Abstract
Protein synthesis is catalyzed by the ribosome, a two-subunit enzyme comprised of four ribosomal RNAs and, in Arabidopsis (Arabidopsis thaliana), 81 ribosomal proteins (r-proteins). Plant r-protein genes exist as families of multiple expressed members, yet only one r-protein from each family is incorporated into any given ribosome, suggesting that many r-protein genes may be functionally redundant or development/tissue/stress specific. Here, we characterized the localization and gene-silencing phenotypes of a large subunit r-protein family, RPL23a, containing two expressed genes (RPL23aA and RPL23aB). Live cell imaging of RPL23aA and RPL23aB in tobacco with a C-terminal fluorescent-protein tag demonstrated that both isoforms accumulated in the nucleolus; however, only RPL23aA was targeted to the nucleolus with an N-terminal fluorescent protein tag, suggesting divergence in targeting efficiency of localization signals. Independent knockdowns of endogenous RPL23aA and RPL23aB transcript levels using RNA interference determined that an RPL23aB knockdown did not alter plant growth or development. Conversely, a knockdown of RPL23aA produced a pleiotropic phenotype characterized by growth retardation, irregular leaf and root morphology, abnormal phyllotaxy and vasculature, and loss of apical dominance. Comparison to other mutants suggests that the phenotype results from reduced ribosome biogenesis, and we postulate a link between biogenesis, microRNA-target degradation, and maintenance of auxin homeostasis. An additional RNA interference construct that coordinately silenced both RPL23aA and RPL23aB demonstrated that this family is essential for viability.
The ribosome is a massive enzyme (2.5–4.5 MD) responsible for catalyzing protein synthesis. It consists of two subunits of unequal size that exist freely in the cell but assemble together on mRNA to become translationally competent. Plant cytoplasmic ribosomes synthesize the majority of cellular proteins (Bogorad, 1975; Bailey-Serres, 1998), and in Arabidopsis (Arabidopsis thaliana) are comprised of four ribosomal RNAs (rRNAs; small subunit [SSU] 18S and large subunit [LSU] 26S, 5.8S, and 5S) and 81 ribosomal proteins (r-proteins; 33 SSU, 48 LSU; Barakat et al., 2001; Chang et al., 2005). Ribosomes contain only a single copy of nearly all r-proteins (Ban et al., 2000; Wimberly et al., 2000; Guarinos et al., 2003; Hanson et al., 2004; Schuwirth et al., 2005), yet in plants r-proteins are encoded by large multigene families containing more than one transcriptionally active member (Barakat et al., 2001; Popescu and Tumer, 2004; Hulm et al., 2005; McIntosh and Bonham-Smith, 2005; Ouyang et al., 2007). For example, the Arabidopsis genome contains 254 genes for the 81 r-proteins, with families of between two and five expressed members (Barakat et al., 2001; Chang et al., 2005). This high degree of paralogy suggests that many r-protein genes may be functionally redundant or development/tissue/stress specific.
Arabidopsis RPL23a is part of a universally conserved r-protein family (Lecompte et al., 2002) that binds directly to LSU rRNA and is essential for ribosome biogenesis (El-Baradi et al., 1984, 1985, 1987; Rutgers et al., 1991). Structural studies have mapped its yeast (Saccharomyces cerevisiae) and bacterial counterparts (L25 and L23, respectively) to the LSU, adjacent to the polypeptide exit tunnel (Nissen et al., 2000; Spahn et al., 2001). This position suggests a role for RPL23a in protein translocation and secretion, and this role has been validated in both prokaryotes and eukaryotes (Beckmann et al., 1997; Morgan et al., 2002; Pool et al., 2002; Halic et al., 2004; Maier et al., 2005; Menetret et al., 2005). There are two members of the RPL23a family in Arabidopsis, RPL23aA and RPL23aB. Transcript expression profiles have shown that RPL23aA is more abundant than RPL23aB in all tissues, and that accumulation of each differs in response to cold, wounding, and copper stress (McIntosh and Bonham-Smith, 2005). Both isoforms are incorporated into the ribosome (Chang et al., 2005; Giavalisco et al., 2005; Carroll et al., 2008), and RPL23aA functionality was confirmed by its ability to complement a yeast l25 mutant (McIntosh and Bonham-Smith, 2001).
The consequences of overlapping r-protein expression in plants have yet to be fully elucidated. In yeast, 59 of 79 r-proteins are encoded by two expressed paralogs, producing identical or near-identical r-protein isoforms. Although these paralogs can be transcribed at divergent levels (Tornow and Santangelo, 1994; Planta, 1997), a high transcription rate from one paralog is compensated for by a low rate from the other, resulting in a consistent transcript level for each r-protein (Warner et al., 1985; Planta, 1997). As with yeast, most of the Arabidopsis r-protein paralogs encode very similar proteins. However, unlike yeast, there is no evidence suggesting that transcript levels among paralogs are linked in a regulated manner. For example, hormone and stress treatment of Arabidopsis seedlings leads to changes in transcript levels of single paralogs from the RPS15a and RPL23a families but has no affect on the levels of the other expressed paralogs (Hulm et al., 2005; McIntosh and Bonham-Smith, 2005). Further, despite overlapping transcript expression, phenotypic characterization of single paralog silencing or knockout lines suggest that paralogs are not functionally equivalent. In tobacco (Nicotiana tabacum), for example, silencing RPL3A led to an increase in RPL3B transcript levels, but this increase was unable to compensate for the reduced RPL3A levels, resulting in abnormal growth and development (Popescu and Tumer, 2004). In Arabidopsis, rps13b and rps18a T-DNA insertional mutants develop narrow, pointed first leaves and have stunted root growth and delayed flowering; in both cases, northern analyses confirmed that transcript levels from paralogs were unaffected (Van Lijsebettens et al., 1994; Ito et al., 2000). It remains to be determined whether these findings result from a biological shortage of r-proteins for ribosome biogenesis caused by the loss of one copy from a redundant family, or if they are indicative of specialized functions for specific paralogs.
In this work, we investigated whether the two Arabidopsis RPL23a paralogs are equivalent with respect to cellular localization and phenotypic response to gene knockdowns. Accordingly, C- and N-terminal RPL23a-fluorescent protein fusions were made, and their localization followed in vivo in a heterologous tobacco system. We found that the two isoforms have different affinities for nucleolar accumulation, with RPL23aA predominating. We then designed estrogen-inducible, RNA interference (RNAi)-mediated silencing constructs targeting RPL23aA and RPL23aB, independently and coordinately. RPL23aA silencing resulted in growth retardation and morphological abnormalities, while RPL23aB silencing had no affect. We also showed, for the first time in plants, that coordinate silencing of both RPL23aA and RPL23aB is lethal. Our results indicate that the two RPL23a isoforms are not of equivalent importance for normal plant development.
RESULTS
RPL23a Isoform Comparison
RPL23aA and RPL23aB share 68.8% identity at the transcript level (83.7% between open reading frames [ORFs]) but encode proteins exhibiting 94.8% amino acid identity. They also share a high degree of primary sequence conservation with other eukaryotic orthologs (Fig. 1A), especially within the C-terminal domain that binds LSU rRNA (Rutgers et al., 1991). The eukaryotic L23a/L25 r-proteins possess an N-terminal extension region that is absent from prokaryotic counterparts and contains the nuclear localization signal (NLS; Rutgers et al., 1990; Schaap et al., 1991; Jakel and Gorlich, 1998). Although the NLS of plant RPL23a proteins has yet to be experimentally determined, all residue differences between Arabidopsis RPL23a isoforms reside within the N-terminal region (Fig. 1A). Further, two of these differences occur within a classical monopartite NLS [Fig. 1A; consensus (K/R)2XK/R, where X denotes any residue] that purportedly also functions as a nucleolus localization signal (NoLS; Kalderon et al., 1984; Dingwall and Laskey, 1991; Weber et al., 2000; Horke et al., 2004).
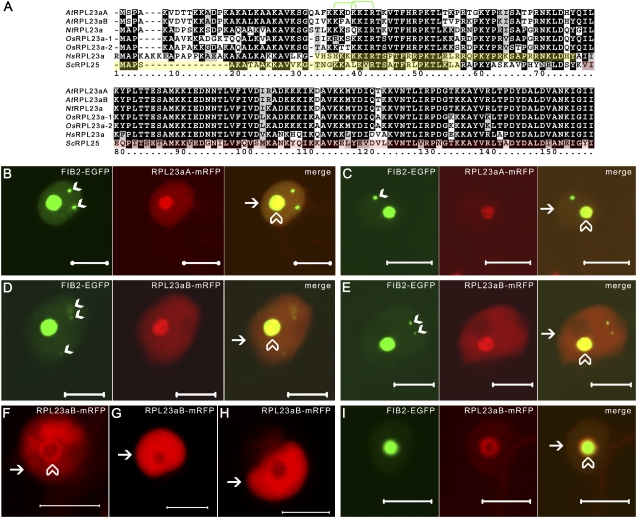
Figure 1.
Alignment and localization of the Arabidopsis RPL23a isoforms. A, Clustal alignment of the two Arabidopsis RPL23a isoforms (AtRPL23aA and AtRPL23aB) with tobacco RPL23a (NtRPL23a), two rice RPL23a isoforms (OsRPL23a-1, LOC_Os01g24690; OsRPL23a-2, LOC_Os04g42270), human RPL23a (HsRPL23a), and yeast RPL25 (ScRPL25). Identical and similar residues shared by four or more orthologs are shaded black and gray, respectively, while those shared by three or fewer orthologs are shaded white. Experimentally determined domains responsible for nuclear localization in human (Jakel and Gorlich, 1998) and yeast (Schaap et al., 1991) are highlighted in yellow. The C-terminal domain, which contains rRNA-binding capacity, is highlighted red (Rutgers et al., 1991; Kooi et al., 1994). Putative monopartite NLSs [(K/R)XK/R] in Arabidopsis are delineated by green brackets. Numbering is for reference only. B to I, CLSM images of tobacco epidermal cells transiently coexpressing FIB2-EGFP and RPL23aA-mRFP (B and C) or FIB2-EGFP and RPL23aB-mRFP (D, E, and I), or expressing RPL23aB-mRFP alone (F–H). Nucleolar and nucleoplasmic signals are indicated by transparent white arrowheads and small white arrows, respectively (B–I). White arrowheads point to cajal bodies (Beven et al., 1995; B–E, left). Images of the same optical slice were merged to show signal overlap (B–E and I, right). Bars = 10 μm.
C-Terminally Tagged RPL23a Isoforms Localize to the Nucleolus
To investigate whether differences in putative RPL23a NLS/NoLS domains have any impact on localization patterns, we designed RPL23aA/B C-terminal fusions with monomeric red fluorescent protein (mRFP) separated by a glutathione S-transferase (GST) linker. The GST linker was added to increase translational fusion mass beyond the size exclusion limit of nuclear pore complexes (>60 kD) and to enable affinity purification of bound proteins (Grebenok et al., 1997; Merkle, 2003); it did not interfere with localization in any noticeable way (see also Ookata et al., 1995; Kishi et al., 1996). C-terminal fusions were investigated because it has previously been shown that attachment of a small (approximately 2 kD) FLAG-His6 tag to the C terminus of RPL23aA did not disrupt the ability of the fusion protein to incorporate into ribosomes and form polysomes (Zanetti et al., 2005). RPL23a orthologs are known to be involved in LSU biogenesis within the nucleolus (Jeeninga et al., 1996; van Beekvelt et al., 2001), and hence we hypothesized that Arabidopsis RPL23a would accumulate to the greatest extent in the nucleolus. To enable definitive identification of nucleolar accumulation, we used a nucleolar marker, Arabidopsis FIBRILLARIN2 (FIB2), with a C-terminal enhanced GFP (EGFP) tag (Barneche et al., 2000). FIB2 specifically localizes to the nucleolus, where it directs a requisite step in rRNA processing and ribosome assembly (Tollervey et al., 1993; Barneche et al., 2000). RPL23aA/B-mRFP chimerics were transiently coexpressed with the FIB2-EGFP marker in tobacco epidermal cells via infiltration with transformed Agrobacterium tumefaciens (Sparkes et al., 2006). Cells imaged with a confocal laser scanning microscope (CLSM) 72 h postinfiltration showed strong nucleolar accumulation of FIB2-EGFP (Fig. 1, B–E and I, left) and weaker but clearly discernible nucleolar signals for RPL23aA-mRFP (Fig. 1, B and C, middle) and RPL23aB-mRFP (Fig. 1, D and E, middle). FIB2-EGFP also localized to mobile cajal bodies (Fig. 1, B–E; see Supplemental Results S1 for details on cajal bodies and nucleoli properties), which are nonmembrane-bound inclusions associated with nucleoli (Beven et al., 1995; Kim et al., 2007).
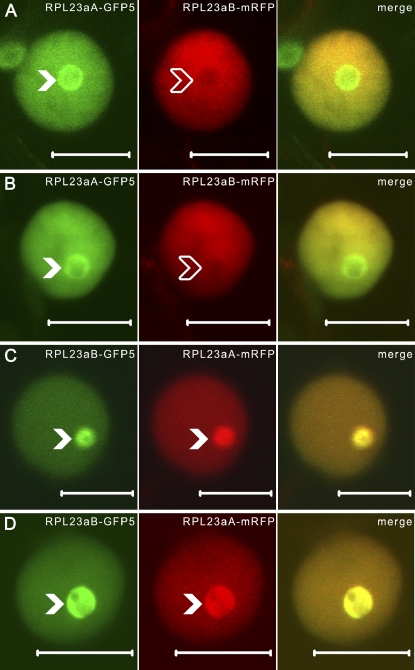
While both RPL23a isoforms were capable of localizing to the nucleolus, RPL23aB-mRFP was occasionally unable to target the core of the nucleolus (Fig. 1, F–I), instead accumulating only at the periphery of the nucleolus (13.6% of cells, n = 66; Fig. 1, F and I) or being excluded altogether (19.7% of cells, n = 66; Fig. 1, G and H). Further, although nonnucleolar targeting of RPL23aA-mRFP was observed (10.4% of cells, n = 67), it was a significantly more common occurrence with RPL23aB-mRFP (33.3% of cells, P = 0.001). Alignment of Arabidopsis RPL23a isoforms with tobacco RPL23a (Gao et al., 1994) showed that RPL23aB had two nonconservative K substitutions within putative NLS/NoLS domains (Fig. 1A, reference numbering positions 38 and 56) relative to RPL23aA and NtRPL23a. To examine whether these substitutions might confer a competitive advantage for nucleolar accumulation to RPL23aA, we coexpressed both RPL23aA and RPL23aB in the same tobacco epidermal cells. Accordingly, two additional fusion-protein constructs were made where the C-terminal mRFP tag of RPL23aA and RPL23aB was replaced with a GFP variant modified for plant expression, GFP5 (Haseloff et al., 1997). Coexpression of RPL23aA-GFP5 with RPL23aB-mRFP resulted in a high rate of coexclusion from the nucleolus (40% of cells, n = 25; data not shown). Yet in all instances where nucleolar localization was observed, RPL23aA-GFP5 successfully targeted the nucleolus (100% of cells, n = 13), whereas accumulation of RPL23aB-mRFP was inconsistent, as it was frequently excluded or peripherally localized (40 and 20% of cells, respectively, n = 13; Fig. 2, A and B). However, when the two isoforms were coexpressed with the converse set of fluorescent protein tags, RPL23aA-mRFP and RPL23aB-GFP5 showed conucleolar accumulation (100% of cells, n = 13; Fig. 2, C and D). These contradictory findings suggest that the fluorescent protein tags may be affecting RPL23aA/B localization (discussed below) but nevertheless indicate that both isoforms accumulate in the nucleolus.
Figure 2.
Nucleolar colocalization of C-terminally tagged RPL23aA and RPL23aB in tobacco epidermal cells is fluorophore dependent. A to D, CLSM images of tobacco epidermal cells transiently coexpressing RPL23aA-GFP5 and RPL23aB-mRFP (A and B) or RPL23aA-mRFP and RPL23aB-GFP5 (C and D). Solid white arrowheads point to nucleoli; transparent white arrowheads point to nucleolus exclusion zones. Images of the same optical slice were merged to show signal overlap (A–D, right). Bars = 10 μm. [See online article for color version of this figure.]
N-Terminally Tagged RPL23aB Is Excluded from the Nucleolus
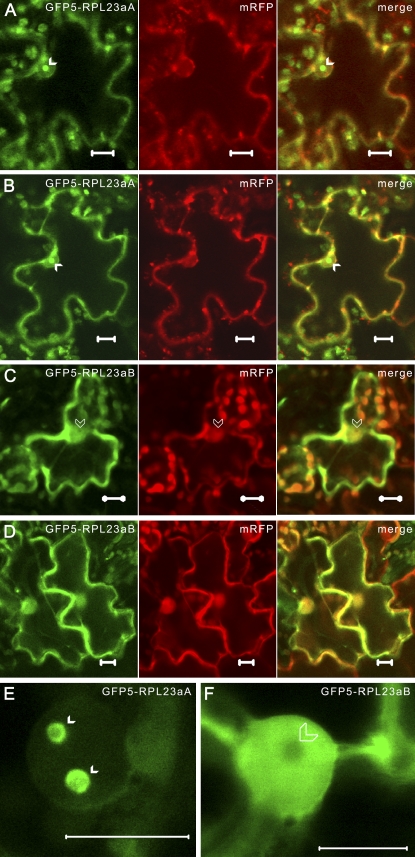
The N-terminal domain of the yeast RPL23a ortholog is necessary for both nuclear localization and LSU biogenesis (van Beekvelt et al., 2001). To determine if we could interfere with the ability of the N-terminal domain of Arabidopsis RPL23a to direct nuclear/nucleolar accumulation, the RPL23a fusion protein constructs were redesigned such that the GFP5 tag was N terminal, separated from RPL23aA and RPL23aB by the GST linker. As a control, we coexpressed GFP5-RPL23aA/B with free mRFP, which labels the cytoplasm and nucleoplasm but is actively excluded from the nucleolus (Campbell et al., 2002). Shifting the 466-residue fluorescent protein tag to the N terminus completely disrupted GFP5-RPL23aB nucleolar localization (0% of cells showed nucleolar localization, n = 50), resulting in a solely cytoplasmic and nucleoplasmic distribution (Fig. 3, C, D, and F). However, the N-terminal tag did not interfere with nucleolar targeting of RPL23aA (100% of cells showed nucleolar localization, n = 50), which accumulated within the nucleolus at levels indistinguishable from RPL23aA-GFP5 (Fig. 3, A, B, and E; compare with Fig. 2, A and B).
Figure 3.
RPL23aB with an N-terminal GFP5 tag is unable to accumulate in the nucleolus in tobacco epidermal cells. A to D, CLSM images of tobacco epidermal cells transiently coexpressing GFP5-RPL23aA and free mRFP (A and B) or GFP5-RPL23aB and free mRFP (C and D). E and F, Close-up images of cells expressing GFP5-RPL23aA (E) or GFP5-RPL23aB (F). Solid white arrowheads point to nucleoli (A, B, and E) and transparent white arrowheads point to nucleolar exclusion zones (C and F). Free mRFP delineates the cytoplasm and nucleoplasm (A–D, middle). Images of the same optical slice were merged to show signal overlap (A–D, right). Bars = 10 μm. [See online article for color version of this figure.]
Silencing of RPL23aA Produces a Strong pointed first leaf Phenotype
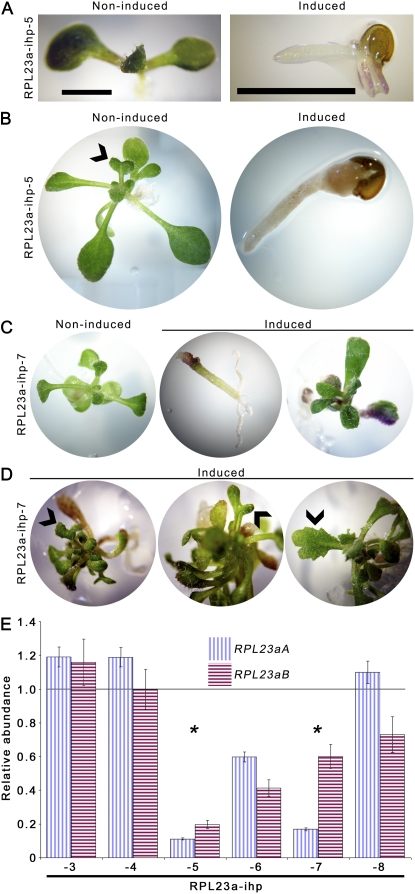
As our previous results suggested that the two RPL23a isoforms are differentially accumulated in the nucleolus, we wanted to investigate the resulting phenotypical consequences of paralog knockdowns in Arabidopsis. Correspondingly, we individually silenced RPL23aA and RPL23aB by engineering paralog-specific self-complementary segments of 3′ untranslated regions within an estradiol-regulated vector, pER8 (Zuo et al., 2000; Guo et al., 2003). Consistent with our results from the characterization of an rpl23ab T-DNA knockout line (R. Degenhardt and P. Bonham-Smith, unpublished data), none of the three T3 transgenic RPL23aB-silencing lines (RPL23aB-ihp-1 to RPL23aB-ihp-3) showed any abnormal phenotype when grown on inductive media (Fig. 4A). However, T3 transgenic RPL23aA-silencing lines (RPL23aA-ihp-1 to RPL23aA-ihp-5) showed retarded growth and developmental abnormalities when plated on inductive media, which were most severe in line RPL23aA-ihp-4. Initially, this line developed narrow, pointed first leaves on short, stubby petioles (Fig. 4, A and C). With further development, it produced leaf or leaf-like organs prolifically (Fig. 4B), with an average of 26 leaf organs at bolting (SEM = 0.88, n = 7) compared to an average of 11 (SEM = 0.24, n = 30) on the induced empty vector control pER8-ihp transgenics. RPL23aA-ihp-4 transgenics also produced irregularly shaped or fused older leaves (Fig. 4, B and D), showed delayed transition to reproductive growth when plated on inductive media (some induced seedlings had not flowered at 7 weeks of age; data not shown), and were shorter than pER8-ihp controls at maturity (15.3 cm, SEM = 1.8 cm, versus 20.4 cm, SEM = 0.9; Fig. 4F). Apical dominance was lost, with older RPL23aA-ihp-4 plants showing substantially increased rosette branching (RPL23aA-ihp-4 = 8.8 rosette reproductive shoots, SEM = 1.4; pER8-ihp-4 = 3.3, SEM = 0.6; Fig. 4F). The RPL23aA-ihp-4 line also had substantially reduced root growth and an abnormal root phenotype characterized by short root hairs and malformed lateral roots (Fig. 4D). The atypical leaf phenotype of RPL23aA-ihp-4 resembled that of pointed first leaf (pfl) mutants characterized for RPS13B and RPS18A knockouts (Van Lijsebettens et al., 1994; Ito et al., 2000), but was unique with respect to a greater delay in the transition to reproductive growth, reduced seed production (data not shown), increased leaf number, longer persistence of the pointed morphology (Fig. 4B), and abnormal root growth (Fig. 4D). Nishimura et al. (2005) have reported that an RPL24B knockout also develops a pfl phenotype, as well as additional defects in patterning of cotyledon vasculature and the gynoecium, which were attributed to a specific role for RPL24 in regulating expression of auxin polar transport and response factors. As RPL23aA-ihp-4 has aberrant phyllotaxis, apical dominance, and root development, all of which are auxin-mediated processes (Reed et al., 1998; Chatfield et al., 2000; Reinhardt, 2005), we investigated whether vascular patterning was also disrupted. Chloral hydrate-cleared RPL23aA-ihp-4 seedlings were examined and found to have venation that deviated dramatically from the closed, reticulate venation of pER8-ihp controls (Fig. 4E). Mutant leaves had substantially reduced venation, little to no tertiary or quaternary veins, open vein loops with distal segments ending freely in the lamina, and aberrant anastamosis that was most apparent in the first leaves where the midvein bifurcated close to the hypocotyl-petiole junction (Fig. 4E). Veins also exhibited reduced lateral orientation, with a predominance for base to tip alignment. We did not observe any defects in gynoecium patterning of RPL23aA-ihp-4 plants that had been transferred at bolting to fresh media augmented with estradiol. However, the inducible system we utilized was not readily amenable to analysis of reproductive organs or fertility due to the half-life of the inducer and the requirement of growing transgenics on sealed plates for both sterility and to prevent rapid oxidation of estradiol.
Figure 4.
Characterization of Arabidopsis RPL23aA- and RPL23aB-silencing mutants. A, The 10-d-old seedlings grown on basal media without or with 10 to 50 μm estradiol (noninduced and induced, respectively). Images shown are representative of T3 transgenics expressing the RPL23aA- or the RPL23aB-silencing cassette (RPL23aA-ihp and RPL23aB-ihp, respectively) or the empty vector control (pER8-ihp). Misshapen leaves with a pointed morphology are indicated by arrowheads. B, The 13-, 18-, and 21-d-old pER8-ihp-2 and RPL23aA-ihp-4 grown on inductive media as above. Bar = 2 mm. C, Comparison of the first four leaves from 13-d-old pER8-ihp-2 and RPL23aA-ihp-4 T3 transgenic seedlings grown on inductive media. Leaves are ordered chronologically (oldest to youngest) left to right. Bars = 2 mm. D, Abnormal leaf and root morphology of 18-d-old RPL23aA-ihp-4 transgenics grown on inductive media. A segment of root from an 18-d-old pER8-ihp-2 transgenic seedling is shown for comparison. Roots are stained with toluidine blue. Arrowheads point to lateral roots. E, Dark-field micrographs showing leaf venation of 14- to 18-d-old pER8-ihp-2 and RPL23aA-ihp-4 T3 transgenic seedlings grown on inductive media and cleared with chloral hydrate. Shown are the first, second, and third leaves and the hypocotyl/petiole junction. Arrowhead shows bifurcation of midveins in the petiole of RPL23aA-ihp-4 transgenics. F, The 38- and 74-d-old (top and bottom, respectively) pER8-ihp-2 and RPL23aA-ihp-4 T3 transgenic seedlings grown on inductive media. Arrowheads in top segments point to rosette flowering branches in RPL23aA-ihp-4 transgenics. Bars = 2 cm. G, qRT-PCR quantification of RPL23aA and RPL23aB transcript abundance in RPL23aA-ihp-1 to -5, RPL23aB-ihp-1 to -3, and pER8-ihp-2 to -4 T3 Arabidopsis transgenic seedlings plated on inductive media. Vertical and horizontal hatched lines represent the ACT7 standardized level of RPL23aA and RPL23aB transcripts, respectively, in transgenic lines on inductive media normalized to the transcript level in noninduced wild-type seedlings (set to 1). Black bars represent SEM. Gray bar represents 100% of wild-type transcript abundance. Asterisks highlight the lines depicted in A to F. [See online article for color version of this figure.]
To ascertain whether the observed phenotypes corresponded to RPL23aA transcript levels, quantitative reverse transcription (qRT)-PCR was conducted on RNA from transgenic seedlings (10–13 d old) grown on inductive and noninductive media. The greatest down-regulation in the RPL23aA transcript was recorded for induced RPL23aA-ihp-4 seedlings (approximately 40% reduction in transcript level; Fig. 4G), while transcript levels in induced empty vector control pER8-ihp transgenics (Fig. 4G) and in induced wild-type seedlings (data not shown) were unaltered. Silencing of RPL23aB was observed in all RPL23aB-ihp transgenic lines (approximately 30%–45% reduction in transcript level). A small degree of cross-silencing was also observed in some of the RPL23aA/B-ihp transgenic lines (A-ihp-2 and -3, B-ihp-1) but did not lead to development of an observable phenotype. Our results suggest that the pfl phenotype is strongly correlated to RPL23aA transcript level.
The RPL23a Family Is Essential for Viability
The purported involvement of RPL23a orthologs in numerous critical ribosomal functions suggests that RPL23a should be essential for plant viability. To test this hypothesis, we designed an RNAi construct that targets a highly conserved region of the ORFs. T3 seed from six transgenic lines (RPL23a-ihp-3 to RPL23a-ihp-7) were screened on inductive media, and the majority of lines showed acute growth defects, characterized by severely retarded development, reduced root growth, atypical leaf and root morphology, accumulation of anthocyanins, prolific leaf organ development, delayed transition to reproductive growth, flower abortion, and early senescence (Fig. 5, C and D; reproductive defects data not shown). Two lines (RPL23a-ihp-5 and -6) were nonviable on inductive media and died postgermination (Fig. 5, A and B). Line RPL23a-ihp-5 was grown for 14 d on noninductive media and transferred to inductive media; thereafter, it began to show symptoms consistent with protein synthesis inhibition (impeded growth, chlorosis, necrosis), which progressively worsened and proved lethal 14 to 21 d postinduction (data not shown). To confirm that observed phenotypes were a direct result of silencing RPL23aA and RPL23aB, we conducted qRT-PCR analyses on RNA from transgenic seedlings (10–13 d old) grown on inductive and noninductive media. Transcript levels of RPL23aA and RPL23aB were dramatically lower in induced seedlings of lines RPL23a-ihp-5 to -7 relative to wild type (Fig. 5E). Line -7, which had a survival rate of approximately 10% to 20% when plated on inductive media, had decreased levels of RPL23aA and increased levels of RPL23aB relative to line -6, which had a survival rate of <5% on inductive media. This suggests that RPL23aB may be capable of functionally compensating for RPL23aA. Overall, findings show that, consistent with its orthologs, the Arabidopsis RPL23a family is essential for viability.
Figure 5.
The Arabidopsis RPL23a family is essential for viability. A to C, Representative images of 11-d-old (A) and 18-d-old (B and C) T3 transgenics expressing the RPL23a family-silencing cassette (RPL23a-ihp) germinated on basal media without or with 10 to 50 μm estradiol (noninduced and induced, respectively). Bars = 2 mm. Abnormal leaf morphology of RPL23a-ihp-5 transgenics on noninductive media, indicated by black arrowhead, suggests leaky expression. D, Aberrant leaf morphology apparent on 29-d-old RPL23a-ihp-7 transgenics grown on inductive media. Black arrowheads indicate leaves with particular anomalous form. E, qRT-PCR quantification of RPL23aA and RPL23aB transcript abundance in RPL23a-ihp-3 to -8 T3 Arabidopsis transgenic seedlings plated on inductive media. Vertical and horizontal hatched lines represent the ACT7 standardized level of RPL23aA and RPL23aB transcripts, respectively, in transgenic lines on inductive media normalized to the transcript level in noninduced wild-type seedlings (set to 1). Black bars represent SEM. Gray bar represents 100% of wild-type transcript abundance. Asterisks highlight the lines depicted in A to D. [See online article for color version of this figure.]
DISCUSSION
We have shown that both RPL23a isoforms accumulate in the nucleolus when transiently expressed in tobacco, providing further support for the hypothesis that this family contributes to ribosome heterogeneity (see Supplemental Results S1 for further discussion; Chang et al., 2005; Giavalisco et al., 2005; Carroll et al., 2008). We found that only RPL23aA is able to tolerate N-terminal GFP5 fusions and C-terminal fusion to mRFP without disrupting localization. Nucleolar targeting of RPL23aB was completely disrupted by N-terminal fusions with GFP5, yet these fusions did not impede nuclear localization, as this construct readily accumulated in the nucleoplasm (Fig. 3, C, D, and F) despite being larger than the size exclusion limit of the plant nuclear pore complex (Merkle, 2003). It could also be argued that nuclear localization of GFP5-RPL23aB results from overexpression of this construct and its slow diffusion into the nucleus (Haasen et al., 1999; Bohnsack et al., 2002), but this is not supported by our observations of nuclear localization in cells with only low expression levels (data not shown). Nucleolar targeting of RPL23aB was also impaired by C-terminal fusion with mRFP but not by an equivalent fusion to GFP5. This may be explained, at least partially, by the biochemical properties of the GST-mRFP/GFP5 tags. Using the EMBOSS toolbox (Rice et al., 2000), we determined that the GST-GFP5 tag is more positively charged (pI = 6.31) and more basic than the GST-mRFP tag (pI = 6.16). RPL23a, like most r-proteins associating with negatively charged rRNA within the nucleolus (Brodersen et al., 2002; Klein et al., 2004), is positively charged (pI = 10.91–10.94) and basic. Thus, the GST-GFP5 tag, attached to RPL23aA and RPL23aB, would be less likely to impede nucleolar localization via charge repulsion than the GST-mRFP tag. Nonetheless, we found that only RPL23aB localization was significantly disrupted by the mRFP tag, and thus we propose that RPL23aA has a stronger NoLS(s) or greater affinity for nucleolus-localized ligands.
We have shown that there are differences in nucleolar targeting of tagged isoforms but not of nuclear accumulation, suggesting that the nine residue differences between RPL23aA and RPL23aB disrupt one or more NoLSs or reduce efficiency of NoLS(s). One divergent region between the RPL23a isoforms occurs within a stretch of basic amino acids containing a putative NoLS/NLS (Kalderon et al., 1984; Dingwall and Laskey, 1991; Weber et al., 2000; Horke et al., 2004). In RPL23aA, this putative NoLS conforms to the core consensus sequence for nucleolin binding in mammals [(K/R)2XK; Xue et al., 1993; Lee et al., 1998; Intine et al., 2004; Wang et al., 2005]. Nucleolin is a major nucleolar protein involved in RNA polymerase I transcription of rDNA, rRNA processing, and nucleocytoplasmic trafficking of RNA and ribonucleoprotein particles (RNPs; Bouvet et al., 1998; Ginisty et al., 1998; Roger et al., 2003; Mongelard and Bouvet, 2007). It has recently been reported that nucleolin also functions in rDNA condensation and maintaining the nucleolus's structure in plants (Pontvianne et al., 2007). A nucleolin-binding motif in RPL23aA could explain why this isoform shows greater affinity for nucleolar accumulation than RPL23aB, which lacks a consensus nucleolin-binding motif. Nucleolin could function as a nucleolar anchor for the r-protein prior to its assembly into preribosomal particles, in the same manner as nucleolin-containing U3 small nucleolar RNPs bind to rDNA to prepare for processing of nascent rRNA transcripts (Caparros-Ruiz et al., 1997). A similar mechanism may function in the monocot rice (Oryza sativa), which also has two members to its RPL23a family. Only one rice isoform has a putative nucleolin-binding motif (OsRPL23a-1; Fig. 1A), suggesting that one member of both the rice and Arabidopsis RPL23a families may have lost (or gained) its nucleolin-binding capacity following the respective duplication events that created the paralogs.
RPL23aA silencing results in development of a pleiotropic phenotype with symptoms similar to other characterized r-protein mutants: RPS5B, RPS13B, RPS18A, and RPL24B (Van Lijsebettens et al., 1994; Ito et al., 2000; Weijers et al., 2001; Nishimura et al., 2005). In each case, loss/reduction of an r-protein leads to reduced cell division, retarded growth, morphological abnormalities, and late flowering. Further, vascular patterning is disrupted in all r-protein mutants (Ito et al., 2000; Weijers et al., 2001; Nishimura et al., 2005), except possibly for RPS18A, where vasculature was not examined (Van Lijsebettens et al., 1994). This phenotype has been replicated in a recently characterized Arabidopsis nucleolin gene knockout mutant, Atnuc-l1 (Kojima et al., 2007; Petricka and Nelson, 2007; Pontvianne et al., 2007). As described above, nucleolin has an established role in rDNA transcription and rRNA processing (Ginisty et al., 1999). For the latter, it assembles with the U3 small nucleolar RNP complex that cleaves nascent rRNA at the 5′ external transcribed spacer primary processing site (Caparros-Ruiz et al., 1997; Saez-Vasquez et al., 2004). In the Atnuc-l1 mutant, pre-rRNA precursor accumulation is decreased, and primary cleavage and subsequent pre-rRNA processing are disrupted (Kojima et al., 2007; Petricka and Nelson, 2007; Pontvianne et al., 2007). In yeast, the RPL23a ortholog is required for efficient pre-rRNA processing (van Beekvelt et al., 2001), and both RPS13 and RPS18 are universally conserved, core structural components of the SSU that bind 18S rRNA and are associated with the pre-rRNA processing complex (Xiang and Lee, 1989; Yusupov et al., 2001; Brodersen et al., 2002; Pontvianne et al., 2007). The striking similarity of phenotype between rpl23aa, rps5b, rps13b, rps18a, rpl24b, and Atnuc-l1 mutants may thus represent the in vivo response to decreased production of processed rRNAs and the concomitant effects on ribosome biogenesis and protein production, with the observed severity gradient being a measure of the extent of the reduction in mature rRNAs. In this scenario, r-proteins likely act indirectly on pre-rRNA processing, with their binding required to induce conformational changes that facilitate pre-rRNA processing or allow the association of other r-proteins and factors involved in processing (Mandiyan et al., 1991; Ban et al., 2000; Wimberly et al., 2000; van Beekvelt et al., 2001).
It is interesting that the phenotype obtained by silencing RPL23aA is not only similar to other r-protein and ribosome biogenesis mutants, but also to plants with disrupted auxin-responsiveness/polar auxin transport. For example, vascular patterning abnormalities similar to those resulting from impaired ribosome biogenesis have also been reported for ettin/auxin response factor3 (arf3) and monopteros/arf5, which are auxin-regulated transcription factor mutants (Przemeck et al., 1996; Sessions et al., 1997; Nemhauser et al., 2000); pin-formed1, an auxin efflux transporter mutant (Goto et al., 1991; Bennett et al., 1995; Petrasek et al., 2006); auxin resistant1, a mutant with reduced expression of a subunit of an enzyme complex that activates the auxin-regulated SCFTIR ubiquitin-ligase (del Pozo et al., 2002; Deyholos et al., 2003); hve1/cand1, a mutant with reduced levels of a regulator of SCFTIR assembly (Alonso-Peral et al., 2006); and lop1/tornado1 and tornado2, which are mutant in establishing/maintaining auxin homeostasis (Carland and McHale, 1996; Cnops et al., 2000, 2006). Moreover, treatment of wild-type plants with inhibitors of polar auxin transport has also been reported to disrupt vasculature, causing veins to be restricted to leaf margins and disrupting the midvein (Mattsson et al., 1999; Sieburth, 1999). Petricka and Nelson (2007) determined that auxin response maxima (detected with a synthetic reporter construct, DR5-GUS; Ulmasov et al., 1997; Sabatini et al., 1999), which predict future sites of higher order venation (primary, secondary, tertiary, etc.; Mattsson et al., 2003), were mislocalized in developing leaves of their Atnuc-l1 mutant, such that response maxima were not observed in leaf margins or at sites predicting tertiary or quaternary vein development. This reduction in marginal auxin response maxima was maintained when Atnuc-l1 leaves were treated with an inhibitor of polar auxin transport, which in wild-type plants restricts response maxima to leaf margins (Mattsson et al., 2003; Petricka and Nelson, 2007). It is suggested that the observed pointed/narrow leaf and abnormal venation phenotypes result from a lack of marginal auxin sources/response factors, causing a reduction in auxin-mediated laterally directed cell division and negating procambial differentiation at leaf margins, leading to a decrease in lateral expansion of the leaf and reduced vasculature with predominantly base to tip vein orientation (Petricka and Nelson, 2007). A more general reduction of auxin response would also explain our observation of a loss of apical dominance, as auxin/auxin-responsiveness at the shoot apex is responsible for preventing the formation of lateral buds (Lincoln et al., 1990; Cline, 1991). Thus, perhaps the simplest explanation for the observed relationship between ribosome biogenesis and auxin transport/responsiveness is that synthesis of specific response/trafficking proteins is decreased below a certain required spatiotemporal threshold, disrupting auxin-regulated development and altering patterning. However, this explanation would seem to require a very specific reduction in the translation of auxin-response/-trafficking genes.
Another possibility is that auxin homeostasis is linked to cellular translational status through microRNAs (miRNAs). miRNAs are a class of small RNAs (approximately 22 nucleotides) that originate from long single-stranded RNAs transcribed from endogenous genes by RNA polymerase II (Xie et al., 2005). The single-stranded RNAs possess internal complementarity allowing fold-back and formation of double-stranded RNA that is processed by DICER-LIKE1 (DCL1) in conjunction with the double-stranded RNA-binding protein HYL1 (for review, see Mallory and Vaucheret, 2006). The double-stranded miRNAs are subsequently methylated by HEN1 and loaded onto ARGONAUTE1 (ARG1), which facilitates miRNA maturation and directs cleavage of transcripts with near-perfect complementarity to the miRNAs. Support for a link between miRNAs and auxin homeostasis is 2-fold. First, mutants in miRNA biogenesis (dcl1, hyl1, hen1, arg1) have a range of severe pleiotropic symptoms consistent with disrupted auxin responses, including altered leaf morphology, reduced stature, aberrant gynoecium patterning, decreased apical dominance, atypical phyllotaxis, and irregular vasculature (Bohmert et al., 1998; Jacobsen et al., 1999; Lu and Fedoroff, 2000; Chen et al., 2002). Secondly, several experimentally and computationally determined targets of miRNAs are involved in the auxin response, including TIR1, a component of the SCFTIR complex; NAC1/NAM, an auxin signal transducer; and ARF6, ARF8, ARF10, ARF16, and ARF17 (Bonnet et al., 2004; Vazquez et al., 2004; Xie et al., 2007; for review, see Eckardt, 2005; Mallory and Vaucheret, 2006). How then are miRNA-mediated processes dependent on translational status? A possible answer comes from recent experiments in fruit fly (Drosophila melanogaster), where Eulalio et al. (2007) fused the 3′ untranslated region of different miRNA targets to the firefly luciferase ORF and quantified luciferase transcript levels in cell culture following incubation with or without cognate miRNAs. Transcript levels were shown to decrease significantly following incubation with miRNAs; however, treatment of cells with translation inhibitors (cycloheximide, homoharringtonine, and hippuristanol) caused the stabilization of a subset of miRNA targets. Eulalio et al. (2007) attributed the translational dependency of miRNA target degradation to the possibility that silencing of some targets is initiated after translation commences. A similar mechanism operating in plants would provide an elegant model to explain our findings. Under normal conditions, translation in mitotically active tissues is occurring at a very high rate, allowing the accurate degradation of miRNA targets (which include a large number of auxin response transcripts) and the maintenance of auxin homeostasis. However, when ribosome biogenesis and, consequently, translation are impaired, some miRNA targets are stabilized, resulting in the disruption of auxin feedback cycles, leading to an array of developmental abnormalities. This model could, for example, explain our finding of reduced apical dominance; it has previously been shown that transgenic Arabidopsis expressing an engineered miRNA-resistant version of ARF17 have increased levels of ARF17 transcript (Mallory et al., 2005). This line had a variety of auxin-related developmental defects and showed an increase in transcript levels of two auxin-conjugating enzymes, GH3.2/YDK1 and GH3.3, suggesting that ARF17 functions as a repressor of the conjugating enzymes. Overexpression of YDK1 has previously been shown to result in reduced apical dominance (Takase et al., 2004), presumably because of a reduction in free auxin acting on the meristem to inhibit the formation of lateral buds. It is clear from the phenotypes of auxin response, miRNA synthesis, and ribosome biogenesis mutants that the regulation of auxin homeostasis is very complex, but it is an intriguing possibility that a common link relates all three processes.
We observed that no phenotype was detected as a result of silencing RPL23aB or in a T-DNA insertion rpl23ab knockout line (R. Degenhardt and P. Bonham-Smith, unpublished data). This adds to the mounting body of evidence indicating that, despite often overlapping transcript accumulation patterns, disparity exists in the requirement of r-protein paralogs for normal development (Barakat et al., 2001; McIntosh and Bonham-Smith, 2006). For example, expression profiles of Arabidopsis RPS5A and RPS5B (Weijers et al., 2001), RPS18A to RPS18C (Van Lijsebettens et al., 1994; Vanderhaeghen et al., 2006), and RPL23aA and RPL23aB (McIntosh and Bonham-Smith, 2005) indicate that transcripts from all paralogs accumulate to the highest levels in mitogenic tissues (e.g. meristems and young leaves) and the lowest in nondividing tissues. In all cases, the absolute transcript level varies, but the relative contribution from each paralog remains fairly constant. This is exemplified by transcript profiling of RPS18A to RPS18C, indicating that RPS18A, RPS18B, and RPS18C represent approximately 27%, 16%, and 57%, respectively, of the total RPS18 transcript pool (Vanderhaeghen et al., 2006). Yet, for each of these families, it has been demonstrated that knockout or knockdown mutants of a single paralog triggers development of the atypical phenotype (RPS5B, RPS18A, and RPL23aA; Van Lijsebettens et al., 1994; Weijers et al., 2001; this work). These findings could easily be reconciled if the phenotype-triggering mutation disrupted the predominantly expressed gene or if a corresponding mutation in another paralog induced the same phenotypic response, but this is not supported by experimental data. For example, RPS18A represents roughly only one-quarter of the total RPS18 transcript, and yet its loss produces the pfl phenotype (Van Lijsebettens et al., 1994). Similarly, this phenotype was reproduced in a knockout of RPS13B, which also makes only a small contribution to the RPS13 transcript pool (Ito et al., 2000). It could also be the case that differential spatiotemporal expression of paralogs leads to observed knockout/knockdown phenotypes. It has been shown that RPS5B is predominately localized to cell division zones, while RPS5A is found in less mitotically active regions (Weijers et al., 2001). Yet, this too would be an oversimplified explanation given our findings of overlapping GUS-staining directed by RPL23aA and RPL23aB 5′ regulatory regions (K. McIntosh, R. Degenhardt, and P. Bonham-Smith, unpublished data). Moreover, evidence to date suggests that plant r-protein paralogs are differentially required for normal development, with ancillary paralogs functioning only under exceptional conditions, perhaps mediated through spatiotemporal regulation, stimulus-induced expression, assumption of extra-ribosomal roles (for review, see Wool, 1996), or via disparity in cognate binding partners (e.g. affinity for nucleolin).
We have demonstrated that the RPL23a family is essential for viability in Arabidopsis. This is in agreement with previous findings in numerous prokaryotes, yeast, and Caenorhabditis elegans (Kamath et al., 2003; Zhang et al., 2004). Induction of the RPL23a family-silencing construct results in a range of severe pleiotropic phenotypes characterized by developmental defects, reduced shoot and root growth, flower abortion, and, in the most severe case, death upon germination. Given the established importance of RPL23a, our inducibly lethal RPL23a-silencing lines could be useful tools for the in vivo analysis of rRNA processing, LSU assembly, and protein synthesis defects. This adds RPL23a to a growing list of r-proteins from higher eukaryotes that are essential for viability, including RPS5 from Arabidopsis (Weijers et al., 2001), and RPS2, RPS3, RPS4, RPS5, RPS6, RPS13, RPS14, RPL5, RPL9, RPL14, RPL19, and RPL38 from fruit fly (for review, see Saboe-Larssen et al., 1998; Marygold et al., 2005).
Herein, we have demonstrated that the Arabidopsis RPL23a paralogs are differentially targeted to the nucleolus. Disparity within a putative NoLS appears to be responsible, but future work is necessary to determine whether this directly affects their respective abilities to bind to the nucleolus structural protein, nucleolin. We have shown by RNAi-mediated silencing that RPL23aB is phenotypically dispensable, while RPL23aA knockdown leads to a severe pfl phenotype that is possibly due to impaired pre-rRNA processing and consequential effects on miRNA target stability and auxin homeostasis. How directly r-proteins, and particularly RPL23a, are involved in pre-rRNA processing has yet to be elucidated. We have determined that the RPL23a family is essential for survival by the nonviability of transgenic lines that silence both paralogs. These lines have a normal lifecycle on noninductive media and thus may be useful tools for studying ribosome biogenesis at different developmental stages.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) ‘Columbia-0’ and tobacco (Nicotiana tabacum) ‘Petit Havana’ were used for all experiments. Unless otherwise stated, Arabidopsis was plated on one-half-strength Murashige and Skoog media (Sigma-Aldrich) supplemented with 0.8% phytagar (Invitrogen) and 1.5% Suc (hereafter called basal media). Growth conditions are provided in Supplemental Materials and Methods S1.
Fluorescent Protein and RNAi Constructs
Standard techniques were followed for all molecular cloning (Sambrook et al., 1989). All cloning products were verified by automated sequencing (National Research Council-Plant Biotechnology Institute). The fluorescent proteins used were monomeric GFP5 modified for plants (Haseloff et al., 1997), mRFP (Campbell et al., 2002), and EGFP (ClonTech). Details of cloning methodology are available in the Supplemental Materials and Methods S1.
For RNAi-mediated gene silencing, targeted regions of RPL23aA and RPL23aB were cloned in sense and antisense orientation, separated by an intron, into the binary vector pER8 (Zuo et al., 2000), creating the estrogen-inducible, hairpin RNA-forming cassettes RPL23aA-ihp, RPL23aB-ihp, and RPL23a-ihp. The pER8 vector system is strictly regulated by estrogen and shows no nonspecific effects on plant growth or development (Zuo et al., 2000). Details of cloning methodology are available in the Supplemental Materials and Methods S1.
Transient Expression in Tobacco
Fluorescent protein constructs within binary vectors were used to transform Agrobacterium tumefaciens strain LBA4404 (Hoekema et al., 1983) via electroporation. All constructs in pGREEN were coelectroporated with pSOUP, which must be coresident in A. tumefaciens to provide the replication functions, in trans, for pGREEN (Hellens et al., 2000). Tobacco infiltrations were carried out following previously described protocols (Batoko et al., 2000; Brandizzi et al., 2002; Sparkes et al., 2006). Following infiltration, tobacco plants were returned to the growth chamber for 48 to 72 h prior to visualization with the CLSM.
Confocal Microscopy
Live cell imaging was conducted with an inverted Zeiss LSM 510 META CLSM using previously described settings (Brandizzi et al., 2002; Runions et al., 2006), with the exception that a 585- to 615-nm bandpass filter was used to detect mRFP. These settings prevented spectral bleed-through of fluorescence emission from EGFP/GFP5 + mRFP during coexpression experiments. Acquired images were processed with Zeiss LSM Image Browser software and exported to Adobe Photoshop 7.0 software for figure preparations.
Generation and Induction of Stable RNAi Transgenics
Stable transgenics carrying the inducible silencing cassettes were generated via the floral dip protocol (Clough and Bent, 1998). As a negative control, Arabidopsis was transformed with the pER8 empty vector using the same technique. T3 transgenic lines, created by selecting each successive generation on basal media supplemented with 25 μg mL−1 hygromycin (Invivogen) and 200 μg mL−1 cefotaxime (Sanofi-Aventis), were used for RNAi studies. Seed from independently transformed lines was plated on basal media supplemented with 0 to 100 μm estradiol. Transgenics grown on noninductive media (0 μm estradiol) were occasionally transferred to inductive media (2–100 μm estradiol) to observe responses.
Dark-Field Microscopy
Digital images of transgenics were taken with a Zeiss Stemi 2000-C stereomicroscope. Root sections were observed following staining with toluidine blue. Vasculature was examined by fixing 14- to 18-d-old seedlings overnight in 3:1 ethanol:acetic acid. Fixed seedlings were processed through an ethanol series (80%, 90%, 95%, and 100% ethanol) and cleared by incubation overnight in saturated chloral hydrate. Images were taken at 100 to 200× zoom with a Zeiss Axioskop microscope equipped with a dark-field diaphragm. Approximately eight to 10 cleared seedlings were analyzed for each genotype (pER8-ihp-4 and RPL23aA-ihp-4).
qPCR
qPCR was performed using RNA extracted from 10- to 18-d-old whole seedlings with an iQ5 real-time PCR detection system (Bio-Rad). For each sample, amplifications of RPL23aA, RPL23aB, and ACT7 were performed in triplicate, within the same qPCR run, and only one amplicon was produced per reaction. The ACT7 gene was used as an internal control to standardize RPL23aA/B levels in induced and noninduced transgenics, and threshold cycle changes were compared to standardized levels in noninduced wild-type seedlings of equivalent age using the comparative threshold cycle method (Livak and Schmittgen, 2001). The entire procedure (RNA extraction, first-strand synthesis, qPCR) was repeated for a minimum of three biological replicates. Data was obtained using the iQ5 Optical System software (Bio-Rad) and subsequently exported to Microsoft Excel for summarizing. Detailed methodology for RNA extractions, first-strand synthesis, primer selection and validation, and qPCR optimization are available in the Supplemental Materials and Methods S1.
Accession Numbers
GenBank accession numbers for materials/sequences described in this manuscript are X65305 (pGEM4), U13855 (pGEX-4T-3), AF234306 (pCAMBIA1381z), AF234301 (pCAMBIA1380), AF309825 (pER8), X52327 (pBSKS+), U87973 (GFP5), AF506027 (mRFP), Q07761 (tobacco RPL23a), EAW51131 (human RPL23a), NP_014514 (yeast RPL25), NP_001042974 (rice RPL23a-1), CAC09511 (rice RPL23a-2), AC004218 (Arabidopsis F12L6 BAC), At2g39460 (Arabidopsis RPL23aA), At3g55280 (Arabidopsis RPL23aB), At4g25630 (Arabidopsis FIB2), and AT5G09810 (Arabidopsis ACT7).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleoli of tobacco epidermal cells exhibit spatial separation.
Supplemental Figure S2. Fluorescent protein-tagged FIB2, RPL23aA, and RPL23aB accumulate outside the nucleolus in the cytoplasm and nucleoplasm.
Supplemental Results S1. Ribosome heterogeneity and properties of tobacco nucleoli.
Supplemental Materials and Methods S1.
Supplementary Material
Acknowledgments
We are grateful to Federica Brandizzi (Michigan State University) for the pVKH18En6 binary vector and the monomeric GFP5 and mRFP fluorescent protein constructs, Manuel Echeverría (Université de Perpignan) for the ppk100-FIB2-EGFP construct, and Nam-Hai Chua (Rockefeller University) for the pER8 binary vector and pSK-int cloning vector. We are indebted to Sally Hanton and Loren Matheson for their assistance with tobacco infiltrations and confocal microscopy. We thank Jacqueline Hulm, Donna Lindsay, and Heather Wakely for technical assistance. Special thanks to Kerri McIntosh for cloning the RPL23a paralogs and for helpful discussions.
This work was supported by the Natural Sciences and Engineering Research Council of Canada.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Peta C. Bonham-Smith (peta.bonhams@usask.ca).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso-Peral MM, Candela H, del Pozo JC, Martinez-Laborda A, Ponce MR, Micol JL (2006) The HVE/CAND1 gene is required for the early patterning of leaf venation in Arabidopsis. Development 133 3755–3766 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J (1998) Cytoplasmic ribosomes of higher plants. In J Bailey-Serres, DR Gallie, eds, A Look Beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. American Society of Plant Physiologists, Rockville, MD, pp 125–144
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289 905–920 [DOI] [PubMed] [Google Scholar]
- Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J (2001) The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 127 398–415 [PMC free article] [PubMed] [Google Scholar]
- Barneche F, Steinmetz F, Echeverria M (2000) Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J Biol Chem 275 27212–27220 [DOI] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I (2000) A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12 2201–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science 278 2123–2126 [DOI] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J 8 505–520 [Google Scholar]
- Beven AF, Simpson GG, Brown JWS, Shaw PJ (1995) The organization of spliceosomal components in the nuclei of higher-plants. J Cell Sci 108 509–518 [DOI] [PubMed] [Google Scholar]
- Bogorad L (1975) Evolution of organelles and eukaryotic genomes. Science 188 891–898 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C (1998) AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J 17 170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Gorlich D (2002) Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J 21 6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouze P, Van de Peer Y (2004) Detection of 91 potential in plant conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci USA 101 11511–11516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F (1998) Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem 273 19025–19029 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N (2002) The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM, Carter AP, Wimberly BT, Ramakrishnan V (2002) Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol 316 725–768 [DOI] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparros-Ruiz D, Lahmy S, Piersanti S, Echeverria M (1997) Two ribosomal DNA-binding factors interact with a cluster of motifs on the 5′ external transcribed spacer, upstream from the primary pre-rRNA processing site in a higher plant. Eur J Biochem 247 981–989 [DOI] [PubMed] [Google Scholar]
- Carland FM, McHale NA (1996) LOP1: a gene involved in auxin transport and vascular patterning in Arabidopsis. Development 122 1811–1819 [DOI] [PubMed] [Google Scholar]
- Carroll AJ, Heazlewood JL, Ito J, Millar AH (2008) Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol Cell Proteomics 7 347–369 [DOI] [PubMed] [Google Scholar]
- Chang IF, Szick-Miranda K, Pan S, Bailey-Serres J (2005) Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol 137 848–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield SP, Stirnberg P, Forde BG, Leyser O (2000) The hormonal regulation of axillary bud growth in Arabidopsis. Plant J 24 159–169 [DOI] [PubMed] [Google Scholar]
- Chen XM, Liu J, Cheng YL, Jia DX (2002) HEN1 functions plelotropically in Arabidopsis development and acts in C function in the flower. Development 129 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MG (1991) Apical dominance. Bot Rev 57 318–358 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cnops G, Neyt P, Raes J, Petrarulo M, Nelissen H, Malenica N, Luschnig C, Tietz O, Ditengou F, Palme K, et al (2006) The TORNADO1 and TORNADO2 genes function in several patterning processes during early leaf development in Arabidopsis thaliana. Plant Cell 18 852–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnops G, Wang X, Linstead P, Van Montagu M, Van Lijsebettens M, Dolan L (2000) TORNADO1 and TORNADO2 are required for the specification of radial and circumferential pattern in the Arabidopsis root. Development 127 3385–3394 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos MK, Cavaness GF, Hall B, King E, Punwani J, Van Norman J, Sieburth LE (2003) VARICOSE, a WD-domain protein, is required for leaf blade development. Development 130 6577–6588 [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA (1991) Nuclear targeting sequences: a consensus. Trends Biochem Sci 16 478–481 [DOI] [PubMed] [Google Scholar]
- Eckardt NA (2005) MicroRNAs regulate auxin homeostasis and plant development. Plant Cell 17 1335–1338 [Google Scholar]
- El-Baradi TTAL, de Regt VCHF, Planta RJ, Nierhaus KH, Raue HA (1987) Interaction of ribosomal proteins L25 from yeast and EL23 from E. coli with yeast 26S and mouse 28S rRNA. Biochimie 69 939–948 [DOI] [PubMed] [Google Scholar]
- El-Baradi TTAL, Raue HA, de Regt VCHF, Planta RJ (1984) Stepwise dissociation of yeast 60S ribosomal subunits by LiCl and identification of L25 as a primary 26S rRNA binding protein. Eur J Biochem 144 393–400 [DOI] [PubMed] [Google Scholar]
- El-Baradi TTAL, Raue HA, de Regt VCHF, Verbree EC, Planta RJ (1985) Yeast ribosomal protein L25 binds to an evolutionary conserved site on yeast 26S and E. coli 23S rRNA. EMBO J 4 2101–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E (2007) Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev 21 2558–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Kim SR, Chung YY, Lee JM, An G (1994) Developmental and environmental regulation of two ribosomal protein genes in tobacco. Plant Mol Biol 25 761–770 [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Wilson D, Kreitler T, Lehrach H, Klose J, Gobom J, Fucini P (2005) High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol Biol 57 577–591 [DOI] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P (1998) Nucleolin functions in the first step of ribosomal RNA processing. EMBO J 17 1476–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Roger B, Bouvet P (1999) Structure and functions of nucleolin. J Cell Sci 112 761–772 [DOI] [PubMed] [Google Scholar]
- Goto N, Katoh N, Kranz AR (1991) Morphogenesis of floral organs in Arabidopsis: predominant carpel formation of the PIN-FORMED mutant. Jpn J Genet 66 551–567 [Google Scholar]
- Grebenok RJ, Pierson E, Lambert GM, Gong FC, Afonso CL, Haldeman-Cahill R, Carrington JC, Galbraith DW (1997) Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J 11 573–586 [DOI] [PubMed] [Google Scholar]
- Guarinos E, Santos C, Sánchez A, Qiu DY, Remacha M, Ballesta JPG (2003) Tag-mediated fractionation of yeast ribosome populations proves the monomeric organization of the eukaryotic ribosomal stalk structure. Mol Microbiol 50 703–712 [DOI] [PubMed] [Google Scholar]
- Guo HS, Fei JF, Xie Q, Chua NH (2003) A chemical-regulated inducible RNAi system in plants. Plant J 34 383–392 [DOI] [PubMed] [Google Scholar]
- Haasen D, Kohler C, Neuhaus G, Merkle T (1999) Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J 20 695–705 [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, Frank J, Beckmann R (2004) Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427 808–814 [DOI] [PubMed] [Google Scholar]
- Hanson CL, Videler H, Santos C, Ballesta JPG, Robinson CV (2004) Mass spectrometry of ribosomes from Saccharomyces cerevisiae: implications for assembly of the stalk complex. J Biol Chem 279 42750–42757 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42 819–832 [DOI] [PubMed] [Google Scholar]
- Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA (1983) A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303 179–180 [Google Scholar]
- Horke S, Reumann K, Schweizer M, Will H, Heise T (2004) Nuclear trafficking of La protein depends on a newly identified nucleolar localization signal and the ability to bind RNA. J Biol Chem 279 26563–26570 [DOI] [PubMed] [Google Scholar]
- Hulm JL, McIntosh KB, Bonham-Smith PC (2005) Variation in transcript abundance among the four members of the Arabidopsis thaliana ribosomal protein S15a gene family. Plant Sci 169 267–278 [Google Scholar]
- Intine RV, Dundr M, Vassilev A, Schwartz E, Zhao Y, Zhao Y, DePamphilis ML, Maraia RJ (2004) Nonphosphorylated human La antigen interacts with nucleolin at nucleolar sites involved in rRNA biogenesis. Mol Cell Biol 24 10894–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Kim GT, Shinozaki K (2000) Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J 22 257–264 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM (1999) Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126 5231–5243 [DOI] [PubMed] [Google Scholar]
- Jakel S, Gorlich D (1998) Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J 17 4491–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeeninga RE, Venema J, Raue HA (1996) Rat RL23a ribosomal protein efficiently competes with its Saccharomyces cerevisiae L25 homologue for assembly into 60 S subunits. J Mol Biol 263 648–656 [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE (1984) A short amino-acid sequence able to specify nuclear location. Cell 39 499–509 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231–237 [DOI] [PubMed] [Google Scholar]
- Kim SH, Ryabov EV, Kalinina NO, Rakitina DV, Gillespie T, MacFarlane S, Haupt S, Brown JWS, Taliansky M (2007) Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J 26 2169–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi F, Yoshida T, Aiso S (1996) Location of NRAMP1 molecule on the plasma membrane and its association with microtubules. Mol Immunol 33 1241–1246 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA (2004) The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol 340 141–177 [DOI] [PubMed] [Google Scholar]
- Kojima H, Suzuki T, Kato T, Enomoto K, Sato S, Kato T, Tabata S, Saez-Vasquez J, Manuel E, Nakagawa T, et al (2007) Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J 49 1053–1063 [DOI] [PubMed] [Google Scholar]
- Kooi EA, Rutgers CA, Kleijmeer MJ, Vantriet J, Venema J, Raue HA (1994) Mutational analysis of the C-terminal region of Saccharomyces cerevisiae ribosomal protein-L25 in-vitro and in-vivo demonstrates the presence of 2 distinct functional elements. J Mol Biol 240 243–255 [DOI] [PubMed] [Google Scholar]
- Lecompte O, Ripp R, Thierry JC, Moras D, Poch O (2002) Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res 30 5382–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Chang SC, Chen CJ, Chang MF (1998) The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J Biol Chem 273 7650–7656 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lu C, Fedoroff N (2000) A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T, Ferbitz L, Deuerling E, Ban N (2005) A cradle for new proteins: trigger factor at the ribosorne. Curr Opin Struct Biol 15 204–212 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17 1360–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet 38 S31–S36 [DOI] [PubMed] [Google Scholar]
- Mandiyan V, Tumminia SJ, Wall JS, Hainfeld JF, Boublik M (1991) Assembly of the Escherichia coli 30s ribosomal-subunit reveals protein-dependent folding of the 16S ribosomal-RNA domains. Proc Natl Acad Sci USA 88 8174–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Coelho CMA, Leevers SJ (2005) Genetic analysis of RpL38 and RpL5, two minute genes located in the centric heterochromatin of chromosome 2 of Drosophila melanogaster. Genetics 169 683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J, Ckurshumova W, Berleth T (2003) Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol 131 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J, Sung ZR, Berleth T (1999) Responses of plant vascular systems to auxin transport inhibition. Development 126 2979–2991 [DOI] [PubMed] [Google Scholar]
- McIntosh KB, Bonham-Smith PC (2001) Establishment of Arabidopsis thaliana ribosomal protein RPL23A-1 as a functional homologue of Saccharomyces cerevisiae ribosomal protein L25. Plant Mol Biol 46 673–682 [DOI] [PubMed] [Google Scholar]
- McIntosh KB, Bonham-Smith PC (2005) The two ribosomal protein L23A genes are differentially transcribed in Arabidopsis thaliana. Genome 48 443–454 [DOI] [PubMed] [Google Scholar]
- McIntosh KB, Bonham-Smith PC (2006) Ribosomal protein gene regulation: what about plants? Can J Bot 84 342–362 [Google Scholar]
- Menetret JF, Hegde RS, Heinrich SU, Chandramouli P, Ludtke SJ, Rapoport TA, Akey CW (2005) Architecture of the ribosome-channel complex derived from native membranes. J Mol Biol 348 445–457 [DOI] [PubMed] [Google Scholar]
- Merkle T (2003) Nucleo-cytoplasmic partitioning of proteins in plants: implications for the regulation of environmental and developmental signalling. Curr Genet 44 231–260 [DOI] [PubMed] [Google Scholar]
- Mongelard F, Bouvet P (2007) Nucleolin: a multiFACeTed protein. Trends Cell Biol 17 80–86 [DOI] [PubMed] [Google Scholar]
- Morgan DG, Menetret JF, Neuhof A, Rapoport TA, Akey CW (2002) Structure of the mammalian ribosome-channel complex at 17 angstrom resolution. J Mol Biol 324 871–886 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Feldman LJ, Zambryski PC (2000) Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127 3877–3888 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K (2005) The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA (2000) The structural basis of ribosome activity in peptide bond synthesis. Science 289 920–930 [DOI] [PubMed] [Google Scholar]
- Ookata K, Hisanaga S, Bulinski JC, Murofushi H, Aizawa H, Itoh TJ, Hotani H, Okumura E, Tachibana K, Kishimoto T (1995) Cyclin-B interaction with microtubule-associated protein-4 (Map4) targets P34(Cdc2) kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J Cell Biol 128 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35 D883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, et al (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312 914–918 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Nelson TM (2007) Arabidopsis nucleolin affects plant development and patterning. Plant Physiol 144 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planta RJ (1997) Regulation of ribosome synthesis in yeast. Yeast 13 1505–1518 [DOI] [PubMed] [Google Scholar]
- Pontvianne F, Matia I, Douet J, Tourmente S, Medina FJ, Echeverria M, Saez-Vasquez J (2007) Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol Biol Cell 18 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B (2002) Distinct modes of signal recognition particle interaction with the ribosome. Science 297 1345–1348 [DOI] [PubMed] [Google Scholar]
- Popescu SC, Tumer NE (2004) Silencing of ribosomal protein L3 genes in N. tabacum reveals coordinate expression and significant alterations in plant growth, development and ribosome biogenesis. Plant J 39 29–44 [DOI] [PubMed] [Google Scholar]
- Przemeck GKH, Mattsson J, Hardtke CS, Sung ZR, Berleth T (1996) Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200 229–237 [DOI] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK (1998) Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D (2005) Regulation of phyllotaxis. Int J Dev Biol 49 539–546 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16 276–277 [DOI] [PubMed] [Google Scholar]
- Roger B, Moisand A, Amalric F, Bouvet P (2003) Nucleolin provides a link between RNA polymerase I transcription and pre-ribosome assembly. Chromosoma 111 399–407 [DOI] [PubMed] [Google Scholar]
- Runions J, Brach T, Kuhner S, Hawes C (2006) Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J Exp Bot 57 43–50 [DOI] [PubMed] [Google Scholar]
- Rutgers CA, Rientjes JMJ, van't Riet J, Raue HA (1991) rRNA binding domain of yeast ribosomal protein L25: identification of its borders and a key leucine residue. J Mol Biol 218 375–385 [DOI] [PubMed] [Google Scholar]
- Rutgers CA, Schaap PJ, van't Riet J, Woldringh CL, Raue HA (1990) In vivo and in vitro analysis of structure-function relationships in ribosomal protein L25 from Saccharomyces cerevisiae. Biochim Biophys Acta 1050 74–79 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472 [DOI] [PubMed] [Google Scholar]
- Saboe-Larssen S, Lyamouri M, Merriam J, Oksvold MP, Lambertsson A (1998) Ribosomal protein insufficiency and the minute syndrome in Drosophila: a dose-response relationship. Genetics 148 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Vasquez J, Caparros-Ruiz D, Barneche F, Echeverria M (2004) A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol Cell Biol 24 7284–7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schaap PJ, van't Riet J, Woldringh CL, Raue HA (1991) Identification and functional analysis of the nuclear localization signals of ribosomal protein L25 from Saccharomyces cerevisiae. J Mol Biol 221 225–237 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD (2005) Structures of the bacterial ribosome at 3.5 A resolution. Science 310 827–834 [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC (1997) ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124 4481–4491 [DOI] [PubMed] [Google Scholar]
- Sieburth LE (1999) Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol 121 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CMT, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J (2001) Structure of the 80S ribosome from Saccharomyces cerevisiae: tRNA-ribosome and subunit-subunit interactions. Cell 107 373–386 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protocols 1 2019–2025 [DOI] [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, Shimada H, Manabe K, Matsui M (2004) ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J 37 471–483 [DOI] [PubMed] [Google Scholar]
- Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC (1993) Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-ribosomal-RNA processing, pre-ribosomal-RNA methylation, and ribosome assembly. Cell 72 443–457 [DOI] [PubMed] [Google Scholar]
- Tornow J, Santangelo GM (1994) Saccharomyces cerevisiae ribosomal protein L37 is encoded by duplicate genes that are differentially expressed. Curr Genet 25 480–487 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beekvelt CA, de Graaff-Vincent M, Faber AW, van 't Riet J, Venema J, Raue HA (2001) All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res 29 5001–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, Van Montagu M (1994) An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J 13 3378–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhaeghen R, De Clercq R, Karimi M, Van Montagu M, Hilson P, Van Lijsebettens M (2006) Leader sequence of a plant ribosomal protein gene with complementarity to the 18S rRNA triggers in vitro cap-independent translation. FEBS Lett 580 2630–2636 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crete P, Vaucheret H (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14 346–351 [DOI] [PubMed] [Google Scholar]
- Wang YH, Chen YH, Lu JH, Tsai HJ (2005) A 23-amino acid motif spanning the basic domain targets zebrafish myogenic regulatory factor Myf5 into nucleolus. DNA Cell Biol 24 651–660 [DOI] [PubMed] [Google Scholar]
- Warner JR, Mitra G, Schwindinger WF, Studeny M, Fried HM (1985) Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating messenger-RNA splicing, translational initiation, and protein-turnover. Mol Cell Biol 5 1512–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Kuo ML, Bothner B, DiGiammarino EL, Kriwacki RW, Roussel MF, Sherr CJ (2000) Cooperative signals governing ARF-Mdm2 interaction and nucleolar localization of the complex. Mol Cell Biol 20 2517–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hookaas P, Offringa R (2001) An Arabidopsis minute-like phenotype caused by a semi-dominant mutation in a ribosomal protein S5 gene. Development 128 4289–4299 [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V (2000) Structure of the 30S ribosomal subunit. Nature 407 327–339 [DOI] [PubMed] [Google Scholar]
- Wool IG (1996) Extraribosomal functions of ribosomal proteins. Trends Biochem Sci 21 164–165 [PubMed] [Google Scholar]
- Xiang RH, Lee JC (1989) Identification of proteins crosslinked to RNA in 40S ribosomal-subunits of Saccharomyces-cerevisiae. Biochimie 71 1201–1204 [DOI] [PubMed] [Google Scholar]
- Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM (2007) Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett 581 1464–1474 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue ZX, Shan XY, Lapeyre B, Melese T (1993) The amino-terminus of mammalian nucleolin specifically recognizes SV40 T-antigen type nuclear-localization sequences. Eur J Cell Biol 62 13–21 [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science 292 883–896 [DOI] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong FC, Galbraith DW, Bailey-Serres J (2005) Immunopurification of polyribosomal complexes of arabidopsis for global analysis of gene expression. Plant Physiol 138 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Ou HY, Zhang CT (2004) DEG: a database of essential genes. Nucleic Acids Res 32 D271–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo JR, Niu QW, Chua NH (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.