Abstract
The control of potassium (K+) acquisition is a critical requirement for plant growth. Although HAK1 (high affinity K+ 1) transporters provide a pathway for K+ acquisition, the effect exerted by the ionic environment on their contribution to K+ capture remains essentially unknown. Here, the influence of the ionic environment on the accumulation of transcripts coding for the barley (Hordeum vulgare) HvHAK1 transporter as well as on HvHAK1-mediated K+ capture has been examined. In situ mRNA hybridization studies show that HvHAK1 expression occurs in most root cells, being augmented at the outermost cell layers. Accumulation of HvHAK1 transcripts is enhanced by K+ deprivation and transiently by exposure to high salt concentrations. In addition, studies on the accumulation of transcripts coding for HvHAK1 and its close homolog HvHAK1b revealed the presence of two K+-responsive pathways, one repressed and the other insensitive to ammonium. Experiments with Arabidopsis (Arabidopsis thaliana) HvHAK1-expressing transgenic plants showed that K+ deprivation enhances the capture of K+ mediated by HvHAK1. A detailed study with HvHAK1-expressing Saccharomyces cerevisiae cells also revealed an increase of K+ uptake after K+ starvation. This increase did not occur in cells grown at high Na+ concentrations but took place for cells grown in the presence of NH4+. 3,3′-Dihexyloxacarbocyanine iodide accumulation measurements indicate that the increased capture of K+ in HvHAK1-expressing yeast cells cannot be explained only by changes in the membrane potential. It is shown that the yeast protein phosphatase PPZ1 as well as the halotolerance HAL4/HAL5 kinases negatively regulate the HvHAK1-mediated K+ transport.
Potassium (K+) is the most abundant essential cation in almost all living cells. Besides having several major functions in normal physiology, K+ plays an important role protecting plants during acclimation to saline-rich and ammonium-rich environments (Flowers and Läuchli, 1983; Cao et al., 1993). Acquisition of K+ from the soil solution is primarily dependent on the activity of transport proteins located in the plasma membrane of soil bacteria, fungi, and root epidermal cells of plants. Early on, it was observed that the kinetics and energetics of K+ transport in most fungi and plants share some common features that are not found in animal cells (Kochian and Lucas, 1988; Rodríguez-Navarro, 2000). This observation led to the concept that in most cases K+ uptake is mediated by similar transport proteins in roots and fungal cells. Providing support for this claim, homologues of the fungal HAK (high affinity K+) and TRK (transport of K+) transporters have been identified in plants (Véry and Sentenac, 2003). The possibility that regulatory elements involved in alkali cation homeostasis are functional in both plants and fungi has also been proposed (Gisbert et al., 2000; Quintero et al., 2002). Unlike their fungal counterparts, the plant homologues of TRK transporters, named HKT (high-affinity K+ transporter), appear to be involved mainly in Na+ transport rather than in K+ acquisition (Garcíadeblás et al., 2003; Rus et al., 2004; Horie et al., 2007). On the other hand, members of the HAK1 subgroup of HAK-KUP-KT proteins have been found to play a major role in K+ uptake in plants (Santa-María et al., 1997; Rubio et al., 2000; Gierth et al., 2005), being an additional route for K+ uptake in roots provided by a Shaker-like inward rectifying K+ channel, the Arabidopsis (Arabidopsis thaliana) K+ transporter AKT1 (Sentenac et al., 1992; Hirsch et al., 1998). Studies in yeasts indicate that K+ transport mediated by HAK1 transporters is sensitive to the presence of NH4+, while studies with akt1 mutant plants demonstrate that the AKT1 channel is involved in the capture of K+ for plants grown at high NH4+ concentrations (Hirsch et al., 1998; Santa-María et al., 2000). Other studies support the view that the NH4+-sensitive pathway involves the activity of more than a single K+ transporter (Spalding et al., 1999; Gierth et al., 2005; Vallejo et al., 2005).
It has been shown that the acquisition of K+ by plants is a tightly regulated process. Perception of K+ deprivation rapidly occurs after K+ removal from the growth medium, leading to an enhancement of the K+ uptake capacity from diluted K+ solutions (Glass, 1976; Kochian and Lucas, 1982), which probably requires the accumulation of reactive oxygen species in a discrete root zone (Shin and Schachtman, 2004). Additionally, the pharmacological properties and thermodynamic constrains of K+ uptake from diluted K+ solutions in plants depend on the abundance of K+, sodium, and ammonium in the medium encountered by roots during growth (Spalding et al., 1999; Santa-María et al., 2000; Kronzucker et al., 2003; Nieves-Cordones et al., 2007). When plants are grown in the presence of high NH4+ or Na+ concentrations, the enhancement of the K+ uptake capacity resulting from K+ deprivation is affected and a change in the contribution of the NH4+-sensitive and NH4+-insensitive components becomes evident. These results suggest a regulatory effect of the ionic environment on the contribution of AKT1 and HAK1 transporters. A clear influence of K+ starvation on the control of AKT1 activity was reported recently (Li et al., 2006; Xu et al., 2006). However, the influence of the ionic environment found by plants during growth on the contribution of a HAK1 transporter remains essentially unknown. Here, we introduce evidence for the presence of long-term regulatory mechanisms affecting the abundance of transcripts and the contribution to K+ capture of the canonical member of the HAK1 subgroup, the barley (Hordeum vulgare) HvHAK1 transporter.
RESULTS
HvHAK1 Is Preferentially Expressed in Cells of the Root Outer Layers
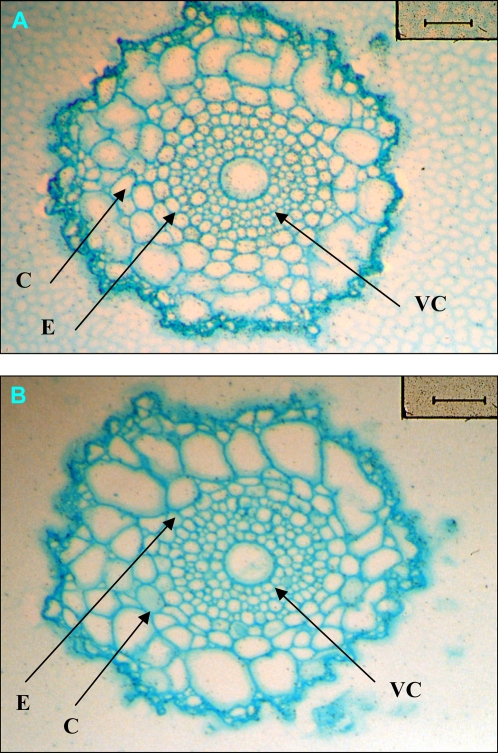
As root cells expressing K+ transporters carry out the primary uptake of K+ from the soil, the site of HvHAK1 expression in barley roots was determined. Because of the existence of several HAK1 genes in Triticeae genomes and the high similitude among them (Santa-María et al., 1997), we used a specific probe mainly containing the 3′ untranslated region to study HvHAK1 expression in transversal sections of roots. In situ hybridization studies in seminal barley roots showed that the accumulation of HvHAK1 transcripts occurs in all cell layers, with the expression augmented in the outermost layers (Fig. 1A, Supplemental Fig. S1). No signal was detected with the sense probe, showing the suitability of the technique used (Fig. 1B, Supplemental Fig. S1).
Figure 1.
In situ hybridization of HvHAK1 mRNA in barley roots. Cross sections of seminal barley roots located 10 to 20 mm from the tip are shown. The expression of HvHAK1 is indicated by dark granules. A, Antisense probe. B, Sense probe. C, cortex; E, endodermis; VC, vascular cylinder. Bars, 100 μm.
The Accumulation of Transcripts Coding for HAK1 Transporters Involves Cross Talk between NH4+-Sensitive and NH4+-Insensitive Mechanisms
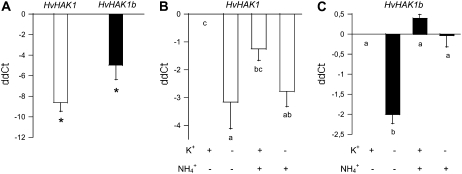
The specific accumulation of HvHAK1 mRNA was estimated by real-time PCR coupled with reverse transcription (RT). Long-term K+ deprivation (no K+ added to the complete aerated culture solution) led to increased accumulation of HvHAK1 transcripts (Fig. 2A), with a similar pattern observed for plants grown in a MES-Ca2+ solution in the absence of aeration (Supplemental Fig. S2). Experiments with plants grown at combined levels of NH4+ and K+ showed that the accumulation of HvHAK1 transcripts following K+ deprivation occurs to the same extent in the presence and absence of a high NH4+ concentration during plant culture (Fig. 2B).
Figure 2.
Expression of K+ transporter-coding genes is enhanced by K+ starvation through an ammonium-sensitive and an ammonium-insensitive pathway. A, Accumulation of HvHAK1 and HvHAK1b transcripts in 1-week-old plants grown with or without K+ since germination (n = 3). ddCt corresponds to the difference in transcript accumulation between the presence of K+ (control condition) and its absence (no K+ added). Asterisks denote significant effects of K+ deprivation. B and C, Effects of the presence or absence of K+ and NH4+ (100 μm and 5 mm, respectively) during a 3-d period on the accumulation of HvHAK1 and HvHAK1b transcripts (n = 6). Error bars represent se. Different letters indicate significantly different values (P < 0.05 for A and B, P < 0.001 for C).
The accumulation of transcripts coding for HvHAK1b, a very close relative of HvHAK1, was also enhanced by long-term K+ deprivation (Fig. 2A). However, the accumulation of HvHAK1b transcripts following K+ deprivation was significantly lower for plants grown at a high NH4+ concentration compared with those grown in the absence of NH4+ (Fig. 2C). These results indicate that following K+ deprivation, the accumulation of transcripts coding for HAK1 transporters involves two different routes, one of them sensitive and the other insensitive to NH4+.
HvHAK1 Transcript Accumulation Is Up-Regulated by NaCl Salinization
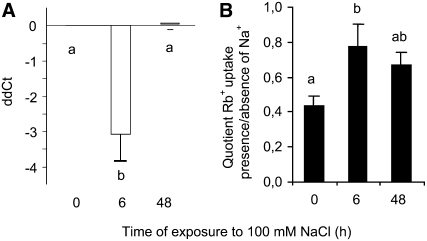
Given that exposure to high Na+ concentrations interferes with K+ nutrition, the effect of NaCl on the accumulation of HvHAK1 was analyzed. A 6-h exposure to 100 mm NaCl, in the presence of 1 mm K+, led to a significant increase in the amount of HvHAK1 transcripts compared with that measured for control plants, an effect that was reversed at 48 h after salinization (Fig. 3A). These results indicate a strong and transient NaCl-triggered HvHAK1 up-regulation at the transcript level, consistent with a possible role of HvHAK1 during the fast response of plants to salinity. With this possibility in mind, we studied the influence of Na+ on the uptake of Rb+, a good analog of K+ for HAK1 transporters, following salinization. The quotient between the uptake of Rb+ measured in the presence of 100 mm NaCl and that measured in the absence of this salt increased significantly at 6 h after salinization (Fig. 3B). These results argue for a parallel control of K+ uptake properties and HvHAK1 mRNA accumulation under saline conditions.
Figure 3.
Expression of HvHAK1 is transiently enhanced by salinization and parallels changes in the reduction of Na+-inhibitory effects on Rb+ uptake in barley roots. Plants were exposed to 100 mm NaCl in the presence of 1 mm KCl. A, Accumulation of HvHAK1 transcripts measured at 0, 6, and 48 h since the beginning of salt stress (n = 5). B, Rb+ uptake from a 100 μm Rb+ solution, after those periods, was measured in both the presence and the absence of 100 mm NaCl, and the quotient between the uptake measured in those conditions is shown (n = 6). Error bars represent se. Different letters indicate significantly different values (P < 0.005 for A, P < 0.05 for B).
Because of the possibility that the rapid up-regulation of HvHAK1 transcript accumulation could be the result of an osmotic shock, the effect of sorbitol was explored. A 6-h exposure to 200 mm sorbitol led to an accumulation of HvHAK1 transcripts not significantly different from that determined for control plants and lower than that determined for 100 mm NaCl-treated plants (Supplemental Fig. S3). While the contribution of an osmotic component cannot be ruled out since it could account for 44% of that observed in NaCl-stressed plants, these data indicate that nonosmotic components could be involved in the transient response. In turn, the 6-h effect of 100 mm NaCl on the accumulation of HvHAK1 transcripts was not accompanied by a change in the total concentration of K+ in roots. Instead, the concentration of Na+ in roots clearly increased over that period (Supplemental Fig. S3).
HvHAK1 Transgenic Plants Display an Enhanced Rb+ Transport When Subjected to K+ Deprivation
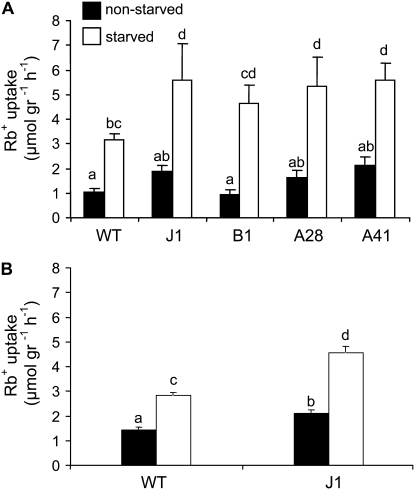
The results shown above indicate that K+ deprivation exerts a strong effect on the accumulation of transcripts coding for two HAK1 transporters (Fig. 2). It seems likely that these transporters, and others not examined here but homologous with those already studied in Arabidopsis (Hirsch et al., 1998; Ahn et al., 2004; Gierth et al., 2005), contribute to the capture of K+ by roots. Therefore, in order to study the effect of K+ deprivation on the specific contribution of HvHAK1 to Rb+ capture, we performed a stable transformation of Arabidopsis plants with a construct containing HvHAK1 under the control of the 35S promoter. All of the HvHAK1-expressing transgenic lines assayed displayed a similar pattern (Fig. 4A). A more detailed study performed with a selected transgenic line (J1) showed that the uptake of Rb+ was just slightly higher for HvHAK1-expressing plants than for HvHAK1-nonexpressing plants grown in the presence of 1 mm KCl. When plants were deprived of K+ for 48 h, the subsequent transport of Rb+ was markedly higher in HvHAK1-expressing plants than in HvHAK1-nonexpressing plants (Fig. 4B). These results indicate that K+ availability controls the contribution of HvHAK1 to K+ capture in transgenic Arabidopsis plants.
Figure 4.
Arabidopsis plants expressing HvHAK1 display enhanced Rb+ uptake following K+ withdrawal. A, Four homozygous HvHAK1-expressing Arabidopsis lines (named J1, B1, A28, and A41) display an enhanced uptake of Rb+ from a 100 μm Rb+ solution relative to wild-type (WT) plants when deprived of K+ for 48 h (on average, n = 7). B, Detailed analysis for the HvHAK1-expressing J1 line. Results are means of 13 experiments (each one consisting of seven independent replicates, on average). Error bars represent se. Different letters indicate significantly different values (P < 0.05 for A, P < 0.01 for B).
Rb+ Uptake in Yeast Cells Expressing HvHAK1 Is Modulated by K+ Supply
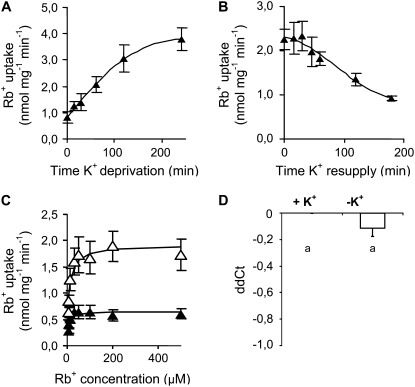
In order to gain further insight on how the contribution of HvHAK1 is modulated by the composition of the environment, we performed a detailed study of Rb+ transport in Saccharomyces cerevisiae mutants compromised for K+ uptake. Expression of HvHAK1 in a yeast mutant lacking the TRK1 and TRK2 K+ transporters shows that after K+ deprivation, the uptake of Rb+ mediated by HvHAK1 increased progressively until reaching a plateau (Fig. 5A). After attaining this plateau, the uptake of Rb+ was 3- to 4-fold higher in K+-deprived cells than in cells never subjected to K+ starvation. A reciprocal experiment showed that K+ resupply led to a progressive decline of Rb+ uptake (Fig. 5B), indicating that the modulation of HvHAK1-mediated Rb+ transport is reversible. In measuring Rb+ uptake under these experimental conditions, special care should be taken to avoid the masking effect caused by the release of K+ from cells to the medium during the Rb+-loading procedure, which may be particularly pronounced at the very beginning of K+ deprivation. We found that 15 and 210 min after K+ deprivation, the concentration of K+ in the loading solution was 8 and 4 μm, respectively, indicating the presence of only minor interference of K+ on Rb+ uptake measurements made from a 100 μm Rb+ solution after 15 min of K+ deprivation. Consequently, further comparisons between K+-nonstarved and K+-starved cells were done by comparing cells deprived of K+ for 15 and 210 min, respectively.
Figure 5.
Rb+ uptake is enhanced by K+ starvation in HvHAK1-expressing yeast cells. A, Rb+ uptake from a 100 μm Rb+ solution by HvHAK1-expressing yeast cells grown overnight at 30 mm KCl and deprived of K+ (no K+ added) for different time periods (n = 6). B, Reciprocal experiment (n = 5) with 210-min K+-deprived cells transferred to a 30 mm KCl medium. C, Rb+ uptake (n = 6) from a wide range of micromolar Rb+ concentrations for K+-starved cells (white triangles) and K+-nonstarved cells (black triangles). D, Effect of K+ deprivation on the accumulation of HvHAK1 transcripts in yeast cells (n = 7). Error bars represent se.
It has been shown that some HAK-KUP-KT transporters could mediate the biphasic transport of K+ (Fu and Luan, 1998; Kim et al., 1998). No evidence for the modulation of HvHAK1 transport involving a switch between monophasic and biphasic modes of transport was found (Supplemental Fig. S4). Consistently, a single and saturable component, operative in the micromolar range of Rb+ concentrations, was observed (Fig. 5C). Values of Vmax were 0.66 ± 0.12 and 1.92 ± 0.32 nmol mg−1 min−1 for K+-nonstarved and K+-starved cells, respectively, indicating a 3-fold enhancement of the K+ transport capacity. On the other hand, Rb+ Km increased significantly from 5.6 ± 1.0 to 9.6 ± 1.3 μm in HvHAK1-expressing cells during the course of K+ starvation. Given that the K+ concentration in the loading solution is somewhat higher for K+-nonstarved than for K+-starved cells (due to K+ loss), the actual difference in Rb+ Km may be even higher. An important question to address is whether this increased Km could be just a by-product of a response to K+ starvation with no acclimation value. Therefore, we explored whether changes in the Rb+ Km of HvHAK1-mediated transport are accompanied by a change in the sensitivity of Rb+ transport to Na+. The inhibitory effect of 100 mm NaCl on the transport of Rb+ was more pronounced for cells deprived of K+ for 210 min than for K+-nonstarved cells (Supplemental Fig. S4), a result compatible with a decreased K+/Na+ selectivity of the HvHAK1 transporter under conditions of K+ starvation. Unfortunately, attempts to detect differences in Na+ transport between K+-starved and K+-nonstarved HvHAK1-expressing cells in different yeast mutants failed because of the presence of yeast low-affinity transporters that dominate the transport of Na+ over that attributed to the plant transporter.
An important question to be considered is whether the changes in Vmax reported above are linked, at least in part, to changes in the accumulation of HvHAK1 transcripts during the course of K+ deprivation. RT-PCR studies showed that the amount of HvHAK1 transcripts in HvHAK1-expressing yeasts was not significantly different between K+-starved and K+-nonstarved cells (Fig. 5D).
The Contribution of HvHAK1 to K+ Capture in Yeast Cells Is Modulated by the Concentration of K+ and Na+, But Not by NH4+, in the Growth Medium
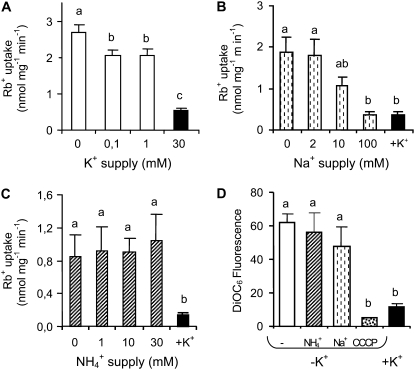
We also tried to gain insight into the S. cerevisiae mechanisms leading to the enhanced HvHAK1 K+ transport contribution in K+-starved cells. For this purpose, we exposed HvHAK1-expressing cells to a wide range of external alkali cation concentrations for 15 or 210 min and measured the subsequent uptake of Rb+ in a solution with no addition of K+, Na+, and NH4+. We observed a similar internal K+ concentration and a similar uptake of Rb+ for cells exposed for 15 min to different external K+ concentrations (data not shown). For cells exposed for 210 min, the internal K+ concentration was similar at different external K+ concentrations (data not shown) but the rate of Rb+ uptake depended on the new external K+ concentration in the growth medium (Fig. 6A). These results are consistent with a role of K+ trafficking across the membrane in controlling HvHAK1 transport. In order to perform an assessment of the specificity of K+ supply on changes in Rb+ uptake, we next investigated whether or not the inclusion of Na+ and NH4+ in the growth medium interferes with the course of K+ deprivation. The addition of Na+ during the first 15 min of K+ starvation did not exert any effect on the subsequent Rb+ transport (data not shown). However, the addition of NaCl during a 210-min K+ deprivation period resulted in a decrease of the subsequent Rb+ uptake (Fig. 6B), indicating that the presence of high Na+ concentrations during growth interferes with the stimulatory effect of K+ starvation on the HvHAK1 contribution to Rb+ transport. However, not all monovalent cations possessed this capacity. Complementary experiments highlighted that culture at high NH4+ concentrations did not interfere with the enhancement of HvHAK1-mediated Rb+ transport observed in yeast cells after 210 min of K+ deprivation (Fig. 6C).
Figure 6.
High sodium, but not high ammonium, concentrations interfere with the increased contribution of HvHAK1 following K+ starvation in yeast cells. HvHAK1-expressing yeast cells were grown overnight at 30 mm KCl and then exposed for 210 min to different external concentrations of alkali cations. A, Rb+ uptake from a 100 μm Rb+ solution after cells were exposed to different external K+ concentrations (n = 4). B and C, Rb+ uptake for cells exposed for 210 min to the absence of K+ in the presence of different Na+ concentrations (n = 3) or NH4+ concentrations (n = 5), respectively. D, Accumulation of the fluorescent dye DiOC6 measured in yeast cells grown for 210 min in the absence of K+ with or without 30 mm NH4+, 100 mm Na+, or 20 μm carbonyl cyanide m-chlorophenylhydrazone (CCCP) uncoupler as well as in the presence of 30 mm KCl (n = 5). Error bars represent se. Different letters indicate significantly different values (P < 0.05 for A to C, P < 0.005 for D).
The observed dependence of Rb+ uptake on the external concentration of K+ used for yeast culture is consistent with the possibility that changes in HvHAK1 transport are associated with changes in membrane potential. In order to provide an estimate of changes in membrane potential following K+ deprivation, we measured the accumulation of the fluorescent dye 3,3′-dihexyloxacarbocyanine iodide (DiOC6) by flow cytometry, which has been validated as an indicator of membrane potential for yeast cells grown under the same conditions (Madrid et al., 1998). HvHAK1-expressing cells deprived of K+ for 210 min accumulated more DiOC6 than K+-nonstarved cells, indicating that hyperpolarization takes place during K+ starvation (Fig. 6D). Addition of the uncoupler carbonyl cyanide m-chlorophenylhydrazone abolished this high accumulation of DiOC6, providing a control on the reliability of membrane potential measurements performed here. The presence of high NH4Cl concentrations during long-term K+ deprivation, in turn, affected neither the subsequent membrane potential difference nor the transport of Rb+ (Fig. 6, D and C, respectively). If the contribution of HvHAK1 were strictly associated with increased hyperpolarization, it would be expected that any condition leading to a more negative membrane potential must result in higher Rb+ uptake. However, we found that when cells were long-term deprived of K+ in the presence of 100 mm NaCl, they became hyperpolarized and exhibited low Rb+ transport (Fig. 6, D and B, respectively). Therefore, other factors should contribute to the transport of K+ mediated by HvHAK1.
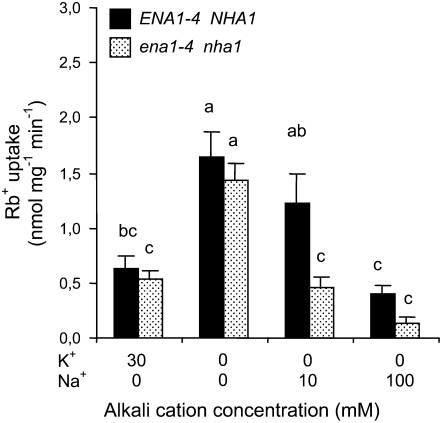
Results obtained for HvHAK1-expressing yeast cells grown in the presence of Na+ prompted us to analyze the role of Na+ exclusion on HvHAK1 activity. For this purpose, we transformed HvHAK1 into yeast cells carrying a disruption of the genes coding for the Na+-ATPases ENA1 to ENA4 and the Na+/H+ antiporter NHA1 in addition to the disruption of genes coding for the TRK1 and TRK2 K+ transporters. Pretreatment with moderate NaCl concentrations during the course of K+ deprivation led to a higher relative reduction of the subsequent Rb+ uptake by cells lacking the systems involved in Na+ exclusion than by cells in which active Na+ exclusion takes place (Fig. 7).
Figure 7.
Culture at moderate NaCl concentrations affects Rb+ transport mediated by HvHAK1 in yeast cells lacking Na+ exclusion systems. Rb+ uptake from a 100 μm Rb+ solution (n = 4) was measured after 210 min of K+ starvation in the presence of different NaCl concentrations for HvHAK1-expressing trk1Δtrk2Δ cells (black columns) and trk1Δtrk2Δena1-4Δnha1Δ cells, which lack the Na+-excluding systems ENA1 to ENA4 and NHA1 (dotted columns). Data for K+-nonstarved cells are included for comparative purposes. Error bars represent se. Different letters indicate significantly different values (P < 0.01).
Modulation of the HvHAK1 Contribution to K+ Capture in Yeast Cells Involves PPZ1 and HAL4/5 Proteins
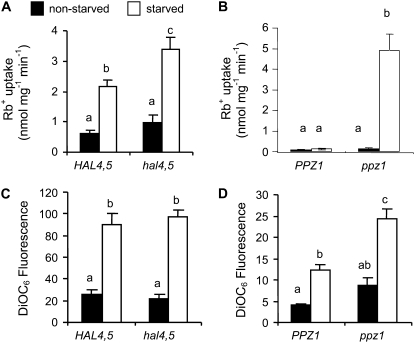
The results shown above indicate a complex control of the contribution of HvHAK1 to K+ transport. In yeast cells, regulation of K+ influx mediated by TRK transporters involves, among other components, protein phosphatases PPZ1/2 as well as halotolerance HAL4/5 kinases. HvHAK1-expressing yeast cells lacking HAL4 and HAL5 in addition to the lack of TRK1 and TRK2 showed higher Rb+ transport than that displayed by cells only lacking TRK1 and TRK2 when deprived of K+ for 210 min (Fig. 8A). Since hal4Δhal5Δ disruption in a TRK1-TRK2 background causes membrane hyperpolarization (Mulet et al., 1999), we next tried to determine whether the results obtained in Rb+ uptake experiments could be explained by differences in membrane potential between trk1Δtrk2Δhal4Δhal5Δ and trk1Δtrk2ΔHAL4HAL5 cells expressing HvHAK1. Ruling out this possibility, we found a similar DiOC6 accumulation in both kinds of cells (Fig. 8C). As such, we concluded that HAL4/HAL5 proteins down-regulate the contribution of HvHAK1 to K+ transport.
Figure 8.
The PPZ1 phosphatase and the HAL4/5 kinases control the inducible transport of Rb+ mediated by HvHAK1 in yeast cells. A, Rb+ uptake from a 100 μm Rb+ solution measured in K+-starved (210 min) and K+-nonstarved trk1Δtrk2Δ and trk1Δtrk2Δhal4Δhal5Δ HvHAK1-expressing yeast cells (n = 5). B, Under the same experimental conditions, Rb+ uptake was also measured in trk1Δtrk2Δ and trk1Δtrk2Δppz1Δ HvHAK1-expressing yeast cells derived from the DBY746 strain (n = 5). C and D, Accumulation of the DiOC6 fluorescent dye for cells grown in the presence or absence of K+ (n = 3). Error bars represent se. Different letters indicate significantly different values (P ≤ 0.01 for A and D, P < 0.001 for B and C).
To study the role of PPZ1, we transformed trk1Δtrk2Δ and trk1Δtrk2Δppz1Δ cells with a p424 plasmid containing the HvHAK1 cDNA. Expression of HvHAK1 into p424 did not restore the growth of yeast cells lacking TRK1 and TRK2. However, in a background also lacking PPZ1, expression of HvHAK1 into p424 restored the capacity of yeast cells to grow at low external K+ concentrations, indicating a control of the HvHAK1 contribution to K+ capture (Supplemental Fig. S5). The role of PPZ1 was tested directly by measuring Rb+ uptake after 15 or 210 min of K+ starvation in trk1Δtrk2Δ or trk1Δtrk2Δppz1Δ HvHAK1-expressing cells. A strong response of Rb+ uptake to K+ starvation was clearly found in cells lacking PPZ1 (Fig. 8B), suggesting a role of the encoded protein in down-regulating the HvHAK1 contribution to K+ capture. Studies of the accumulation of DiOC6 revealed that after a 210-min period of K+ starvation, HvHAK1-expressing ppz1Δtrk1Δtrk2Δ cells were hyperpolarized compared with HvHAK1-expressing trk1Δtrk2Δ cells (Fig. 8D).
DISCUSSION
The HAK-KT-KUP-type transporters constitute a large family of proteins thought to play a pivotal role in the maintenance of K+ homeostasis in plants. Here, we introduced evidence demonstrating that the ionic environment regulates the contribution of the HvHAK1 K+ transporter to K+ movement. This regulation may be critical to ensure K+ capture in K+-deficient environments and during the early response to salinity.
It has been shown previously that HAK1 transporters provide a route for K+ uptake in HAK1-expressing yeast cells that is similar to that found previously in plants suffering from K+ deficiency (Epstein et al., 1963; Santa-María et al., 1997; Rubio et al., 2000). The contribution of HAK1 transporters to high-affinity K+ transport was later confirmed in an Arabidopsis mutant lacking the HvHAK1 homolog, AtHAK5, which displays reduced capture of Rb+ under conditions of K+ starvation (Gierth et al., 2005). The evidence that HvHAK1 accumulation is augmented in the outermost layers of the root (Fig. 1) supports a role of the encoded transporter at the boundary between the external medium and the roots. This tissue location (Fig. 1), the enhanced accumulation of HvHAK1 transcripts upon K+ deprivation in barley roots (Fig. 2), as well as the enhanced capture of K+ displayed by HvHAK1 transgenic plants following K+ withdrawal (Fig. 4), are consistent with a role of this transporter in the inducible component of K+ uptake from diluted K+ solutions.
Studies with transgenic Arabidopsis plants expressing HvHAK1 indicate a strong effect of K+ deprivation on the contribution of HvHAK1. Studies with yeast cells expressing HvHAK1 allowed us to dissect the way in which K+ deprivation sets the contribution of a HAK-KUP-KT transporter in a model organism in which the endogenous transport of K+ from diluted K+ solutions is nil. In this organism, changes in the contribution of HvHAK1 to Rb+ capture following K+ starvation are not linked to changes in the accumulation of HvHAK1 transcripts (Fig. 5D), indicating that K+ deprivation should act on the amount and/or the activity of the HvHAK1 transporter. A primary phenomenon that takes place in plants and yeasts suffering K+ deficiency is plasma membrane hyperpolarization (Maathuis and Sanders, 1994; Walker et al., 1996; Hirsch et al., 1998; Madrid et al., 1998). In this context, an important question to address is whether the enhancement of Rb+ uptake observed here (Figs. 5A and 6A) results only from a long-term change in the membrane potential. Measurements of DiOC6 accumulation indicate that K+ starvation in HvHAK1-expressing yeast cells also leads to plasma membrane hyperpolarization (Figs. 6D and 8). These data are consistent with a possible role of membrane potential on changes in the transport of Rb+ mediated by HvHAK1 by affecting either the driving force or a signaling cascade. The evidence offered here (Figs. 6D and 8C) indicates that changes in the contribution of HvHAK1 to K+ capture cannot be entirely accounted for by changes driven by membrane potential. Therefore, other mechanisms modulating the HvHAK1 contribution to K+ capture in yeasts should be considered.
In most fungi, K+ transport results from the activity of TRK and HAK transporters (Benito et al., 2004). The mechanisms controlling K+ homeostasis through TRK1 have been deeply explored in S. cerevisiae, revealing that TRK1 is activated by the HAL4 and HAL5 kinases and inhibited by the PPZ1 and PPZ2 phosphatases (Mulet et al., 1999; Yenush et al., 2002). Interestingly, genetic evidence obtained with this work supports the hypothesis that HAL4/5 kinases act as down-regulators of the HvHAK1 contribution (Fig. 8A), which is opposite to their effect on TRK1. On the other hand, current evidence indicates that PPZ1 is involved in determining the upper limits of K+ accumulation in yeast cells, mainly in a TRK1-dependent manner (Yenush et al., 2005). Our results indicate that PPZ1 could exert this regulatory role, critical for K+ homeostasis, by setting the contribution of a HAK1 transporter to K+ capture (Fig. 8B). The fact that both HAL4/5 kinases and the PPZ1 phosphatase act as down-regulators of the contribution of HvHAK1 to K+ transport could be explained by their action at different levels of regulation. Although the precise mode by which PPZ1 and HAL4/5 modulate the HvHAK1 contribution is still uncertain, our results illustrate that both phosphorylation and dephosphorylation processes act in concert to determine the uptake of K+ mediated by a HAK1 transporter in yeasts. In this context, the search for plant functional counterparts of PPZ1 phosphatase and HAL4/5 kinases could help to determine whether or not the mechanisms setting the contribution of HvHAK1 in yeast cells are operative in plants. Besides, the finding that a HAK1 transporter is modulated by PPZ1 and HAL4/5 in S. cerevisiae, in which no HAK genes have been found, poses important questions regarding the conservation of the regulatory network controlling K+ homeostasis in fungi.
Transcriptome studies revealed the existence of a cross talk of signals associated with the perception of nitrogen and K+ status in plants (Wang et al., 2001; Armengaud et al., 2004). Additionally, the presence of high NH4+ concentrations has been used as a tool to dissect the components involved in K+ transport (Hirsch et al., 1998; Spalding et al., 1999; Nieves-Cordones et al., 2007). In barley, the presence of high NH4+ concentrations in the growth medium generates a switch on the properties of Rb+ uptake from diluted K+ solutions (Santa-María et al., 2000). This switch involves two parallel processes, the first one being the dominance of channel-like features, partially explained by a thermodynamic gradient favorable to channel participation (Kronzucker et al., 2003). The second process involved in that switch is the reduced size of the inducible component of Rb+ transport sensitive to NH4+. Our results indicate that the reduction of this inducible component does not involve a long-term down-regulation of HvHAK1 transcript accumulation mediated by high ammonium levels (Fig. 2B). Therefore, it seems possible that the reduced relative contribution of the NH4+-sensitive component of Rb+ transport after long-term growth at high NH4+ supplies could be, at least in part, the result of a down-regulation of the accumulation of transcripts coding for other transporters. Gene expression studies (Fig. 2C) suggest that HvHAK1b could be a target for such a regulation. Therefore, the accumulation of transcripts coding for HAK1 transporters presumably involved in the NH4+-sensitive pathway of K+ transport occurs through two signaling routes: one NH4+-sensitive, acting on HvHAK1b, and the other insensitive to NH4+, acting on HvHAK1.
The effect of sodium salts on K+ homeostasis has been a main subject in salinity studies (Flowers and Läuchli, 1983). In Arabidopsis, the current evidence indicates that, for plants grown in the presence of ammonium, the inhibitory effect of Na+ on K+ uptake results from an effect of Na+ on the activity of the AKT1 K+ channel (Qi and Spalding, 2004). The evidence contributed here indicates that culturing at high NaCl concentrations precludes the subsequent enhancement of K+ transport mediated by a HAK1 transporter in K+-deprived yeasts (Fig. 6B and 7). While this effect tends to diminish the contribution of HvHAK1 to K+ capture, the transient enhanced accumulation of HvHAK1 transcripts observed in barley roots after a 6-h exposure to a high external NaCl concentration (Fig. 3) could lead to the opposite outcome and is likely to play a role in determining the low inhibitory effect of Na+ on Rb+ transport observed in barley roots at 6 h after salinization (Fig. 3). Whereas our data (Supplemental Fig. S3) do not allow us to rule out the contribution of an osmotic component, they indicate a role of nonosmotic components on the peak of accumulation of HvHAK1 transcripts. Evidence for cross talk between osmotic and nonosmotic signals during the early transcriptome response to salt stress has been offered for barley plants (Ueda et al., 2004). The enhancement and decline in the amount of HvHAK1 transcripts observed here (Fig. 3) is similar to that reported for a member of the group II HAK transporters in Mesembryanthemum crystallinum (Su et al., 2002) and essentially mimics that observed formerly for the genes implicated in the early coordinated gene response to salt stress described in the barley close relative species Lophopyrum elongatum (Gulick and Dvořák, 1992; Galvez et al., 1993). Since a similar response is elicited by abscisic acid (Galvez et al., 1993) and a transient change in the balance between methyl jasmonate and abscisic acid has been observed during the first hours of exposure to high NaCl concentrations (Moons et al., 1997), with a prominent role of jasmonic acid in the K+-dependent transcriptome also advanced (Armengaud et al., 2004), the possibility that hormone signaling plays a role in setting the response of HvHAK1 to salt stress should not be discarded.
In conclusion, this study indicates the presence of mechanisms driven by the ionic environment that determine the contribution of a HAK1 transporter to K+ capture. Furthermore, studies with yeast cells reveal a role of phosphorylation and dephosphorylation processes in setting this contribution.
MATERIALS AND METHODS
Plant Materials and Culture
Seeds of barley (Hordeum vulgare ‘Golden Promise’) were germinated in the dark on moistened filter paper for 48 h. Seedlings were then transferred to a 0.8-L plastic pot filled with a complete nutrient solution of the following composition: 1.0 mm Ca(NO3)2, 0.5 mm MgSO4, 0.5 mm H3PO4, 50 μm FeEDTA, 50 μm CaCl2, 25 μm H3BO3, 2 μm ZnSO4, 2 μm MnSO4, 0.5 μm CuSO4, 0.5 μm molybdic acid, and 2.5 mm MES, with or without the addition of 1 mm K+, except where indicated. The pH was brought to 6.0 by the addition of Ca(OH)2, and the solution was aerated. When added, K+, Na+, and NH4+ were provided as chloride salts. Temperature in the growth chamber was set to 22°C (day/night), and the relative humidity was kept at 85%. The photon flux density at the plant level was set at 70 μmol m−2 s−1 over a photoperiod of 14 h. Experiments were carried out with 1-week-old plants, except for the experiment reported in Figure 2, B and C, in which 13-d-old plants were used. Root and shoot samples were extracted with 0.5 n HCl to release free cations, and K+ (or Rb+ in Rb+ uptake experiments) was determined with a Perkin-Elmer AA 100 spectrophotometer in emission mode. Arabidopsis (Arabidopsis thaliana ecotype Columbia) seeds were sowed on plates containing the medium described above plus 0.8% agar. After 10 d, seedlings were transferred to 0.125-L plastic pots in which only roots were in contact with the nutrient solution. Plants grew for another 2 weeks, until the experiments were performed.
Rb+ Uptake Measurements in Plants
Roots of intact plants were transferred for 5 min to a solution with the same composition as used for growth but without K+. This allows elution of this chemical species from the root apoplast and thus helps to minimize the effect of ionic perturbations on the subsequent measurement. Loading was performed in 50-mL plastic pots containing the complete nutrient solution with no K+ added. This solution, heavily aerated, contained 100 μm Rb+. Loading was extended for 60 min for barley and for 120 min for Arabidopsis. Subsequently, roots were washed two times for a total of 6 min with the same solution used for loading but without Rb+. Results are expressed on a fresh weight basis.
Quantitation of Plant mRNAs
For the quantitation of plant mRNAs, RNA extracted from whole barley roots was used. Extraction of total RNA was performed with the use of the RNeasy Plant Mini kit (Qiagen). After extraction, total RNA was treated with RQ1 DNase (Promega), and the presence of contamination from a genomic origin was specifically tested for by PCR. For each DNA-free RNA sample, several independent RT reactions were performed. RT was carried out with SuperScript II (Gibco-BRL) on 20 ng of total RNA using Oligonucleotide-dT18 as 3′ primer. Real-time PCR was performed in duplicate for each RT reaction using the ABI Prism 5700 sequence detection system (Perkin-Elmer Applied Biosystems) and SYBR Green PCR Master Mix (Perkin-Elmer Applied Biosystems). Primers used for PCR for each gene, as well as amplicon sizes, have been described (Vallejo et al., 2005). Amplification was carried out with an initial step at 50°C for 2 min followed by 1 cycle at 95°C for 10 min and then by 40 amplification cycles. After each PCR, the dissociation curve of the PCR product was analyzed. The cDNA content of β-tubulin for each run was also estimated. In addition, two negative controls were included to exclude the possibility of genomic DNA contamination: one of them consisted of a reaction without cDNA, while the other contained an aliquot of the DNA-free RNA sample. Subsequently, the accumulation of transcripts was estimated by the use of the ΔΔCt method.
In Situ Hybridization
One-week-old barley plants, grown without aeration in a 10 mm MES solution brought to pH 6.0 ± 0.1 with Ca(OH)2, were used for in situ hybridization studies. Roots were cut into 10-mm-long fragments, from 10 to 60 mm from the root apex. For the fixation protocol, roots were treated with 3% (v/v) paraformaldehyde and 0.25% (v/v) glutaraldehyde in 0.1 m sodium phosphate buffer, pH 7.2, at room temperature, dehydrated in graded ethanol and xylene series, and embedded in Paraplast. Sections (2 μm thick) were attached to poly-l-Lys-coated slides. Sections were deparaffinized with xylene and rehydrated through a graded ethanol series. They were subsequently pretreated with 1 μg mL−1 proteinase K in 200 mm Tris-HCI, pH 7.5, and 2 mm CaCl2 at 37°C for 30 min and with 0.25% (v/v) acetic anhydride in 0.1 m triethanolamine, pH 8.0, at room temperature for 10 min, dehydrated in a graded ethanol series, and air dried. Sections were hybridized with either sense or antisense RNA 35S-labeled probes.
A pGEM-T Easy vector (Promega) containing a 325-bp cDNA of the HvHAK1 3′ untranslated region was linearized with PstI or NcoI to be used as DNA template for the in vitro synthesis of the sense or antisense RNA probe, respectively. Sense and antisense RNA probes were synthesized by the incorporation of [α-35S]UTP using an RNA labeling kit following the manufacturer's instructions (Boehringer Mannheim). Southern-blot analyses with barley genomic DNA previously showed a single hybridization band with this 325-bp probe. Hybridization and detection of the radioactive signal were performed as described previously (Langdale et al., 1988). Photographs were taken using an automatic camera coupled to a light microscope (Axiophot; Zeiss).
Yeast Strains and Growth Conditions
The Saccharomyces cerevisiae strains used in this work were WΔ3 (W303.1A trk1Δ∷LEU2 trk2Δ∷HIS3), which is deficient in the endogenous K+ uptake systems TRK1 and TRK2; MA5 (W303.1A trk1Δ∷LEU2 trk2Δ∷HIS3 nha1Δ∷LEU2 ena1-4Δ∷HIS3), which is derived from WΔ3 and also deficient in the Na+ efflux systems ENA1 to ENA4 and NHA1 (Benito et al., 2004); and JM110 (W303.1A trk1Δ∷LEU2 trk2Δ∷HIS3 hal4Δ∷TRP1 hal5Δ∷KanMX), also derived from WΔ3, which carries a disruption on the genes coding for the protein kinases HAL4 and HAL5 (Mulet et al., 1999). These strains were the recipient of the pYPGE15 plasmid or its derivative containing the HvHAK1 cDNA, pGF718. Strains ESV212 (DBY746 trk1Δ∷LEU2 trk2Δ∷HIS3) and its derivative MAR70 (DBY746 trk1Δ∷LEU2 trk2Δ∷HIS3 ppz1Δ∷URA3), which carries a disruption of the gene coding for the PPZ1 phosphatase in addition to the disruption of TRK1 and TRK2 (Ruiz et al., 2004), were also used. In this case, the HvHAK1 cDNA was cloned into the p424 plasmid. Yeast cells were grown in Arg (AP) medium supplemented with 30 mm KCl (Santa-María et al., 1997). Cells were washed twice and transferred to AP medium without added K+. The basal concentration of K+ in that medium was 2.5 μm. At different times from the beginning of K+ deprivation, Rb+ uptake measurements were performed. Cells were suspended in 2% Glc and 10 mm MES buffer brought to pH 6.0 with Ca(OH)2. Unless indicated specifically (Supplemental Fig. S4B), Rb+ uptake measurements were performed in the absence of K+, Na+, or NH4+ in the loading solution. Cells were taken at intervals, filtered through a 0.8-μm-pore nitrocellulose membrane (Millipore), and washed with 20 mm MgCl2. Filters were incubated overnight in 0.5 n HCl, and Rb+ was determined by atomic emission spectrophotometry. Results are expressed on a cell dry weight basis. Vmax and Km values were estimated by nonlinear regression.
Accumulation of DiOC6 was estimated for yeast cells grown for 210 min in the presence or absence of different alkali cations. These cells did not receive any other treatment and were suspended in MES-Ca2+ and exposed to 1 nm DiOC6 for 30 min at 28°C in the dark. To test cell viability, propidium iodide was used. Flow cytometry analysis was performed in a FACSCalibur (Becton-Dickinson).
Plant Transformation
The HvHAK1 718 cDNA was cloned into the plant binary vector pB112 containing the nptII kanamycin resistance marker. Agrobacterium tumefaciens pgv3101 was the recipient of the HvHAK1-containing plasmid, which was later used to transform Arabidopsis plants, ecotype Col-0, by the floral dip procedure (Clough and Bent, 1998). Plants displaying resistance to the marker were selected for further analysis. The presence of the transgene was evaluated by PCR on genomic DNA. To identify homozygous plants, studies of the segregation of kanamycin resistance were performed. Expression of HvHAK1 for each homozygous line was determined by RT-PCR using DNA-free RNA.
Except where indicated, results obtained in this work were analyzed by two-factor ANOVA, with post-hoc comparisons made by Duncan's test. The analysis was performed using the Statistica 6.0 program (StatSoft*).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. In situ hybridization studies show the accumulation of HvHAK1 transcripts in most root cells.
Supplemental Figure S2. Effect of K+ deprivation, in a nonaerated MES-Ca2+ solution, on the accumulation of HvHAK1 transcripts.
Supplemental Figure S3. Comparative effect of sorbitol and NaCl on the accumulation of HvHAK1 transcripts, and effect of NaCl exposure on the concentration of K+ and Na+ in roots.
Supplemental Figure S4. Rb+ uptake properties of yeast cells expressing HvHAK1 following K+ withdrawal.
Supplemental Figure S5. HVHAK1-expressing yeast cells lacking the PPZ1 phosphatase display an enhanced growth capacity at low K+ concentrations.
Supplementary Material
Acknowledgments
We express our thanks to Dr. Joaquín Ariño and Dr. Amparo Ruíz (Universitat Autónoma de Barcelona), to Dr. Begonia Benito and Dr. Rosario Haro (Universidad Politécnica de Madrid), and to Dr. Ramón Serrano (Universidad Politécnica de Valencia, Spain) for the gift of the yeast strains used in this work. Thanks are also given to Dr. Fernando Bravo-Almonacid (Instituto de Investigaciones en Ingeniería Genética y Biología Molecular), Dr. Victoria Busi (Instituto Tecnológico Chascomús), and Dr. Diego Gomez-Casati (Instituto Tecnológico Chascomús) for generous help with transgenic plants. We also express gratitude to Dr. Amparo Monfort and Dr. Matilde José-Estanyol (Departament de Genètica Molecular, Centro de Investigaciones y Desarrollo-Consejo Superior de Investigaciones Científicas) for technical assistance during in situ hybridization studies.
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica (grant nos. PICT 080005/2000 and PICT 20138/2004) of Argentina as well as by a project from the Universidad Politécnica de Madrid of Spain to G.E.S.-M. F.R.F. is the recipient of a fellowship from the Universidad Nacional de San Martín. M.L.P. and S.M. are fellows of the Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Guillermo E. Santa-María (gsantama@iib.unsam.edu.ar).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ahn SJ, Shin R, Schachtman DP (2004) Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol 134 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P, Breitling R, Amtmann A (2004) The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol 136 2556–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito B, Garciadeblás B, Schreier P, Rodríguez-Navarro A (2004) Novel P-type ATPases mediate high-affinity potassium or sodium uptake in fungi. Eukaryot Cell 3 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Glass ADM, Crawford NM (1993) Ammonium inhibition of Arabidopsis thaliana root growth can be reversed by potassium and by auxin resistance mutations aux1, axr1 and axr2. Plant Physiol 102 983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Epstein E, Rains DW, Elzam OE (1963) Resolution of dual mechanisms of potassium absorption by barley roots. Proc Natl Acad Sci USA 49 684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers TJ, Läuchli A (1983) Sodium versus potassium: substitution and compartmentation. In A Láuchli, RL Bieleski, eds, Encyclopedia of Plant Physiology, New Series, Vol 15B, Inorganic Plant Nutrition. Springer-Verlag, Berlin, pp 651–681
- Fu HH, Luan S (1998) AtKup1: a dual-affinity K+ transporter from Arabidopsis thaliana. Plant Cell 10 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez AF, Gulick PJ, Dvořák J (1993) Characterization of the early stages of genetic salt-stress responses in salt-tolerant Lophopyrum elongatum, salt-sensitive wheat, and their amphiploid. Plant Physiol 103 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcíadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A (2003) Sodium transport and HKT transporters: the rice model. Plant J 34 1–14 [DOI] [PubMed] [Google Scholar]
- Gierth M, Mässer P, Schroeder JI (2005) The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisbert C, Rus AM, Bolarín C, López-Coronado M, Arrillaga I, Montesinos C, Caro M, Serrano R, Moreno V (2000) The yeast HAL1 gene improves salt tolerance of transgenic tomato. Plant Physiol 123 393–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass ADM (1976) Regulation of potassium absorption in barley roots: an allosteric model. Plant Physiol 58 33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick PJ, Dvořák J (1992) Coordinate gene response to salt stress in Lophopyrum elongatum. Plant Physiol 100 1384–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR (1998) A role for the AKT1 potassium channel in plant nutrition. Science 280 918–921 [DOI] [PubMed] [Google Scholar]
- Horie T, Costa A, Houn Kim T, Jung Han M, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI (2007) Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J 26 3003–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Kwak JM, Uozumi N, Schroeder JI (1998) AtKup1: an Arabidopsis thaliana gene encoding high-affinity potassium transporter activity. Plant Cell 10 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV, Lucas WJ (1982) Potassium transport in corn roots: resolution of kinetics into saturable and linear components. Plant Physiol 70 1723–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV, Lucas WJ (1988) Potassium transport in roots. Adv Bot Res 15 93–178 [Google Scholar]
- Kronzucker HJ, Szczerba MW, Britto DT (2003) Cytosolic potassium homeostasis revisited: 42K-tracer analysis in Hordeum vulgare L. reveals set-point variations in K+. Planta 217 540–546 [DOI] [PubMed] [Google Scholar]
- Langdale JA, Rothermel BA, Nelson T (1988) Cellular pattern of photosynthetic gene expression in developing maize leaves. Genes Dev 2 106–115 [DOI] [PubMed] [Google Scholar]
- Li L, Kim BG, Cheong YH, Pandey GK, Luan S (2006) A Ca2+-signaling pathway regulates a K-channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA 103 12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJM, Sanders D (1994) Mechanism of high-affinity potassium uptake in roots of Arabidopsis thaliana. Proc Natl Acad Sci USA 91 9272–9276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid R, Gómez MJ, Ramos J, Rodríguez-Navarro A (1998) Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J Biol Chem 273 14838–14844 [DOI] [PubMed] [Google Scholar]
- Moons A, Prinsen E, Bauw G, Van Montagu M (1997) Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell 9 2243–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulet JM, Leube MP, Kron SJ, Rios G, Fink GR, Serrano R (1999) A novel mechanism of ion homeostasis and salt tolerance in yeast: The Hal4 and Hal5 protein kinases modulate the Trk1-Trk2 potassium transporter. Mol Cell Biol 19 3328–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves-Cordones M, Martínez-Cordero MA, Martínez V, Rubio F (2007) An NH4+-sensitive component dominates high-affinity K+ uptake in tomato plants. Plant Sci 172 273–280 [Google Scholar]
- Qi Z, Spalding EP (2004) Protection of plasma membrane K+ transport by the salt overly sensitive 1 Na+-H+ antiporter during salinity stress. Plant Physiol 136 2548–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro A (2000) Potassium transport in fungi and plants. Biochim Biophys Acta 1469 1–30 [DOI] [PubMed] [Google Scholar]
- Rubio F, Santa-María GE, Rodríguez-Navarro A (2000) Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant 109 34–44 [Google Scholar]
- Ruiz A, Ruiz MC, Sánchez-Garrido MA, Ariño J, Ramos J (2004) The Ppz protein phosphatases regulate Trk-independent potassium influx in yeast. FEBS Lett 578 58–62 [DOI] [PubMed] [Google Scholar]
- Rus A, Lee BH, Muñoz-Mayor A, Sharkhuu A, Zhu JK, Bressan RA, Hasegawa PM (2004) AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol 136 2500–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-María GE, Danna CH, Czibener C (2000) High-affinity potassium transport in barley roots: ammonium-sensitive and -insensitive pathways. Plant Physiol 123 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-María GE, Rubio F, Dubcovsky J, Rodríguez-Navarro A (1997) The HAK1 gene of barley belongs to a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9 2281–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentenac H, Bonneaud N, Minet M, Lacroute F, Salmon JM, Gaymard F, Grignon C (1992) Cloning and expression in yeast of a plant potassium ion transport system. Science 256 663–665 [DOI] [PubMed] [Google Scholar]
- Shin R, Schachtman DP (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD (1999) Potassium uptake supporting plant growth in the absence of AKT1 channel activity: inhibition by ammonium and stimulation by sodium. J Gen Physiol 113 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Golldack D, Zhao C, Bohnert HJ (2002) The expression of HAK-type K+ transporters is regulated in response to salinity stress in common ice plant. Plant Physiol 129 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Kathiresan A, Inada M, Narita Y, Nakamura T, Shi W, Takabe T, Bennett J (2004) Osmotic stress in barley regulates expression of a different set of genes than salt stress does. J Exp Bot 55 2213–2218 [DOI] [PubMed] [Google Scholar]
- Vallejo AJ, Peralta ML, Santa-María GE (2005) Expression of potassium-transporter coding genes, and kinetics of rubidium uptake, along a longitudinal root axis. Plant Cell Environ 28 850–862 [Google Scholar]
- Véry AA, Sentenac H (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54 575–603 [DOI] [PubMed] [Google Scholar]
- Walker DJ, Leigh RA, Miller AJ (1996) Potassium homeostasis in vacuolated plant cells. Proc Natl Acad Sci USA 93 10510–10514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Garvin DF, Kochian LV (2001) Nitrate-induced genes in tomato roots. Array analysis reveals novel genes that may play a role in nitrogen nutrition. Plant Physiol 127 345–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125 1347–1360 [DOI] [PubMed] [Google Scholar]
- Yenush L, Merchan S, Holmes J, Serrano R (2005) pH-responsive, posttranslational regulation of the Trk1 potassium transporter by the type 1-related Ppz1 phosphatase. Mol Cell Biol 25 8683–8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenush L, Mulet JM, Ariño J, Serrano R (2002) The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity and cell cycle progression. EMBO J 21 920–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.