In the early 1800s the population of the world was estimated to be around 1 billion. It grew to 2 billion in the 1920s and to 6 billion at the close of the last millennium. We are currently adding approximately 80 million more per year and, at this rate, the global population will increase from the current 6 billion to >10 billion people by the mid-2020s (Pimentel and Pimentel, 2000). Simultaneous decline of arable agricultural land will challenge our ability to meet increasing demands for food, feed, fuel, and fiber. These are the grand challenges of the 21st century. Remarkably, almost 300 years ago, the British poet, Jonathan Swift (1667–1745) noted with extraordinary perception that “Whoever could make two ears of corn (Zea mays) or two blades of grass to grow upon a spot of ground where only one grew before, would deserve better of mankind, and do more essential service to his country, than the whole race of politicians put together.” Fast forward to the 21st century, Mr. Swift would be very pleased. Thanks to years of gradual selection through traditional plant breeding techniques, farmers have been able to achieve crop productivity that is orders of magnitude greater than the esteemed poet could have ever anticipated. Yield increases have come from continual improvements in agricultural practices and selections for improved abiotic and biotic stress tolerance and better pest management practices.
However, faced with a burgeoning world population, competition for space and climatic changes, classical plant breeding alone is insufficient and not rapid enough for improving the characteristics of crops. Whereas traditional plant breeding is limited to accessing genes from closely related species, genome sequencing efforts of the last decade have yielded a plurality of genes of many different functions from many different species. Together with modern techniques in recombinant DNA methodology and plant transformation protocols, we are now postured to introduce any gene with desired function into a crop of interest. In the ideal case, a gene encoding a protein for a given trait from a given species will behave identically in the transgenic crop and faithfully confer the desired trait or phenotype. Often, owing to compromised expression, folding, and stability, the protein will have to be engineered or redesigned to achieve the end goal. In my opinion, we can attribute our current ability to alter function or impart new function into proteins to three significant observations: (1) the extraordinarily rapid growth in the knowledge base of the disparate disciplines of protein biochemistry, plant science, proteomics, genetics, genomics, molecular biology, mathematics, bioinformatics, and computer science and their synergistic value; (2) technological advances in macromolecular structure analysis methods such as NMR, x-ray crystallography, and mass spectrometry; and (3) the active participation of numerous companies in the development of reagents and tools for advancing the goals of basic and applied research. Thus, in the context of a world of limited and rapidly diminishing natural resources, protein engineering of crop species offers promising solutions to meeting the four Fs, i.e. food, feed, fiber, and fuel of societies in both developed and developing countries. In this narrative, I provide a holistic view of the state-of-the-art in protein engineering and potential applications of these strategies to crop improvement for a variety of societal benefits.
STRUCTURE- AND SEQUENCE-BASED ENGINEERING
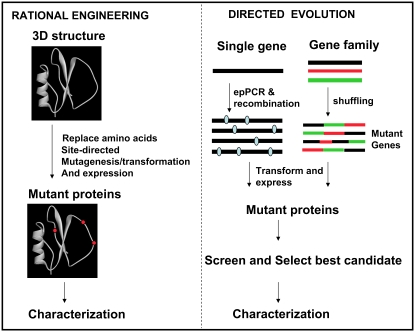
Properties of proteins are determined by their three-dimensional (3D) structure, with the precise configuration of specific amino acid residues contributing to the functional site(s) within the protein. In structure-based protein engineering, appropriate sites are selected for mutation based on an examination of the 3D structure of the protein and the mutants characterized for desired activity (Fig. 1). Despite the recent movement toward directed evolution methods for redesign of proteins (see later section) it would be amiss to discount the power of rational concepts utilizing 3D structures and/or homologous sequences. Indeed, in concert with random mutagenesis and directed evolution methods, structure-based protein engineering is a powerful approach. Over the years, a number of examples of rational engineering for the elucidation of enzyme mechanisms, changing substrate specificity, cofactor specificity, etc., have been described (for review, see Cedrone et al., 2000). One of the best examples of structure-based protein engineering of an enzyme in the plant sciences is the redesign of the substrate specificity of acyl-acyl-carrier protein (ACP) desaturase, Δ6-palmitoyl (16:0)-ACP desaturase, based on the x-ray structure of its homolog, Δ9-stearoyl (18:0)-ACP desaturase (Cahoon et al., 1997). In another elegant example, Van den Burg et al. (1998) took a moderately stable thermolysin-like protease from Bacillus stearothermophilus and made it hyperstable to elevated temperatures by judicious mutations of residues chosen on the basis of the 3D structure of thermolysin.
Figure 1.
A comparison of the steps involved in rational protein engineering (left) and directed protein evolution (right).
Similarly, knowledge-based protein engineering of the family of δ-endotoxins from Bacillus thuringiensis has resulted in the development of novel toxins with enhanced insecticidal activity and specificity (Saraswathy and Kumar, 2004; Mandal et al., 2007). It is pertinent to mention here that the widespread use of B. thuringiensis (Bt) crops has raised alarms regarding the possible development of resistance to the insecticidal protein by various pests (Gould, 1998) and at least two crop pests have been reported to have acquired resistance to Bt sprays outside of the laboratory environment (Soberon et al., 2007). In addition to the “high dose/refuge strategy” recommended by the U.S. Environmental Protection Agency (www.epa.gov/pesticides/biopesticides/white_bt.pdf; Gould, 1998), an emerging strategy is the use of protein engineering approaches to counter the adaptive behavior of insects based on a molecular understanding of the toxin and its interaction with proteins in the insect gut. One hypothesis is that upon ingestion the protease-activated toxin binds to the primary receptor cadherin, followed by protease clipping of the N terminus of the toxin that includes helix α-1 in domain 1. These steps facilitate oligomerization and subsequent binding to secondary receptors that result in the formation of pores in cells and eventual cell death (Bravo et al., 2004). On the basis of these observations, it was hypothesized that engineered Bt toxins (Cry1Ab and Cry1Ac), in which the helix α-1 was deleted, could form pore-forming oligomers without the involvement of cadherin. The engineered proteins were indeed found to be toxic to Manduca sexta in which the expression of cadherin was silenced by RNA interference (Soberon et al., 2007).
Roesler and Rao (2000) chose barley (Hordeum vulgare) chymotrypsin inhibitor-2 to exemplify the rational mutagenesis approach for nutritional enhancement. On the basis of its 3D structure, other homologous sequences, and a large body of literature on its biochemical and biophysical properties, they engineered a thermodynamically stable protein containing significantly elevated levels of the essential amino acids Lys, Trp, Thr, Ile, and Met. The value and success of sequence-/structure-guided protein engineering for proteins with enhanced nutritional benefits and improved insecticidal activity has been particularly well documented in recent reviews (Saraswathy and Kumar, 2004; Beauregard and Hefford, 2006). I will not dwell further on this subject except to state that despite the investment of millions of dollars by the major seed/plant biotechnology companies, the consumer is yet to see a transgenic cereal or legume crop expressing higher levels of essential amino acids such as Lys, Trp, Met, and Thr. Recently, however, Renessen (a joint venture between Cargill Inc. and Monsanto Company) obtained regulatory approval to produce corn engineered to produce high levels of free Lys. Termed Mavera high-Lys corn, the transgenic crop was to be grown on limited acreages in 2007 to produce grain with enhanced Lys levels for the animal feed industry.
The structure-based engineering of strictosidine synthase is an excellent example of redesign of substrate specificity with great potential for changing metabolic pathways and generating novel molecules for health and nutrition. The enzyme catalyzes the condensation of tryptamine and secologanin leading to the synthesis of numerous monoterpenoid indole alkaloids in higher plants. Using the crystallographic structure of strictosidine synthase in complex with strictosidine, Loris et al. (2007) produced and characterized mutants with the capacity to generate novel alkaloid libraries for pharmacological screening. This example augurs well for knowledge-based protein engineering of strategic target molecules to alter pathways and make new products.
A particularly exciting rational method to increase protein stability is the consensus engineering approach (Steipe, 2004). In this method, an improved protein is derived on the basis of a consensus sequence that incorporates favorable amino acid substitutions from a family of homologous sequences, or an existing sequence is modified to more closely match the consensus sequence. A remarkably successful example of this approach is the engineering of thermal stability in phytases, enzymes used quite extensively in animal feed technology. A protein with a consensus sequence derived from 13 homologous sequences displayed an increase in protein stability that was 22°C greater than the best parent (Lehmann et al., 2000, 2002). More recently, Dai et al. (2007) have applied consensus engineering to design a novel fluorescent protein using a family of 31 other fluorescent proteins.
It is expected that a rapidly growing database of protein sequences in conjunction with algorithms to identify sequence homologies, powerful new structure-prediction programs (Zhang and Skolnick, 2004), and worldwide high-throughput structural genomics initiatives (Bhattacharya et al., 2007) will facilitate more extensive applications of such knowledge-based targeted mutations for modified protein function. However, it is important to bear in mind that an inherent limitation of structure-based rational mutagenesis is its inability to factor in the contributions of side chains that may be distal to the functional site of the molecule. Nevertheless, the structure of a protein is invaluable to a mechanistic understanding of protein function and can provide the template for further engineering by rational or combinatorial genetic approaches. Thus, the recent characterization of the x-ray structure of the glyphosate-insensitive form of the enzyme 5-enolpyruvylshikimate 3-P (ESP) synthase and its interaction with glyphosate (Funke et al., 2006), provides an explanation for the molecular basis of herbicide resistance of Roundup Ready crops. Whereas the ESP synthase was an introduced heterologous protein, acetohydroxyacid synthase (also known as acetolactate synthase) was the first example of a rationally engineered enzyme designed to confer resistance to the sulfonylurea and imidazolinone class of herbicides (Ott et al., 1996). Currently, transgenic soybean (Glycine max) resistant to these herbicides is commercially available. Undoubtedly, the recently derived x-ray structure of acetohydroxyacid synthase (Pang et al., 2003) will facilitate further protein engineering and subsequent identification of other novel herbicides.
Phenotypic diversity manifests itself through transcriptional and posttranscriptional regulation of a number of genes. The ability to precisely control gene expression and insert foreign genes in specific sites within the genome has been major quests in molecular biology. In recent years, rapid strides have been made in the design of: (1) zinc finger (ZF) containing artificial transcription factors (TFs) capable of binding to specific DNA sequences (Pabo et al., 2001) and (2) tailored ZF nucleases that can cleave double-stranded DNA at predetermined sites to allow gene insertion by homologous recombination (Durai et al., 2005). The DNA binding modules of the artificial TFs are based on the classic Cys2His2 ZF domains found in many eukaryotic TFs. Importantly, the natural repertoire of ZF domains has been extended by mutant libraries and novel ZF domains binding to specific DNA sequences have been selected by phage display (Dreier et al., 2001; Blancafort et al., 2004). The regulation of plant gene expression by TFs to effect complex phenotypic properties has been successfully demonstrated in plants (for review, see Segal et al., 2003). It is particularly exciting to speculate on the opportunities for controlling metabolic pathways to make novel pharmacological products by turning on or turning off specific genes with designer TFs (Gandet and Memelink, 2002).
COMPUTATIONAL DESIGN
It is evident, therefore, that a variety of improved protein functions can be achieved through experimental protein engineering. However, a major limitation, even for directed evolution, is the highly restricted coverage of sequence space (103–106 sequences) for targeted mutagenesis (Voigt et al., 2001). Following the pioneering work of DeGrado and colleagues in the de novo design of proteins with predefined structure, i.e. helical proteins adopting the four-helix bundle (DeGrado, 1988; DeGrado et al., 1989; Hecht et al., 1990), more recent advances in computational protein design have allowed for a greater exploration of the protein sequence space and ushered in the era of designer synthetic proteins with novel functionalities. Thus, Mayo and colleagues at California Institute of Technology have developed sophisticated computational tools for the design of enzyme-like proteins from protein scaffolds devoid of catalytic activity (Bolon and Mayo, 2001). Although such enzymes have poor kinetic properties, they nevertheless provide the template for adoption of iterative combinatorial mutagenesis strategies and selection schemes for optimizing the desired function. Hellinga and colleagues at the Duke University Medical Center have also used computational tools to specifically focus on redesigning the signaling properties of receptor molecules via changes in ligand recognition sites (Benson et al., 2002). Typically, approximately 12 to 18 amino acid residues on the receptor surface are in direct contact with the wild-type ligand and there is a major challenge associated with experimentally producing and testing libraries for novel ligand-binding functions resulting from mutations of these residues. To overcome this combinatorial challenge, the Hellinga group developed an algorithm that is essentially an in silico evolution and screening program (Looger et al., 2003). In an exciting application, Hellinga's group redesigned the binding specificity of Escherichia coli periplasmic binding protein from native binding to the sugar Ara to high-affinity binding of molecules such as trinitrotoluene, l-lactate, and the neurotransmitter, serotonin. Furthermore, they provided practical evidence for the biosensor properties of the engineered receptors. This pioneering approach to the computational redesign of biomolecular recognition sites has profound implications for the development of the next generation of crop plants that can be specifically regulated by engineered receptor-ligand interactions.
A broader concept of protein engineering is found in the exciting work of Lim and coworkers at the University of California, San Francisco, who are rewiring biochemical circuits in eukaryotic cells to evolve new responses in cell behavior (Bhattacharya et al., 2006). Their elegant approach is built on the extensive biochemical and structural studies of signal transduction mechanisms in eukaryotes that have demonstrated the highly modular nature of participating proteins. For example, in receptors, the input domain is the ligand-binding domain and the output domain typically has kinase activity. Phosphorylation of the kinase domain results in specific interactions with downstream proteins that constitute the wiring diagram for the input-output pair. Synthetic signal proteins consisting of hybrid input-output pairs can be designed to control the specificity of the output signal leading to defined cellular responses. An exquisite demonstration of the use of designer proteins to regulate input-output response affecting cell shape and movement is described in two recent articles by the Wendel group (Dueber et al., 2007; Yeh et al., 2007). It is intriguing to speculate on the application of similar concepts to control plant behavior. The Arabidopsis (Arabidopsis thaliana) genome encodes >600 receptor-like kinases, the vast majority of which are uncharacterized (Shiu and Bleeker, 2001). Ligand identification and direct receptor-ligand binding have been demonstrated only for a select few, with the most well-characterized receptor-like kinase system being the brassinosteroid signaling pathway (Belkhadir and Chory, 2006). A more in-depth understanding of the molecular identity of individual signaling components and the mechanism of their interactions will enable the development of novel crops expressing engineered proteins designed to sense and respond to specific chemical and environmental signals. In addition to turning on engineered pathways in crop plants for disease resistance and stress tolerance, one can envisage the application of designer plants with sensor molecules to detect a variety of signals such as explosives (in war-torn areas) or toxic elements (in contaminated soil).
MOLECULAR EVOLUTION STRATEGIES
The plasticity of proteins and their ability to acquire new functions under selection pressure is well documented by numerous examples in the literature. Perhaps the earliest example of in vivo evolution is that of the E. coli protein, EbgA, with little or no β-galactosidase activity to one with weak but detectable activity (Campbell et al., 1973). This concept of evolving proteins under controlled conditions in the laboratory (spearheaded by the Frances Arnold group at California Institute of Technology and by W.P. Stemmer and colleagues at the company Maxygen in the 1990s), has revolutionized the field of protein engineering both from the standpoint of providing tools for fundamental research in protein structure and function as well as enabling the development of products for the biotechnology industry. Referred to as directed evolution, molecular breeding, or DNA shuffling, the method does not a priori require any structural or mechanistic understanding of the target molecule and has become the most widely used technique for protein evolution. Fundamentally, the process relies on two steps: (1) creation of a library of mutants and (2) a screening protocol for the selection of the protein with the best fit for the desired property from hundreds of thousands of variants (Arnold and Georgiou, 2003a, 2003b). In the simplest approach, libraries of mutants are made by fragmentation and reassembly of proteins generated from a single gene that encodes a protein possessing at least some of the desired biochemical properties (Stemmer, 1994a, 1994b) or using error-prone PCR (Wang et al., 2000) to introduce random mutations (Fig. 1). The randomized genes are then expressed recombinantly and the proteins then screened for improved or modified functions. Typically, the iterative cycle of in vitro recombination and screening is performed five to seven times before the end product is identified. Because beneficial mutations are rare, multiple cycles of mutagenesis and screening are necessary to permit the accumulation of favorable mutants and the weeding out of deleterious mutations.
In the final analysis, the efficiency of in vitro evolution is directly related to the quality of the library construction (Lutz and Patrick, 2004). The greater the expected change in the desired phenotype or function (i.e. novel substrate specificity) the greater the evolutionary sequence space to be traversed to locate the sequence correlating with the best activity (Axe, 2004). Single gene shuffling is inadequate to generate the needed sequence diversity because the members share >90% identity. On the other hand, in multigene shuffling or family shuffling (Fig. 1), recombination of protein segments occurs among a homologous family of proteins that are related in sequence but, more importantly, have a common fold. Because each member of the family has independently evolved to adopt a functional fold, libraries generated through family shuffling are also expected to harbor a significantly high percentage of functionally diverse mutants. A classic application of multigene shuffling is found in the evolution of moxalactamase activity from a family of genes encoding cephalosporinases (Crameri et al., 1998). Currently, a number of different methods have been developed for making libraries in which sequence identity requirements are much less stringent or are not necessary. These include oligonucleotide-directed randomization, whole gene randomization, homology-dependent recombination, and homology-independent combination methods. I refer the reader to many excellent reviews and references therein for more comprehensive coverage (Farinas et al., 2001; Kurtzman et al., 2001; Lutz and Patrick, 2004; Neylon, 2004; Yuan et al., 2005). The success of DNA shuffling notwithstanding, perhaps the greatest disadvantage of the method is the high rate of parental background. Two methods that overcome this problem to a great extent have been described. One is the Recombination Dependent Exponential Amplification PCR, known as RDA-PCR, (Ikeuchi et al., 2003) and the other is Shuffling Using Unpaired Primers, known as SUUPER (Milano and Tang, 2004). In an excellent recent study, Chapparo-Riggers et al. (2007) compare the efficiency of these recombination protocols with that of DNA shuffling and the significant improvements they bring to the field of directed evolution. Regardless of the method, however, there are two other parameters that are critical to the success of directed evolution: (1) screening methods such as phage display (for review, see Arnold and Georgiou, 2003a) and other emerging techniques using synthetic cells (Tawflik and Griffiths, 1998; Doi and Yanagawa, 1999; Bernath et al., 2004) to whittle down the population of variants from >105 to a manageable number and (2) the availability of high-throughput sensitive assays to test the function of the protein and select the best performers (i.e. ligand binding and enzymatic activity).
APPLICATIONS OF DIRECTED EVOLUTION TO CROP IMPROVEMENT
Crops Resistant to Herbicides
The increase in crop productivity in the last couple of decades can be attributed in large part to the development of transgenic crops expressing proteins that render them resistant to herbicides such as glyphosate, sulfonylurea, and imidazolinones. In existing glyphosate-resistant crops, tolerance is derived from the expression of an ESP synthase gene from Agrobacterium that is insensitive to glyphosate. In recent times other successful attempts have been made to identify novel glyphosate-resistant mutants of ESP synthase by protein engineering. He et al. (2001) demonstrated that even a single round of directed evolution yielded variants with vastly superior kinetic properties compared to the parent ESP synthase enzymes isolated from E. coli and Salmonella enterica. More recently, Zhou et al. (2006) isolated a mutant ESP synthase by error-prone PCR mutagenesis of the rice (Oryza sativa) gene. The variant contained a single P106L mutation in the protein sequence, had vastly superior kinetic properties, and was shown to confer glyphosate tolerance in transgenic tobacco (Nicotiana tabacum). As an alternative to evolving an enzyme for tolerance to glyphosate, Castle et al. (2004) described a strategy using directed evolution to design a glyphosate N-acetyltransferase (GAT) that detoxifies the herbicide by acetylating glyphosate to N-acetylglyphosate, a derivative that is a poor inhibitor of ESP synthase. Three genes encoding related GAT enzymes with very poor acetylation activity against glyphosate were isolated from the soil microbe Bacillus licheniformis. They were then subjected to 11 rounds of DNA shuffling to obtain an active enzyme that was 10,000-fold more active than the parental enzymes and was approximately 77% identical in amino acid sequence to the original proteins. The inability to obtain active enzyme by replacing amino acids identified from the shuffling scheme in the wild-type enzyme or the less active variants was an attestation of the power of combinatorial mutagenesis. The recently solved x-ray structure of GAT in complex with acetyl coenzyme A and the competitive inhibitor 3-phosphoglycerate offers insights into the molecular basis of glyphosate resistance (Siehl et al., 2007) and possible further optimization of enzymatic activity by directed evolution. At the commercial level, soybean, corn, and other crops containing the shuffled GAT enzyme are scheduled to be released over the next several years (see the DuPont Web site at http://www2.dupont.com).
Fluorodifen is another example of a herbicide belonging to the diphenylether class that is rapidly detoxified by glutathione-S-transferases in legumes but less efficiently in maize. Dixon et al. (2003) applied a forced evolution strategy to two maize glutathione-S-transferases and isolated seven mutants with enhanced fluorodifen detoxifying activity, the best of which contained a single point mutation and had 19-fold higher activity than the parent enzymes. Further targeted mutagenesis of the point mutant yielded an enzyme that had 29-fold higher activity.
Enhancing Photosynthetic Efficiency
Crop yields are subject to vagaries of the environment and can be dramatically reduced by extreme heat and drought. Numerous studies have indicated that this may, in part, be due to impaired photosynthetic efficiency stemming from a decreased population of catalytically competent Rubisco at high temperatures (Salvucci and Crafts-Brandner, 2004a, 2004b). It is also known that the active state of Rubisco is regulated by Rubisco activase (RCA), an enzyme that is inhibited at temperatures >45°C and recent experiments by Salvucci et al. (2006) in transgenic Arabidopsis plants suggested a strong correlation between RCA expression levels and susceptibility to heat. To test the hypothesis that increasing the thermal stability of RCA can lead to increased photosynthetic activity at elevated temperatures, Kurek et al. (2007) used two rounds of directed evolution of the RCA gene to produce several thermostable variants that were first extensively biochemically characterized in vitro and then expressed in Arabidopsis RCA deletion lines. This study provides unequivocal evidence for higher photosynthetic activity in transgenic Arabidopsis plants expressing thermostable RCA mutants and suggests that protein engineering of RCA, and possibly other proteins in the photosynthetic machinery, may be more appropriate than targeting Rubisco (Sinclair et al., 2004).
Evolution of Bt Toxins
The Cry family of insecticidal proteins from B. thuringiensis has been widely used for host plant resistance to insect pests. By the same token, fears that over time insects will adapt their physiological response to bypass the biochemical route of entomocidal activity, and themselves become resistant, has been borne out by laboratory and field experiments. The diversity of sequences and available 3D structures of toxins (de Maagd et al., 2003), as well as the identification of receptor molecules such as cadherins, glycolipids, and aminopeptidases in the insect gut (Jenkins and Dean, 2001) will only accelerate the use of both rational and directed evolution methods to simultaneously seek novel solutions for host plant resistance and intelligently stay ahead of the adaptive evolution game played by the insect species.
Novel Cellulases for the Biofuel Industry
The development of cellulosic ethanol as part of the biofuel initiative is a national priority. A broad and fundamental understanding of cellulases, the structures of cellulosomes, and engineering-improved cellulases via rational and molecular evolution methods present exciting opportunities for altering plant cell wall architecture to create transgenic crops more amenable to rapid enzymatic breakdown to ethanol. I will stress that research in this area is nascent and we are some years away from engineering crop plants for the biofuel industry. Nevertheless, examples of a number of different cellulases with improved thermal stability and modified enzymatic activities for use in bioreactors (for review, see Zhang et al., 2006), provide a glimpse of the potential opportunities in plant biotechnology.
Directed Evolution for Plant Metabolic Engineering
In an earlier section, the engineering of strictosidine synthase was cited as an example of the potential of plant metabolic engineering for changing the natural repertoire of pharmacological molecules. The diversity of cytochrome P450 sequences in plants and their involvement in complex metabolic pathways represents a treasure house of genes that could be further engineered for advancing the benefits of plant biotechnology for input and output traits, producing pharmaceuticals, and in phytoremediation (Morant et al., 2003; Jackson et al., 2007).
CONCLUSION
The synergistic power of rational design, computation, and directed evolution on the one hand, and parallel advances in plant breeding/plant sciences and the omics technologies on the other, offer unprecedented opportunities for genetic engineering of novel traits into the next generation of crop plants to accrue benefits that go far beyond the four Fs. However, the full impact of agricultural biotechnology for consumers in both developing and developed countries will likely be felt only if the developed countries can put aside their differences over the risk and benefits of genetically modified organisms technology and agree on acceptable regulatory structures for rapidly bringing products to the marketplace (Herrera-Estrella, 2000).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: A. Gururaj Rao (gururao@iastate.edu).
References
- Arnold FH, Georgiou G, editors (2003. a) Directed Enzyme Evolution: Screening and Selection Methods. Methods in Molecular Biology, Vol 230. Humana Press, Totowa, NJ
- Arnold FH, Georgiou G, editors (2003. b) Directed Evolution Library Construction: Methods and Protocols. Methods in Molecular Biology, Vol 231. Humana Press, Totowa, NJ
- Axe DD (2004) Estimating the prevalence of protein sequences adopting functional enzyme folds. J Mol Biol 341 1295–1315 [DOI] [PubMed] [Google Scholar]
- Beauregard M, Hefford MA (2006) Enhancement of essential amino acid contents in crops by genetic engineering and protein design. Plant Biotechnol J 4 561–574 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Chory J (2006) Brassinosteroid signaling: a paradigm for steroid hormone signaling from the cell surface. Science 314 1410–1411 [DOI] [PubMed] [Google Scholar]
- Benson DE, Haddy AE, Hellinga HW (2002) Converting a maltose receptor into a nascent binuclear copper oxygenase by computational design. Biochemistry 41 3262–3269 [DOI] [PubMed] [Google Scholar]
- Bernath K, Hai M, Mastrobattista E, Griffiths AD, Magdassi S, Tawflik DS (2004) In vitro compartmentalization by double emulsions: sorting and gene enrichment by fluorescence activated cell sorting. Anal Biochem 325 151–157 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Tejero R, Montelione GT (2007) Evaluating protein structures determined by structural genomics consortia. Proteins 66 778–795 [DOI] [PubMed] [Google Scholar]
- Bhattacharya P, Remenyi A, Yeh J, Lim WA (2006) Domains, motifs and scaffolds: the role of modular interactions in the evolution and wiring of cell signal circuits. Annu Rev Biochem 75 655–680 [DOI] [PubMed] [Google Scholar]
- Blancafort P, Segal DJ, Barbas III CF (2004) Designing transcription factor architectures for drug discovery. Mol Pharmacol 66 1361–1371 [DOI] [PubMed] [Google Scholar]
- Bolon DN, Mayo SL (2001) Enzyme-like proteins by computational design. Proc Natl Acad Sci USA 98 14274–14279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Gómez I, Conde J, Munoz-Garay C, Sánchez J, Miranda R, Zhuang M, Gill SS, Soberón M (2004) Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim Biophys Acta 1667 38–46 [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Lindquist Y, Schneider G, Shanklin J (1997) Redesign of soluble fatty acid desaturases from plants for altered substrate specificity and double bond position. Proc Natl Acad Sci USA 94 4872–4877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JH, Lengyel JA, Langrdige J (1973) Evolution of a second gene for beta-galactosidase in E. coli. Proc Natl Acad Sci USA 70 1841–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle LA, Siehl DL, Gorton R, Patten PA, Chen YH, Bertain S, Choe HJ, Duck N, Wong J, Liu D, et al (2004) Discovery and directed evolution of a glyphosate tolerance gene. Science 304 1151–1154 [DOI] [PubMed] [Google Scholar]
- Cedrone F, Ménez A, Quéméneur E (2000) Tailoring new enzyme functions by rational redesign. Curr Opin Struct Biol 10 405–410 [DOI] [PubMed] [Google Scholar]
- Chapparo-Riggers JF, Loo BLW, Polizzi KM, Gibbs PR, Tang XS, Nelson MJ, Bommarius AS (2007) Revealing biases inherent in recombination protocols. BMC Biotechnol 7 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crameri A, Raillard SA, Burmudez E, Stemmer WP (1998) DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391 288–291 [DOI] [PubMed] [Google Scholar]
- Dai M, Fisher HE, Temirov J, Kiss C, Phipps ME, Pavlik P, Werner JH, Bradbury ARM (2007) The creation of a novel fluorescent protein by guided consensus engineering. Protein Eng Des Sel 20 69–79 [DOI] [PubMed] [Google Scholar]
- DeGrado WF (1988) Design of peptides and proteins. Adv Protein Chem 39 51–124 [DOI] [PubMed] [Google Scholar]
- DeGrado WF, Wasserman ZR, Lear JD (1989) Protein design, a minimalist approach. Science 243 622–628 [DOI] [PubMed] [Google Scholar]
- de Maagd RA, Bravo A, Berry C, Crickmore N, Schnepf HE (2003) Structure, diversity and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu Rev Genet 37 409–433 [DOI] [PubMed] [Google Scholar]
- Dixon DP, McEwent AG, Lapthorn AJ, Edwards R (2003) Forced evolution of a herbicide detoxifying glutathione transferase. J Biol Chem 278 23930–23935 [DOI] [PubMed] [Google Scholar]
- Doi N, Yanagawa H (1999) STABLE: protein-DNA fusion system for screening of combinatorial protein libraries in vitro. FEBS Lett 457 227–230 [DOI] [PubMed] [Google Scholar]
- Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF III (2001) Development of zinc finger domains for recognition of the 5*-ANN-3* family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem 276 29466–29478 [DOI] [PubMed] [Google Scholar]
- Dueber JE, Mirsky EA, Lim WA (2007) Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat Biotechnol 25 660–662 [DOI] [PubMed] [Google Scholar]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S (2005) Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33 5978–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas ET, Butler T, Arnold F (2001) Directed enzyme evolution. Curr Opin Biotechnol 12 545–551 [DOI] [PubMed] [Google Scholar]
- Funke T, Han H, Healy-Fried ML, Fischer M, Schönbrunn E (2006) Molecular basis for the herbicide resistance of Roundup Ready crops. Proc Natl Acad Sci USA 103 13010–13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandet P, Memelink J (2002) Transcription factors: tools to engineer the production of pharmacologically active plant metabolites. Trends Pharmacol Sci 23 563–569 [DOI] [PubMed] [Google Scholar]
- Gould F (1998) Sustainability of transgenic insecticidal cultivars: integrating pest genetics and ecology. Annu Rev Entomol 43 701–726 [DOI] [PubMed] [Google Scholar]
- He M, Yang ZY, Nie YF, Wang J, Xu P (2001) A new type of class I bacterial 5-enopyruvylshikimate-3-phosphate synthase mutants with enhanced tolerance to glyphosate. Biochim Biophys Acta 1568 1–6 [DOI] [PubMed] [Google Scholar]
- Hecht MH, Richardson JH, Richardson DC, Ogden RC (1990) De novo design, expression and characterization of Felix: a four-helix bundle of native-like sequence. Science 249 884–891 [DOI] [PubMed] [Google Scholar]
- Herrera-Estrella LR (2000) Genetically modified crops and developing countries. Plant Physiol 124 923–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi A, Kawarasaki Y, Shinbata T, Yamane T (2003) Chimeric gene library construction by a simple and highly versatile method using recombination-dependent exponential amplification. Biotechnol Prog 19 1460–1467 [DOI] [PubMed] [Google Scholar]
- Jackson RG, Rylott EL, Fournier D, Hawari J, Bruce NC (2007) Exploring the biochemical properties and remediation applications of the unusual explosive-degrading P450 system XplA/B. Proc Natl Acad Sci USA 104 16822–16826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins JL, Dean DH (2001) Binding specificity of Bt Cry1A for purified, native Bombyx mori aminopeptidase N and cadherin-like receptors. BMC Biochem 2 12–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek I, Chang TK, Bertain SM, Madrigal A, Liu L, Lassner MW, Zhu G (2007) Enhanced thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. Plant Cell 19 3230–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman AL, Govindarajan S, Vahle K, Jones JT, Heinrichs V, Patten PA (2001) Advances in directed protein evolution by recursive genetic recombination: applications to therapeutic proteins. Curr Opin Biotechnol 12 361–370 [DOI] [PubMed] [Google Scholar]
- Lehmann M, Kostrewa D, Wyss M, Brugger R, D'Arcy A, Pasamontes L, Adolphus van Loon PGM (2000) From DNA sequence to improved functionality: using protein sequence comparisons to rapidly design a thermostable consensus phytase. Protein Eng 13 49–57 [DOI] [PubMed] [Google Scholar]
- Lehmann M, Loch C, Middendorf A, Studer D, Lassen SF, Pasamontes L, Adolphus van Loon PGM, Wyss M (2002) The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Eng 15 403–411 [DOI] [PubMed] [Google Scholar]
- Looger LL, Dwyer MA, Smith JJ, Hellinga HW (2003) Computational design of receptor and sensor proteins with novel functions. Nature 423 185–190 [DOI] [PubMed] [Google Scholar]
- Loris EA, Panjrikar S, Ruppert M, Barleben L, Uner M, Schübel H, Stöckigt J (2007) Structure-based engineering of strictosidine synthase: auxiliary for alkaloid libraries. Chem Biol 14 979–985 [DOI] [PubMed] [Google Scholar]
- Lutz S, Patrick WM (2004) Novel methods for directed evolution of enzymes: quality not quantity. Curr Opin Biotechnol 15 291–297 [DOI] [PubMed] [Google Scholar]
- Mandal CC, Gayen S, Basu A, Ghosh KS, Dasgupta S, Maiti K, Sen SK (2007) Prediction-based protein engineering of domain I of Cry2A entomocidal toxin of Bacillus thuringiensis for the enhancement of toxicity against lepidopteran insects. Protein Eng Des Sel 12 599–606 [DOI] [PubMed] [Google Scholar]
- Milano J, Tang XS, inventors (2004) U.S. Patent Application No. 20040014085
- Morant M, Bak S, Møller BL, Werck-Reichhart D (2003) Plant cytochromes P450: tools for pharmacology, plant protection and phytoremediation. Curr Opin Biotechnol 14 151–162 [DOI] [PubMed] [Google Scholar]
- Neylon C (2004) Chemical and biochemical strategies for the randomization of protein encoding DNA sequences: library construction methods for directed evolution. Nucleic Acids Res 32 1448–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott KH, Kwagh JG, Stockton GW, Sidorov V, Kakefuda G (1996) Rational molecular design and genetic engineering of herbicide resistant crops by structure modeling and site-directed mutagenesis of acetohydroxyacid synthase. J Mol Biol 263 359–368 [DOI] [PubMed] [Google Scholar]
- Pabo OC, Peisach E, Grant RA (2001) Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem 70 313–340 [DOI] [PubMed] [Google Scholar]
- Pang SS, Guddat LW, Duggleby RG (2003) Molecular basis of sulfonylurea herbicide inhibition of acetohydroxyacid synthase. J Biol Chem 278 7639–7644 [DOI] [PubMed] [Google Scholar]
- Pimentel D, Pimentel M (2000) Feeding the world's population. Bioscience 50 387 [Google Scholar]
- Roesler KR, Rao AG (2000) A single disulfide bond restores thermodynamic and proteolytic stability to an extremely mutated protein. Protein Sci 9 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ (2004. a) Inhibition of photosynthesis by heat stress: the activation state of Rubisco as a limiting factor in photosynthesis. Plant Physiol 120 179–186 [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ (2004. b) Relationship between the heat tolerance of photosynthesis and the thermal stability of Rubisco activase in plants from contrasting thermal environments. Plant Physiol 134 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci ME, DeRidder BP, Portis AR Jr (2006) Effect of activase level and isoform on the thermotolerance of photosynthesis in Arabidopsis. J Exp Bot 57 5793–5799 [DOI] [PubMed] [Google Scholar]
- Saraswathy N, Kumar PA (2004) Protein engineering of δ-endotoxins of Bacillus thuringiensis. Electron J Biotechnol doi/10.2225/vol7-issue2-fulltext-3
- Segal DJ, Stege JT, Barbas III CF (2003) Zinc fingers and a green thumb: manipulating gene expression in plants. Curr Opin Plant Biol 6 163–168 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siehl DL, Castle LA, Gorton R, Keenan RJ (2007) The molecular basis of glyphosate resistance by an optimized microbial acetyltransferase. J Biol Chem 282 11446–11455 [DOI] [PubMed] [Google Scholar]
- Sinclair TR, Purcell LC, Sneller CH (2004) Crop transformation and the challenge to increase yield potential. Trends Plant Sci 9 70–75 [DOI] [PubMed] [Google Scholar]
- Soberon M, Pardo-Lopez L, Lopez I, Gomez I, Tabashnik BE, Bravo A (2007) Engineering modified Bt toxins to counter insect resistance. Science 318 1640–1642 [DOI] [PubMed] [Google Scholar]
- Steipe B (2004) Consensus-based engineering of protein stability: from antibodies to thermostable enzymes. Methods Enzymol 388 176–186 [DOI] [PubMed] [Google Scholar]
- Stemmer WP (1994. a) Rapid evolution of a protein in vitro by DNA shuffling. Nature 370 389–391 [DOI] [PubMed] [Google Scholar]
- Stemmer WP (1994. b) DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA 91 10747–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawflik DS, Griffiths AD (1998) Man-made cell-like compartments for molecular evolution. Nat Biotechnol 16 652–656 [DOI] [PubMed] [Google Scholar]
- Van den Burg B, Vriend G, Veltman OR, Venema G, Eijsink VG (1998) Engineering an enzyme to resist boiling. Proc Natl Acad Sci USA 95 2056–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt C, Mayo S, Wang ZG (2001) Computational method to reduce the search space for directed protein evolution. Proc Natl Acad Sci USA 98 3778–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Oh MK, Liao JC (2000) Directed evolution of metabolically engineered E. coli for carotenoid production. Biotechnol Prog 16 922–926 [DOI] [PubMed] [Google Scholar]
- Yeh BJ, Rutigliano RJ, Deb A, Bar-Sagi D, Lim WA (2007) Rewiring cellular morphology pathways with synthetic guanine nucleotide exchange factors. Nature 447 596–600 [DOI] [PubMed] [Google Scholar]
- Yuan L, Kurak I, English J, Keenan R (2005) Laboratory-directed protein evolution. Microbiol Mol Biol Rev 69 373–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PYH, Himmel ME, Mielenz JR (2006) Outlook for cellulose improvement: screening and selection strategies. Biotechnol Adv 24 452–481 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J (2004) Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci USA 101 7594–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Xu H, Wei X, Ye Z, Wei Y, Gong W, Wang Y, Zhu Z (2006) Identification of a glyphosate-resistant mutant of rice 5-enolpyruvylshikimate-3-phosphate synthase using a directed evolution strategy. Plant Physiol 140 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]