal acidification as indicated by the colorimetric pH assay were classified as putative mutants. These plants were retested after 1 and 2 weeks of additional salt stress to rule out false positives. Seed (M3) was collected from each plant of a putative mutant line, planted, and M3 plants reassayed as above. To confirm putative mutants, positive and apparent false positives that were identified using the pH screen were subjected to a leaf starch content assay as previously described (Caspar et al., 1985). Leaf discs (0.23 cm2) were placed in 100% methanol and incubated for 30 min with a starch-staining solution containing 5.7 mm iodine, 43.4 mm potassium iodide, and 0.2 n HCl, and followed by a wash with 80% (v/v) ethanol. Starch-containing leaf discs turned purple. Positives were grown to maturity to collect M4 seed.
Net CO2 Exchange
M4 seeds from wild type and two putative CAM-deficient mutant lines (351 and 371) were germinated and propagated as described above. Plants were grown in a controlled environment chamber with a 12-h photoperiod, a day/night temperature regime of 25°C/18°C, and photon flux density of 350 μmol m−2 s−1 at plant height. Five-week-old plants (primary leaf 4 fully emerged) were irrigated daily with 0.5× Hoagland solution containing 0.3 m NaCl for up to 14 d. Gas exchange measurements were made at day 0 (control), day 7, and day 14 of the salinity treatment. Net CO2 uptake was monitored continuously on a single leaf (primary leaf 4) over 24-h periods with three separate runs made for each treatment. The leaf was enclosed in a gas exchange cuvette, which tracked the environmental conditions in the growth chamber with gas exchange parameters measured using an open infrared gas analyzer (H. Walz). Rates of net CO2 exchange were calculated using DIAGAS software provided by Walz.
Leaf Sodium Content
The sodium content of leaves was determined by flame photometry. Dried plant tissue was combusted overnight at 550°C. The ashed samples were placed in ceramic crucibles and moistened with a few drops of distilled water and extracted three times with concentrated HCl while incubated in a steam bath. The sodium was determined in diluted extracts via flame photometry using 0, 0.8, 1.6, 2.4, 3.2, and 4.0 μg mL−1 sodium as standards. Statistical significance was determined using Student's t test, assuming unequal variances (as indicated by a simple test for the homogeneity of variances) using the R statistical software package (http://cran.r-project.org).
Seed Weight and Production
Wild-type and M4 seeds from two putative CAM-deficient mutant lines (351 and 371) were germinated and propagated as described above under greenhouse conditions except that individual plants were grown in 1-L styrofoam pots containing commercial soil mix (MetroMix 200) and irrigated every other day with 0.5× Hoagland solution number 2. Five-week-old plants were irrigated every other day with 0.5× Hoagland solution containing 0.3 m NaCl for up to 3 months until flowering was completed. Plants were then allowed to dry completely. Seed pods were harvested and seeds were cleaned, counted, and weighed from individual plants.
Carbon-Isotope Ratio Analyses
Samples of 1 ± 0.1 mg dry weight were taken from each batch of seeds from wild type and lines 351 and 371 (10 samples from each line) and were placed in tin capsules (D1006; Elemental Microanalysis) before positioning in an autosampler. Samples were then passed through a combustion furnace, reduction furnace, water traps, and gas chromatograph before being transferred to the mass spectrometer. Enrichment in 13C was determined using continuous flow isotope ratio mass spectrometry (ANCA SL preparation unit interfaced with a 20/20 mass spectrometer; PDZ Europa). All samples were analyzed in duplicate and referenced to an internal flour standard. The flour standard had been previously calibrated against NBS22 IAEA standard. Results are given as δ13C (‰).
Leaf Titratable Acidity, Soluble Sugar, and Starch Content
Leaves (primary leaf 4) were harvested from wild type, lines 351 and 371, at 4-h intervals over a 24-h light/dark cycle after the plants had been treated with 0.3 m NaCl for 2 weeks. The sampled leaves were snap frozen, powdered in liquid N2, and stored at −80°C until analyzed. Frozen tissue (0.3 g) was ground completely using a mortar and pestle and placed in 3 mL of 80% ethanol at 80°C for 1 h. Aliquots of the methanol extracts were titrated against 1 mol m−3 NaOH to a neutral endpoint, as indicated by phenolphthalein or direct measure with a pH meter, and leaf titratable acidities were expressed as μmol H+ g−1 fresh weight.
The soluble sugar content of methanol extracts was determined using the colorimetric phenol-sulfuric acid test as described (Dubois et al., 1956). Soluble sugars from the methanol extract were assays for Glc equivalents. The remaining insoluble leaf material was rinsed several times in distilled water, ground in 0.1 m acetate buffer (pH 4.5), and boiled for 30 min. After cooling, each extract was incubated with 5 units of amyloglucosidase and 0.5 units α-amylase (Sigma-Aldrich) overnight at 40°C. After centrifugation at 12,000g for 10 min, the supernatant was assayed for Glc equivalents as described (Dubois et al., 1956).
Sugar Feeding of Detached Leaves
Four-week-old plants were irrigated every other day with 0.5× Hoagland solution (control) or with 0.5× Hoagland solution containing 0.3 m NaCl (salt stressed) for 10 d. Replicates of primary leaf 4 were detached, weighed, and the petiole immersed in water for the controls and 0.2 m Glc, Suc, or mannitol solutions for the treatments. Sugar feeding commenced 2 h into the photoperiod and lasted until the leaves were sampled at the end of the photoperiod (10 h in sugar solution) and at the end of the dark period (22 h in sugar solution). Leaves were frozen in liquid N2 and stored at −80°C until analyzed for titratable acids and soluble sugar content as described above.
In-Gel Enzyme Activity Assays
Five-week-old plants were irrigated every other day with 0.5× Hoagland solution (control) or with 0.5× Hoagland solution containing 0.3 m NaCl (salt stressed) for 14 d. In-gel enzyme activity assays for PGI (EC 5.3.1.9), PGM (EC 5.4.2.2), and AGP (EC 2.7.7.27) were conducted according to previously described methods using 7% (w/v) PAGE with PGI and PGM activity detected as the formation of formazan in enzyme-coupled reactions, whereas AGP activity was detected by calcium pyrophosphate precipitation (Wang et al., 1998).
Total RNA Isolation and Semiquantitative RT-PCR
Total RNA was purified from 150 mg of powdered leaf tissue bulked from leaves of four individual plants using TRIzol reagent (Invitrogen) as described (Taybi and Cushman, 1999). After the assessment of total RNA quality and quantity, 5 μg of total RNA were treated with amplification grade RNase-free DNase I (Invitrogen) to prevent amplification of genomic DNA. DNase-treated RNA was then diluted to 50 ng μL−1 and subjected to RT-PCR amplification. Single-tube RT and PCR reactions were carried out in a final volume of 25 μL using the Qiagen One-Step RT-PCR kit (Qiagen) according to the manufacturer's protocol. The final concentrations for RT-PCR reaction components were as follows: 1× One-Step RT-PCR buffer (2.5 mm MgCl2, Tris-Cl, KCl, (NH4)SO4, dithiothreitol; pH = 8.7), dNTP mix (400 μm each of dATP, dCTP, dGTP, and dTTP), enzyme mix (40 μm dithiothreitol, 4 μm EDTA, Omniscript and Sensiscript reverse transcriptases, and HotStarTaq DNA polymerase), 50 ng total RNA, and 0.6 μm of each forward and reverse gene-specific primer. RT-PCR was conducted as a single-tube reaction and cDNA synthesis was promoted using the reverse primer. PCR primers used to assess gene expression and cycle conditions are listed in Supplemental Table S1. The housekeeping gene encoding Ubq1 (GenBank accession no. AF053563) was used as an internal PCR control for normalizing differences in gene expression as expression of Ubq1 remains constant over the day/night cycle in leaves from well-watered and salted common ice plants (Boxall et al., 2005). PCR products were examined on 1.2% agarose gels stained with ethidium bromide and photographed under UV light. RNA input amount and cycle number were individually optimized for each gene, which ensured a linear relationship between total RNA input and product quantity when separated on gels. The amplification of each cDNA was perfectly specific. Densitometric analysis of stained bands was performed using the National Institutes of Health ImageJ program (http://rsb.info.nih.gov/ij/).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF158091 (Ppck1), X13660 (Ppc1), BG269256 (Pgm2), AB190772 (Gpt2), and AF053563 (Ubq1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The pH curve for the colorimetric indicator dye chlorophenol red (2-chloro-4-[3-(3-chloro-4-hydroxyphenyl)-1,1-dioxobenzo[c]oxathiol-3-yl]phenol).
Supplemental Table S1. Primer sets and conditions used for semiquantitative RT-PCR analyses.
Supplementary Material
Acknowledgments
We thank Mary Ann Cushman and Susan Patterson for technical assistance, Edna Antony and Richard French for help with the carbohydrate analyses, and Karen A. Schlauch for assistance with statistical data analysis. We gratefully acknowledge Davina Bufford, Laura Clay, Monica Dennis, Dennis Shoun, Ryan Snow, Katherine Baumann, Joshua Branco, Monica Orten, and João P. Maroco for technical assistance with plant care, seed cleaning, and routine screening of ice plant mutant collections. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation or the National Institutes of Health.
This work was supported in part by the National Science Foundation (grant nos. IBN–9722285 and IBN–0196070 to J.C.C.), as well as the Nevada Agricultural Experiment Station (publication no. 03087098), the Natural Environment Research Council UK (grant no. NER/A/S/2001/01163 to A.M.B.), and Newcastle University. This publication was also made possible by National Institutes of Health Grant Number P20 RR–016464 from the INBRE Program of the National Center for Research Resources through its support of the Nevada Genomics, Proteomics, and Bioinformatics centers.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: John C. Cushman (jcushman@unr.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agarie S, Shimoda T, Shimizu Y, Baumann K, Sunagawa H, Kondo A, Ueno O, Nakahara T, Nose A, Cushman J (2007) Salt tolerance, salt accumulation, and ionic homeostasis in an epidermal bladder-cell-less mutant of the common ice plant Mesembryanthemum crystallinum. J Exp Bot 58 1957–1967 [DOI] [PubMed] [Google Scholar]
- Bohnert H, Ayoubi P, Borchert C, Bressan R, Burnap R, Cushman J, Cushman M, Deyholos M, Fischer R, Galbraith D, et al (2001) A genomics approach towards salt stress tolerance. Plant Physiol Biochem 39 295–311 [Google Scholar]
- Bohnert H, Cushman J (2000) The ice plant cometh: lessons in abiotic stress tolerance. J Plant Growth Regul 19 334–346 [Google Scholar]
- Borland A, Dodd A (2002) Carbohydrate partitioning in Crassulacean acid metabolism plants: reconciling potential conflicts of interest. J Exp Bot 29 707–716 [DOI] [PubMed] [Google Scholar]
- Borland A, Hartwell J, Jenkins G, Wilkins M, Nimmo H (1999) Metabolite control overrides circadian regulation of phosphoenolpyruvate carboxylase kinase and CO2 fixation in Crassulacean acid metabolism. Plant Physiol 121 889–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland A, Taybi T (2004) Synchronization of metabolic processes in plants with Crassulacean acid metabolism. J Exp Bot 55 1255–1265 [DOI] [PubMed] [Google Scholar]
- Borland A, Tecsi L, Leegood R, Walker R (1998) Inducibility of Crassulacean acid metabolism (CAM) in Clusia species; physiological/biochemical characterisation and intercellular localisation of carboxylation processes in three species which show different degrees of CAM. Planta 205 342–351 [Google Scholar]
- Boxall S, Foster J, Bohnert H, Cushman J, Nimmo H, Hartwell J (2005) Conservation and divergence of circadian clock operation in a stress-inducible Crassulacean acid metabolism species reveals clock compensation against stress. Plant Physiol 137 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann E, Handwerger K, Essex C, Storz G (1996) Analysis of fast-neutron generated mutants at the Arabidopsis thaliana HT4 locus. Plant J 10 755–766 [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber S, Somerville C (1985) Alterations in growth, photosynthesis and respiration in a starchless mutant of Arabidopsis thaliana deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccini E, Mulligan B, Covey S, Milner J (1998) Characterization of gamma irradiation-induced deletion mutations at a selectable locus in Arabidopsis. Mutat Res 401 199–206 [DOI] [PubMed] [Google Scholar]
- Christopher J, Holtum J (1996) Patterns of carbohydrate partitioning in the leaves of Crassulacean acid metabolism species during deacidification. Plant Physiol 112 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crayn D, Smith J, Winter K (2004) Multiple origins of Crassulacean acid metabolism and the epiphytic habit in the neotropical family Bromeliaceae. Proc Natl Acad Sci USA 101 3703–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman J (2001) Crassulacean acid metabolism. A plastic photosynthetic adaptation to arid environments. Plant Physiol 127 1439–1448 [PMC free article] [PubMed] [Google Scholar]
- Cushman J, Borland A (2002) Induction of Crassulacean acid metabolism by water limitation. Plant Cell Environ 25 295–310 [DOI] [PubMed] [Google Scholar]
- Cushman J, Meyer G, Michalowski C, Schmitt J, Bohnert H (1989) Salt stress leads to differential expression of two isogenes of phosphoenolpyruvate carboxylase during Crassulacean acid metabolism induction in the common ice plant. Plant Cell 1 715–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman J, Tillett J, Wood J, Branco J, Schlauch K (2008) Large-scale mRNA expression profiling in the common ice plant, Mesembryanthemum crystallinum, performing C3 photosynthesis and Crassulacean acid metabolism (CAM). J Exp Bot (in press) [DOI] [PubMed]
- Dodd A, Griffiths H, Taybi T, Cushman J, Borland A (2003) Integrating diel starch metabolism with the circadian and environmental regulation of Crassulacean acid metabolism in Mesembryanthemum crystallinum. Planta 6 789–797 [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles K, Hamilton J, Rebus P, Smith F (1956) Colorimetric method for the determination of sugars and related substances. Anal Chem 28 350–356 [Google Scholar]
- Epimashko S, Meckel T, Fischer-Schliebs E, Lüttge U, Thiel G (2004) Two functionally different vacuoles for static and dynamic purposes in one plant mesophyll leaf cell. Plant J 37 294–300 [DOI] [PubMed] [Google Scholar]
- Griffiths H (1988) Crassulacean acid metabolism: a re-appraisal of physiological plasticity in form and function. Adv Bot Res 15 43–92 [Google Scholar]
- Hanson K (1990) Steady-state and oscillating photosynthesis by a starchless mutant of Nicotiana sylvestris. Plant Physiol 93 1212–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson K, McHale N (1988) A starchless mutant of Nicotiana sylvestris containing a modified plastid phosphoglucomutase. Plant Physiol 88 838–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D, Arnon D (1950) The water culture method of growing plants without soil. In California Agricultural Experiment Station Circular No. 347. College of Agriculture, University of California, Berkeley, CA, pp 1–32
- Keeley J (1998) CAM photosynthesis in submerged aquatic plants. Bot Rev 64 121–175 [Google Scholar]
- Kore-eda S, Cushman M, Akselrod I, Bufford D, Fredrickson M, Clark E, Cushman J (2004) Transcript profiling of salinity stress responses by large-scale expressed sequence tag analysis in Mesembryanthemum crystallinum. Gene 341 83–92 [DOI] [PubMed] [Google Scholar]
- Kore-eda S, Noake C, Ohishi M, Ohnishi J, Cushman J (2005) Transcriptional profiles of organellar metabolite transporters during induction of Crassulacean acid metabolism in Mesembryanthemum crystallinum. Funct Plant Biol 32 451–466 [DOI] [PubMed] [Google Scholar]
- Lee S, Hwang S, Han M, Eom J, Kang H, Han Y, Choi S, Cho M, Bhoo S, An G, et al (2007) Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol Biol 65 531–546 [DOI] [PubMed] [Google Scholar]
- Lytovchenko A, Bieberich K, Willmitzer L, Fernie A (2002) Carbon assimilation and metabolism in potato leaves deficient in plastidial phosphoglucomutase. Planta 215 802–811 [DOI] [PubMed] [Google Scholar]
- Nimmo H (2000) The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci 5 75–80 [DOI] [PubMed] [Google Scholar]
- Paul M, Loos K, Stitt M, Ziegler P (1993) Starch-degrading enzymes during the induction of CAM in Mesembryanthemum crystallinum. Plant Cell Environ 16 531–538 [Google Scholar]
- Schulze W, Schulze E-D, Stadler J, Heilmeier H, Stitt M, Mooney H (1994) Growth and reproduction of Arabidopsis thaliana in relation to storage of starch and nitrate in the wild-type and in starch-deficient and nitrate-uptake-deficient mutants. Plant Cell Environ 17 795–809 [Google Scholar]
- Sharkey T, Stitt M, Heineke D, Gerhardt R, Raschke K, Heldt H (1986) Limitation of photosynthesis by carbon metabolism II. Oxygen-insensitive photosynthesis results from limitation of photosynthesis by triose-phosphate limitation. Plant Physiol 81 1123–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera K, Santiago L, Winter K (2005) Distribution of Crassulacean acid metabolism in orchids of Panama: evidence of selection of weak and strong modes. Funct Plant Biol 32 397–407 [DOI] [PubMed] [Google Scholar]
- Tauberger E, Fernie A, Emmermann M, Renz A, Kossmann J, Willmitzer L, Trethewey R (2000) Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose-6-phosphate. Plant J 23 43–53 [DOI] [PubMed] [Google Scholar]
- Taybi T, Cushman J (1999) Signaling events leading to Crassulacean acid metabolism (CAM) induction in the common ice plant, Mesembryanthemum crystallinum. Plant Physiol 121 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taybi T, Patil S, Chollet R, Cushman J (2000) A minimal Ser/Thr protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in CAM-induced leaves of Mesembryanthemum crystallinum. Plant Physiol 123 1471–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lue W, Yu T, Long J, Wang C, Eimert K, Chen J (1998) Characterization of ADG1 and Arabidopsis locus coding for ADPG pyrophosphorylase small subunit, demonstrates that the presence of the small subunit is required for large subunit stability. Plant J 13 63–70 [DOI] [PubMed] [Google Scholar]
- Winter K (1982) Properties of phosphoenolpyruvate carboxylase in rapidly prepared, desalted leaf extracts of the Crassulacean acid metabolism plant Mesembryanthemum crystallinum L. Planta 154 298–308 [DOI] [PubMed] [Google Scholar]
- Winter K, Foster J, Edwards G, Holtum J (1982) Intracellular localization of enzymes of carbon metabolism in Mesembryanthemum crystallinum exhibiting C3 photosynthetic characteristics or performing Crassulacean acid metabolism. Plant Physiol 69 300–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K, Holtum J (2005) The effects of salinity, Crassulacean acid metabolism and plant age on the carbon isotope composition of Mesembryanthemum crystallinum L., a halophytic C3-CAM species. Planta 222 201–209 [DOI] [PubMed] [Google Scholar]
- Winter K, Holtum J (2007) Environment or development? Lifetime net CO2 exchange and control of the expression of Crassulacean acid metabolism in Mesembryanthemum crystallinum. Plant Physiol 143 98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K, Ziegler H (1992) Induction of Crassulacean acid metabolism in Mesembryanthemum crystallinum increases reproductive success under conditions of drought and salinity stress. Oecologia 92 475–479 [DOI] [PubMed] [Google Scholar]
- Yu T-S, Lue W-L, Wang S-M, Chen J (2000) Mutation of Arabidopsis plastid phosphoglucose isomerase affects leaf starch synthesis and floral initiation. Plant Physiol 123 319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotz G (2004) How prevalent is Crassulacean acid metabolism among vascular epiphytes? Oecologia 138 184–192 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.