Abstract
We describe a simple and highly effective means for global identification of genes that are expressed within specific cell types within complex tissues. It involves transgenic expression of nuclear-targeted green fluorescent protein in a cell-type-specific manner. The fluorescent nuclei are then purified from homogenates by fluorescence-activated sorting, and the RNAs employed as targets for microarray hybridization. We demonstrate the validity of the approach through the identification of 12 genes that are selectively expressed in phloem.
Multicellular eukaryotes exist in the form of complex tissues and organs, within which different cell types can be recognized and distinguished (Scheres et al., 2004). Considerable interest exists in establishing an inventory of these different cell types and furthermore in characterizing the global patterns of gene expression that underlie their establishment, function, and maintenance (Galbraith and Birnbaum, 2006). Complicating this characterization is the requirement to separate the different cell types in purified form prior to gene expression measurements. For both animal and plant species, this is most typically done utilizing hydrolytic enzymes to degrade the extracellular structures linking different cells together within tissues and organs, followed by imposition of some cell separation and enrichment method that is specific for the cell type of interest. Particularly productive has been the use of fluorescent proteins (FPs) as transgenic cell-type markers, employing appropriate regulatory sequences to highlight specific cells, followed by the production of protoplasts and the use of fluorescence-activated cell sorting to purify the FP-tagged protoplasts (Birnbaum et al., 2003, 2005; Nawy et al., 2005; Galbraith and Birnbaum, 2006; Brady et al., 2007).
The problems with methods involving tissue and organ dissociation relate, first, to the potential impact that these procedures may have on cellular function, as reflected by gene expression and, second, to the requirement that the methods be applicable to the organ and cell type of interest. This latter problem, common to all multicellular eukaryotes, is exacerbated particularly in higher plants because subtle changes in cell wall structure can render tissues and organs completely recalcitrant to enzymatic dissolution and release of protoplasts (Galbraith, 2007).
A conceptually attractive alternative is to focus not on the cell, but on the transcriptional center of the cell—the nucleus—as the object of selective fluorescent labeling, and then employ gentle tissue homogenization (Galbraith et al., 1983) followed by the use of flow cytometry (Zhang et al., 2005) and fluorescence-activated sorting for purification of these organelles from which transcripts can be amplified and employed for global profiling. This relies on the reasonable assumption that tissue homogenization, performed quickly and on ice, will effectively freeze the transcriptional state of the nucleus at the point of homogenization. Furthermore, because homogenization as a physical process impacts all cells within tissues or organs, we can expect to recover nuclei from all broken cells, including from those that resist enzymatic methods of tissue dissolution and cellular separation.
We have explored this strategy using the model plant Arabidopsis (Arabidopsis thaliana). Utilizing transgenic plants expressing a histone2A-GFP translational fusion within the companion cells of the phloem (Zhang et al., 2005), we simultaneously sorted GFP-positive nuclei and GFP-negative nuclei, and employed microarrays to define those transcripts that were more abundant in GFP-positive nuclei than in nonfluorescent controls. We confirmed the patterns of cell-type-specific expression for 12 genes identified whose transcripts were most highly enriched, through production of transgenic plants in which a GFP marker was placed under control of the cognate upstream regulatory sequences. We extended this work to characterize the general gene set representing transcripts enriched within phloem companion cell (PCC) nuclei. These studies provide a unique view of the genes expressed specifically in PCCs, and a confirmation of the value of this method for the study of the phenotypic characters and their genetic origins in targeted cell types. The application of this technology platform for further dissection of the regulation of gene expression is discussed.
RESULTS AND DISCUSSION
Cell-Type-Specific Analysis of Gene Expression: Platform Description and Implementation
We have previously shown that gentle disruption of plant tissues allows the release and facile analysis of intact nuclei using flow cytometry (Galbraith et al., 1983). We have further demonstrated that nuclei can be highlighted in a cell-type-specific manner by transgenic expression of nuclear-targeted versions of GFP (Zhang et al., 2005), and this allowed flow cytometric characterization of the patterns of endoreduplication occurring within specific cell types of the Arabidopsis root. In extending this work, we aimed to determine whether cell sorting might next be employed to selectively purify nuclei from specific cell types, and thence whether global transcript profiling using microarrays (Galbraith et al., 2004) might allow characterization of gene expression based on the levels of polyadenylated transcripts within these nuclei.
For these experiments, we chose transgenic Arabidopsis plants in which expression of a nuclear-targeted version of GFP was under the control of the SULPHATE TRANSPORTER2;1 (SULTR2;1) promoter (Zhang et al., 2005). Confocal imaging was employed first to characterize the ultrastructural location of the different cell types within the Arabidopsis root, utilizing propidium iodide as a general stain for cell walls, and then to identify which cells contained nuclei that were specifically labeled with GFP. The general ultrastructure of young roots of wild-type Arabidopsis plants has been described by a number of laboratories (Dolan et al., 1993; Baum et al., 2002). Successive cell-type layers, epidermis, cortex, and endodermis, enclose the stele, comprising the central cylindrical vascular core surrounded by the pericycle (Supplemental Fig. S1). The epidermal, cortical, and endodermal cells are readily discernible, being much larger in diameter than the cells within the stele (Supplemental Fig. S1). The vascular core is diarch in organization, with two protophloem elements and two protoxylem elements located along mutually perpendicular axes (Supplemental Fig. S1). Xylem vessels can be readily recognized in the confocal images by the ribbed secondary thickening of the cell walls (Supplemental Fig. S1). Mature sieve elements are less obvious under confocal microscopy because their diagnostic ultrastructure, comprising continuous tubes within the stele (Dolan et al., 1993; Baum et al., 2002), is not readily distinguished from the thin, elongated companion cells that are directly appressed to them, except for an absence of organelles.
In transgenic plants expressing GFP under the control of the SULTR2;1 promoter, nuclear accumulation of GFP is restricted to elongated cells within the stele that fall within the phloem regions of the diarch structure. This is consistent with their identification as companion cells (Fig. 1A; Supplemental Fig. S2); although we cannot formally exclude that they might also include other subsets of the cells of the stele, evidently they comprise a small minority. Biparametric flow cytometric analysis of homogenates, produced from these plants and counterstained using 4′,6-diamidino-phenylindole (DAPI), resolved populations of nuclei with and without GFP fluorescence (Fig. 1, B and D). These were purified by flow sorting, and the RNA was extracted, amplified, and employed for hybridization to whole-genome 70-mer oligonucleotide microarrays.
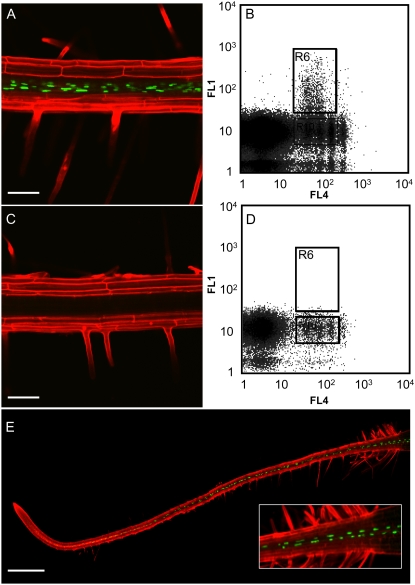
Figure 1.
Confocal and flow cytometric analysis of cell-type-specific expression of nuclear-targeted GFP. A, Confocal image of a primary root of a transgenic Arabidopsis plant expressing pSULTR2;1∷HTA6-GFP. Nuclear fluorescence is restricted to the PCCs. Counterstaining with propidium iodide reveals the positions of the cell walls (bar represents 100 μm). B, Biparametric analysis of homogenates of the pSULTR2;1∷HTA6-GFP plants. FL1, GFP fluorescence; FL4, DAPI fluorescence. R6 represents the region employed for sorting GFP-positive nuclei. C, Confocal image of a primary root of a nontransgenic (wild-type) Arabidopsis plant. No GFP fluorescence is evident (bar represents 100 μm). D, Biparametric analysis of homogenates of the nontransgenic plants. FL1, GFP fluorescence; FL4, DAPI fluorescence. The region below R6 represents that employed for sorting GFP-negative nuclei. E, Confocal image of a primary root of a transgenic plant expressing pNPCC6∷HTB2-GFP-GFP. Nuclear fluorescence is restricted to the PCCs (bar represents 500 μm).
Preliminary comparisons of transcript levels within the nuclear compartment and those of the total RNA of control (nontransgenic plants) revealed general concordance in global gene expression (Supplemental Fig. S3), suggesting that isolated nuclei can be employed as representative sources of transcriptional information concerning cellular states. For the first series of experiments, examination of differences of transcript levels between GFP-positive PCC nuclei and GFP-negative nuclei, utilizing two biological replications, indicated 0.2% were up-regulated and 1% down-regulated by factors of 2-fold or more, at an adjusted P value of less than 0.01 (Supplemental Table S1). A 5-fold enrichment (adjusted P value = 0.045) of the marker gene, the endogenous SULTR2;1, was also observed.
Transgenic Confirmation of Predicted Expression Patterns
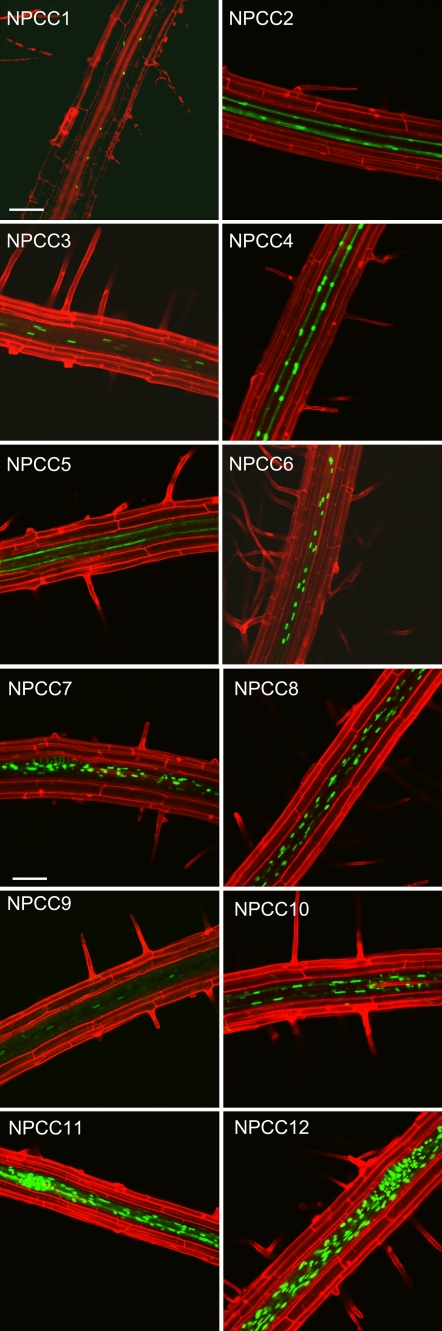
From the list of genes whose transcripts were found to be enriched within GFP-positive nuclei in the first series of experiments, we selected the first 12, for convenience entitled NUCLEAR-ENRICHED PHLOEM COMPANION CELL (NPCC) genes NPCC1 to NPCC12, corresponding to those having the greatest enrichment. Promoter regions, covering approximately 2,000 bp of genomic sequence 5′ to the translational start site (or the entire intergenic region, if shorter than this), were employed to regulate transgenic expression of histone2B-GFP or histone2B-GFP-GFP fusion protein markers (Supplemental Table S2). In all cases, GFP fluorescence was located within the vascular core of transgenic roots, being predominantly associated with the positions of the two phloem sieve tubes and the associated PCCs, confirming the cell-type specificity of expression of the transcripts identified through flow sorting, as well as mapping the basis of this specific regulation to properties of the 5′-cis regions (Figs. 1E and 2; Supplemental Figs. S4–S15). Sieve tube-companion cell localization was strongly implied for NPCC2, NPCC4, and NPCC5 (Fig. 2; Supplemental Figs. S4–S6) given the observation that nonnuclear GFP fluorescence was distributed within two parallel tubes running along the axis of the root, with various degrees of organized nuclear fluorescence being found relatively infrequently along this axis (compare with NPCC4). In two cases (NPCC3 and NPCC6) a spacing of nuclear fluorescence similar to that of NPCC4 was seen but with an absence of nonnuclear GFP fluorescence (Fig. 2; Supplemental Fig. S7). This would be consistent with the hypothesis that GFP is localized solely within the nuclei of companion cells. In other cases, nuclear GFP accumulation was observed within a greater proportion of the cells of the stele, including those of the pericycle (NPCC7, NPCC8, NPCC10–NPCC12), and of the lateral root primordia (NPCC7, NPCC11, and NPCC12), which originate from the pericycle. The differences in GFP accumulation, notably the distribution of GFP within the interior of phloem sieve-tubes (NPCC2, NPCC4, and NPCC5), and within the general vascular core tissues (NPCC7, NPCC8, NPCC10, NPCC11, and NPCC12) suggest that some promoters may be active at different stages of phloem development, and hint at the possibility of selective transport and/or translation of the transgene mRNA in the context of the companion cell/sieve tube interface. As a final point, when these data were obtained, none of the 12 genes had been previously described as exhibiting phloem-specific expression, and none had a well-characterized function, indicating the general utility of the method for gene discovery.
Figure 2.
Confocal images of transgenic plants expressing 12 different transcriptional histone-GFP fusions, utilizing 5′ promoter regions taken from genes whose transcripts were identified as being highly accumulated within sorted GFP-positive nuclei of transgenic plants expressing pSULTR2;1∷HTA6-GFP (bars represent 100 μm).
Comprehensive Classification of Genes Having Transcripts That Are Enriched in Companion Cell Nuclei
For a comprehensive analysis of transcripts enriched in PCC nuclei, we combined the results of the first two sets of experiments with a third biological replicate to permit application of stringent statistical criteria for significance. This analysis, performed using Pointillist (Hwang et al., 2005a, 2005b), yielded a list of genes whose transcripts were significantly enriched in amounts within the PCC nuclei (Supplemental Table S3; the criteria for inclusion in this list were >1.5-fold enrichment and a false discovery rate (FDR) of <0.05. We first examined this list to identify genes having known functions, and then subjected it to an analysis of gene ontology (GO) annotation headings, employing the BiNGO Version 2.0 plug-in (Maere et al., 2005) within Cytoscape (http://www.cytoscape.org) to identify annotational headings that were overrepresented within the list as compared to the genome at large. Further annotational information was retrieved from The Arabidopsis Information Resource database (http://www.arabidopsis.org/).
In terms of genes of established function that are also known to be specifically expressed in the phloem, the entries listed in Supplemental Table S3 include At5g10180, which encodes a sulfate transporter and is the source of the promoter used to drive nuclear expression of GFP within the original transgenic plants employed for flow sorting, as well as At4g14680 and At3g22890, which encode ATP-sulfurylases involved in the initial steps of sulfur assimilation within roots (Lappartient et al., 1999). The entries also include At1g22710, the locus of a Suc/proton symporter (AtSUC2), consistent with a previous report that its protein product is specifically located within PCC plasma membranes (Stadler and Sauer, 1996), the acidic amino acid transporters encoded by the loci At5g49630 (AAP6) and At5g09220 (AAP2; Kwart et al., 1993; Okumoto et al., 2002), and a basic amino acid permease encoded by At1g77380 (AAP3), which has been reported previously to be specifically expressed in root phloem and may function in the distribution of amino acids absorbed by the roots (Okumoto et al., 2004). Finally, the gene list also contains At1g08200, which encodes a UDP-d-apiose/UDP-d-Xyl synthase and is expressed at enhanced levels in the vasculature as visualized by promoter-reporter fusions (Mølhøj et al., 2003).
Comparison, to the genome at large, of the distribution of bioprocess annotational headings for genes whose transcripts were enriched in amounts within the PCC nuclei, indicated overrepresentation of only one classification—that of amino acid transporters (Table I). This is in accord with the well-known physiological function of the phloem in amino acid transport and, by implication, represents a key molecular function for the PCCs. Within this particular classification, the acidic amino acid transporters At5g49630 and At5g09220 participate in the transport of Glu and other amino acids most important to nitrogen metabolism (Kwart et al., 1993; Okumoto et al., 2002). In particular, At5g49630 is essential for Asp transport, but previously was identified primarily in the cells of the xylem parenchyma (Okumoto et al., 2002).
Table I.
GO classifications overrepresented within the NPCC gene list
| Description | Corrected P Value | Cluster Frequency | Genome Frequency | Genes in Test Set |
|---|---|---|---|---|
| Acidic amino acid transport | 2.20E-02 | 2/177 1.13% | 2/20763 0.00% | At5g49630 |
| At5g09220 | ||||
| Basic amino acid transport | 3.28E-02 | 2/177 1.13% | 3/20763 0.00% | At1g44100 |
| At1g77380 | ||||
| Organic acid transport | 3.47E-02 | 4/177 2.26% | 47/20763 0.23% | At5g49630 |
| At5g09220 | ||||
| At1g44100 | ||||
| At1g77380 | ||||
| Amine transport | 3.47E-02 | 4/177 2.26% | 47/20763 0.23% | At5g49630 |
| At5g09220 | ||||
| At1g44100 | ||||
| At1g77380 | ||||
| Amino acid transport | 3.47E-02 | 4/177 2.26% | 47/20763 0.23% | At5g49630 |
| At5g09220 | ||||
| At1g44100 | ||||
| At1g77380 | ||||
| Carboxylic acid transport | 3.47E-02 | 4/177 2.26% | 47/20763 0.23% | At5g49630 |
| At5g09220 | ||||
| At1g44100 | ||||
| At1g77380 | ||||
| Carboxy-lyase activity | 3.32E-02 | 4/186 2.1% | 52/22033 0.24% | At1g08200 |
| At1g08650 | ||||
| At3g26830 | ||||
| At5g57190 | ||||
| Ceramidase activity | 2.69E-02 | 2/186 1.0% | 3/22033 0.01% | At2g38010 |
| At5g58980 | ||||
| Sulfate adenylyltransferase (ATP) activity | 2.69E-02 | 2/186 1.0% | 4/22033 0.0% | At4g14680 |
| At3g22890 |
The α- and β-subunits of Glu dehydrogenase (At5g07440 and At5g18170) have been reported to be expressed uniquely in the companion cells of aerial portions of the plant, and are likely key to amine assimilation and recycling (Kichey et al., 2005; Fontaine et al., 2006). The transcripts for this enzyme and for Gln synthetase (At1g66200) do not appear in our list of NPCC transcripts. Within the root, the transport of Glu is central to the distribution of nitrogen metabolites, which suggests either that root PCCs do not participate directly in Glu/Gln metabolism or that such activity is not characterized by nuclear accumulation of associated transcripts.
Comparison of molecular annotational headings for the NPCC transcripts to those of the whole genome revealed overrepresentation for the terms sulfurylase, ceramidase, and carboxylase (Table I). Notable in the first category are the ATP-sulfurylases, At4g14680 and At3g22890, and coregulation of these genes is of course also consistent with the presence of the sulfate transporter, At5g10180, in the list of NPCC transcripts. The ceramidases, representing the second category, may play important roles in phloem ontology and function given the established involvement of sphingolipids in the regulation of programmed cell death (Liang et al., 2003; Morales et al., 2007). Cell-type-specific regulation of the controlled degradation of cellular contents accompanies differentiation of the vascular system from meristematic cells, and achieving this through invoking programmed cell death mechanisms appears reasonable. Sphingosine is also the immediate metabolic precursor of sphingosine phosphate, a key regulator of cell growth in plants (Imamura et al., 2007) and animals (Tani et al., 2000). Expression of dihydrosphingosine C4 hydroxylase, also involved in sphingosine metabolism, is associated specifically with phloem in rice (Oryza sativa; Imamura et al., 2007), but its Arabidopsis orthologs do not appear in the NPCC list, nor among the genes found associated with phloem cells in studies of root protoplasts (Brady et al., 2007).
Interestingly, the transcript from locus At5g57190 is also enriched within the companion cell nuclei. This transcript encodes a tonoplast-localized phosphatidyl-Ser (PS) decarboxylase (Nerlich et al., 2007), one of three isoforms, whose function, based on the normal phenotypes displayed by pyramided triple knockouts, do not appear related to the role of PS as a precursor to the more abundant phospholipid phosphatidylethanolamine (Nerlich et al., 2007). PS also performs a role in signaling the onset of programmed cell death in animal and plant cells (Weir, 2001), and it therefore may be involved in mechanisms, presumptively also redundant, governing phloem development.
The presence of the various amino acid transporters and permeases within the NPCC transcript list (Supplemental Table S3; Fig. 3) is consistent with the key role the phloem plays in the transport and distribution of amino acids and amines within plants. Along this line, another transcript enriched within the PCC nuclei, from At1g59740, is annotated by TIGR (http://www.tigr.org/tdb/at/atgenome/atgenome.html) as a proton-dependent oligopeptide transporter, and therefore may also participate in amine recycling from peptides derived from protein degradation within senescent tissue.
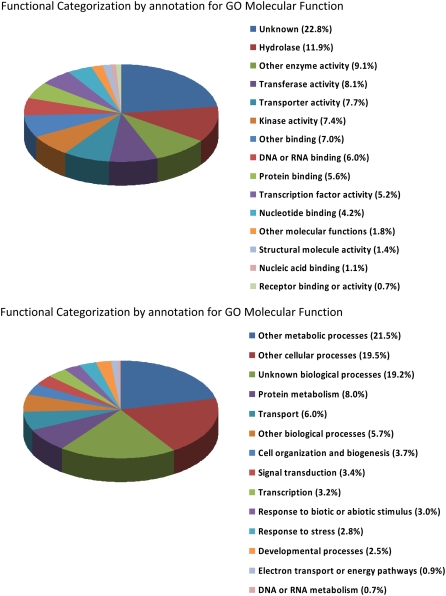
Figure 3.
The 237 genes that were enriched by a factor of 1.5-fold or greater in the PCCs were characterized for GO annotation in the molecular function and bioprocess categories by input to the The Arabidopsis Information Resource Web site (http://www.arabidopsis.org/tools/bulk/go/index.jsp).
A final, general scan of the annotations associated with the NPCC transcripts revealed a large number of entries (15) described as transcription factors (Fig. 3). A further eight are described simply as zinc finger proteins, and may function either as transcription factors or, perhaps, as RNA-binding proteins associated with regulation of phloem function (Lunde et al., 2007). Fourteen of the NPCC transcripts are assigned to the signal transduction category (Fig. 3), implying regulatory roles. Thus, many of the transcripts associated specifically with accumulation in companion cell nuclei have functions consistent with their role in being part of the vasculature, whether that role is in transport or in the sensing of the physiological state of the plant and regulating appropriate responses.
Promoter Motifs
To further explore the regulatory mechanisms associated with cell-type-specific nuclear transcript accumulation, the NPCC gene list (Supplemental Table S3) was divided into quartiles based on the mean raw fluorescent intensity signals recorded for each of the probes representing NPCC transcripts. The groups of genes within each quartile were separately analyzed using Promomer (http://bar.utoronto.ca/ntools/cgi-bin/BAR_Promomer.cgi) for the presence of shared 5-mer sequence motifs within their promoter regions, these regions being empirically defined as the 1,000 bp of sequence upstream of the start of transcription. Table II presents those 5-mer motifs that were significantly overrepresented in the promoter regions for 95% of the genes within that quartile, as compared to the genome at large.
Table II.
Motifs overrepresented within the promoter regions of NPCC genes
| Motif | Background Average | Set Average | Significance | |
|---|---|---|---|---|
| Quartile 1 | AATAA | 240.6 | 340.5 | 0.001 |
| AAATA | 264.6 | 361.0 | 0.001 | |
| ATAAA | 254.4 | 347.5 | 0.001 | |
| AAAAT | 335.4 | 407.1 | 0.005 | |
| TATTT | 267.5 | 332.2 | 0.005 | |
| TTTAT | 240.4 | 298.2 | 0.005 | |
| TTATT | 233.5 | 291.0 | 0.005 | |
| ACAAA | 213.7 | 255.1 | 0.01 | |
| AAAGA | 193.8 | 233.9 | 0.01 | |
| AAAAA | 572.0 | 697.2 | 0.05 | |
| TAAAA | 289.7 | 339.6 | 0.05 | |
| TTAAA | 238.2 | 281.6 | 0.05 | |
| TTTAA | 229.9 | 273.1 | 0.05 | |
| ATTTA | 203.4 | 243.9 | 0.05 | |
| ATATT | 214.6 | 251.8 | 0.05 | |
| AAATG | 138.0 | 160.8 | 0.05 | |
| Quartile 2 | TATAA | 180.1 | 228.0 | 0.005 |
| AAAAC | 205.6 | 249.2 | 0.01 | |
| TATAT | 240.9 | 298.2 | 0.05 | |
| AAATA | 255.4 | 299.4 | 0.05 | |
| ATAAA | 242.2 | 285.4 | 0.05 | |
| TTTAA | 220.5 | 262.4 | 0.05 | |
| AATTA | 199.5 | 241.1 | 0.05 | |
| TAAAA | 277.2 | 317.9 | 0.05 | |
| AAGAA | 208.6 | 248.2 | 0.05 | |
| AAAAG | 187.8 | 223.8 | 0.05 | |
| TTATA | 174.2 | 208.1 | 0.05 | |
| AAAGA | 186.9 | 218.3 | 0.05 | |
| Quartile 3 | TTTAT | 244.0 | 292.2 | 0.01 |
| AAATA | 271.2 | 318.3 | 0.05 | |
| TATTT | 269.9 | 315.6 | 0.05 | |
| TTATT | 235.6 | 279.8 | 0.05 | |
| Quartile 4 | AAGAA | 211.8 | 265.2 | 0.005 |
| AAAAA | 552.8 | 713.4 | 0.005 | |
| ACAAA | 207.5 | 239.9 | 0.05 | |
| TTAAT | 186.2 | 217.7 | 0.05 | |
| AAAGA | 190.5 | 220.8 | 0.05 | |
| AAAAG | 187.8 | 218.1 | 0.05 |
These 5-mer motifs were then compared to plant promoter motifs contained in the PLACE database (http://www.dna.affrc.go.jp/PLACE/signalscan.html). One motif, ROOTMOTIFTAPOX1, identified by the 5-mer ATATT, forms part of the rolD promoter, and is expressed in the elongation zone and vasculature of the root (Elmayan and Tepfer, 1995). ROOTMOTIFTAPOX1 also is present in the upstream regions of >95% of the genes listed in the first quartile. Several of the 5-mer motifs (AAAGT, AAAAG, and AAAGA) overlap with DOFCOREZM (AAAG), a plant motif found in the promoters of the genes encoding phosphoenolpyruvate carboxylase and orthophosphate dikinase in maize (Zea mays; Yanagisawa and Schmidt, 1999). In nonphotosynthetic tissues, phosphoenolpyruvate carboxylase performs the anaplerotic function of maintaining the levels of tricarboxylic acid cycle intermediates (Hartwell et al., 1999), and is involved in regulating pH (Fontaine et al., 2002). Interestingly, PPCK1 (At1g08650) is found in our list of NPCC transcripts (Supplemental Table S3). The DOFCOREZM motif was also overrepresented within the genes of the first, second, and fourth quartiles, and is also overrepresented in the promoter regions for all the genes when analyzed together. Individual probes on microarrays are not thought to produce hybridization signals that can be directly compared in terms of transcript abundance. However, the assumptions inherent to microarray normalization are based on general concordance of this relationship across large numbers of genes. It will therefore be interesting to explore, in further experiments, the potential association of promoter motifs with genes according to gene expression level.
Comparison to Protoplast Sorting
While this work was being prepared for publication, a report appeared that describes a comprehensive analysis of global gene expression patterns in the Arabidopsis root (Brady et al., 2007). This extended the approach of Birnbaum et al. (2003), through sorting of GFP-positive protoplasts, prepared from the roots of transgenic plants, with expression of GFP being driven by many different promoters having known cell-type specificity. As for the previous report, this work employed Affymetrix ATH-1 GeneChips for analysis of global gene expression. Four of the cell-type-specific promoters that were employed (Brady et al., 2007) are described as producing phloem-associated patterns of expression associated with the phloem (those from APL [At1g79430], S17 [At2g22850], S32 [At2g18380], and SUC2 [At1g22710]; Lee et al., 2006).
Of the list of 12 genes (NPCC1–NPCC12) that we selected for confirmation of cell-type specificity tissue promoter tests (Fig. 2; Supplemental Tables S1 and S2), two, NPCC3 (At3g58720) and NPCC11 (At2g18196), are not represented in the probe sets of the Affymetrix ATH-1 GeneChip. Of the remainder, six are found in the phloem-related gene lists as described by Brady et al. (2007): NPCC2 (At3g21600), NPCC4 (At5g64240), NPCC5 (At1g11450), NPCC6 (At5g02600), NPCC8 (At5g54660), and NPCC10 (At5g57190). NPCC1 (At5g18350) and NPCC9 (At1g26930) produced very low signals both in our microarray hybridization experiments and in the confirmatory transgenic plants (Fig. 2), and therefore may not be easily detected in GeneChip analyses.
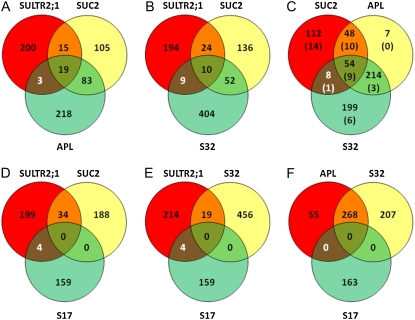
Given that the overall comparison is across platforms, and that the two approaches interrogate very different points within the process of gene expression, this general level of concordance (60%) is both impressive and encouraging. Because NPCC1 to NPCC12 represent genes producing transcripts showing the highest degree of differential accumulation within the nuclei of PCC cells, they may represent a subset of genes most readily identifiable for, and characteristic of, the phloem. Nevertheless, when comparing the complete gene list of NPCC genes (Supplemental Table S4) to those genes defined as being differentially expressed in APL-, S17-, S32-, and SUC2-GFP fusions, considerably less overlap is seen (Fig. 4). The degree of overlap within the results for the APL, S17, S32, and SUC2 gene sets (Brady et al., 2007) likewise is highly variable, the closest concordance being seen between APL and S32. In obvious contrast, no concordance is seen between S17 and either APL, S32, or SUC2. Explanations of the lack of general concordance between the various gene lists include: (1) that general technical differences exist between the experimental manipulations, (2) that the promoters cited as phloem specific are, in fact, active within different cellular subsets of the phloem, (3) that the process of preparing either protoplasts or nuclei likewise selects for specific cellular subsets, and (4) that, for the phloem, nuclear transcript populations are generally different from those of the cytoplasm. All of these explanations appear reasonable; there are a number of differences in the experimental manipulations, such as the method of RNA amplification, and in the ages of plants and their growing conditions. Similarly, the different populations identified by the techniques and promoters may simply represent cells at different developmental stages, different stages of the cell division cycle, or cells that otherwise have differing specialized functions. This avenue that can be directly, if laboriously, tested through production of multiply transgenic plants using combinations of promoters driving expression of different FPs. The third explanation is based on the knowledge that the formation of mature phloem sieve elements and companion cells from meristematic tissue involves a controlled process of programmed cell death, resulting in anucleate sieve elements, which are linked to companion cells by complex plasmodesmatal connections. Despite a recent report (Hafke et al., 2007), the production of transcriptionally functional protoplasts from anucleate tubes is self-inconsistent, and would be problematic for companion cells. Furthermore, the process of protoplast sorting discriminates in favor of protoplasts that are mechanically resilient, whereas that of nuclear sorting automatically excludes anucleate cells or cells in mitosis. The fourth explanation relates to the increasingly popular assumption that the companion cell/sieve element interface provides access to long-distance protein and RNA trafficking within plants. If this assumption is correct, considerable disparities might exist between the compositions of the nuclear and cytoplasmic transcript pools, associated with the requirement for rapid mobilization of these transcripts and/or their protein products in response to environmental or other stimuli. This would be in contrast to the situation observed for all transcripts within all cell types, for which only a small fraction (1.2%) of transcripts are accumulated to significantly different levels in the nucleus as compared to the total RNA pool (Supplemental Fig. S3). Finally, it should be noted that the lack of concordance within the Brady et al. (2007) data set may in part reflect insufficient biological sampling, thereby generating a high FDR, which would also carry over to comparisons with our data.
Figure 4.
Comparison of the degrees of overlap between the NPCC gene list (SULTR2;1) and the four lists of nuclear-encoded genes indicated by Brady et al. (2007) to be enriched in phloem-related cells, respectively, expressing GFP under the control of the SUC2 promoter (SUC2), the APL promoter (APL), the S32 promoter (S32), and the S17 promoter (S17). A, Comparison of SULTR2;1, SUC2, and APL. B, Comparison of SULTR2;1, SUC2, and S32. C, Comparison of SUC2, APL, and S32. D, Comparison of SULTR2;1, SUC2, and S17. E, Comparison of SULTR2;1, S32, and S17. F, Comparison of APL, S32, and S17. Venn diagrams were created with Genevenn (http://mcbc.usm.edu/genevenn/genevenn.htm). The gene loci within each sector are given in Supplemental Table S3. Genes represented on the ATH-1 GeneChips that are encoded within the chloroplast or mitochondrial genomes were omitted from this analysis. In C, the distributions of the NPCC gene list members are indicated within parentheses.
CONCLUSION
We are extending this approach to additional cell types within Arabidopsis, including those found in its aerial organs, as well as to different plant species. We have also established that the approach is applicable to mammalian cells and tissues, through demonstration of general concordance between nuclear and cellular transcript levels (Barthelson et al., 2007). The approach therefore should be of considerable general interest and utility to investigators addressing the molecular basis of cell-type specificity within complex tissues of any eukaryotic species. Finally, given that identification of cell types is achieved simply using promoter sequences of defined specificities, one can envisage extending this analysis to novel cell types identified in transgenics through the expression of multiple combinations of FPs, but otherwise lacking recognizable (morphological or other) phenotypes, thereby leading to a more sophisticated and comprehensive understanding of organogenesis, microanatomy, and the regulation of cell-type specificity of gene expression.
MATERIALS AND METHODS
Biological Materials
Transgenic plants, expressing a translational fusion between GFP and histone H2A (HTA6; At5g59870) under the transcriptional control of the promoter of the SULTR2;1 gene (At5g10180), were used for this study. The genetic background of transgenic Arabidopsis (Arabidopsis thaliana) plants is ecotype Columbia (Col-0). The seeds of 10 to 12 independent transgenic T2 lines were combined and treated as one biological replicate to reduce potential influence of mutations caused by T-DNA insertions. Seeds were sterilized and planted on Murashige and Skoog (MS) agar plates supplemented with 2% Suc, 30 mg/L hygromycin, and 1.2% agar. The plates were maintained at 4°C for 2 d before transfer to a Conviron growth chamber under a 16-h day/8-h night illumination regime, with an incident light flux of 150 to 175 μm m−2 s−1, and temperature of 22°C (day) and 20°C (night). The plates were kept in a vertical orientation so that the roots grew on the surface of, rather than within, the agar medium. The roots of 2-week-old seedlings were sampled during the day, between 9 am and 1 pm (Mountain Standard Time). Three biological replicates were prepared, 1 to 2 weeks apart.
Isolation of Fluorescent Nuclei
Roots were homogenized by chopping (Zhang et al., 2005). The homogenate was stained by addition of DAPI to a final concentration of 2 μg/mL, and biparametric flow analysis of GFP fluorescence versus nuclear DNA content was performed as described previously (Zhang et al., 2005). Sort windows were placed (Fig. 1) such that they included all nuclei having values of green fluorescence between 25 and 1,000. The position of the sort region was established by first determining the baseline of green fluorescence using a GFP-negative control plant, and adjusting the lower boundary of the sort window such that less that 3% of nontransgenic nuclei had fluorescence values that fell above this value. The upper and left- and right-hand boundaries of the sort window were adjusted to include all nuclei produced from GFP-positive controls. Nuclei were sorted onto glass slides and examined under a fluorescence microscope (Olympus BX-50) to verify the presence of DAPI and GFP fluorescence.
Sorting was done using a Dako-Cytomation (now Beckman-Coulter) MoFlo flow cytometer/cell sorter equipped with a 70-μm flow tip and operated at a sheath pressure of 40 psi. Droplets were generated at 60,000 Hz using a piezo drive voltage of 17, and were sorted with a drop-delay setting of 24. Events were triggered on 90° side scatter, and were thresholded to provide an event rate of 30,000/s, with a sort rate for GFP-positive nuclei of 100/s, and 1,000/s for GFP-negative nuclei. The laser illumination power was set to 40 mW for DAPI excitation at 395 nm, and to 200 mW for GFP excitation at 488 nm. The barrier filters were 450/65 nm for DAPI fluorescence, and 530/40 nm for GFP fluorescence.
Extraction and Amplification of RNA
Total cellular RNA from whole roots was isolated using RNeasy Plant Mini Kits (QIAGEN) according to the manufacturer's recommendation. Total RNA was extracted from sorted nuclei using the TRIZOL method, following slight modifications of the manufacturer's recommendations (Invitrogen) as follows: nuclei were flow sorted (a final volume of approximately 120–200 μL) directly into 500 μL of TRIZOL reagent. The mixture was shaken vigorously, and stored at room temperature for 5 min. Chloroform (120 μL) was added and the tube was shaken by hand vigorously for 15 s. The tube was centrifuged at 11,900g for 15 min at 4°C. The upper phase was transferred to a new tube and an equal volume of isopropyl alcohol was added. The sample was mixed and kept overnight at −20°C. RNA was sedimented by centrifugation at 11,900g for 15 min at 4°C. After removing the supernatant, 1 mL of 75% ethanol was added to wash the RNA pellet (invisible). The tube was centrifuged at 11,900g for 10 min at 4°C, and the supernatant was completely removed. The tube was then left open for about 5 min to permit evaporation of residual ethanol. RNAase-free H2O (10 μL) was added to redissolve the RNA. The quantity and quality of the RNA was determined using an Agilent 2100 Bioanalyzer (Agilent).
For the first set of experiments, two consecutive rounds of amplification were done on 5-μL (approximately 2–20 ng RNA) samples, using ExpressArt mRNA amplification kits (Nano plus Version; Artus GmbH) according to the manufacturer's instructions. Aminoallyl-modified UTP was incorporated into the amplified antisense RNA produced during the second round of in vitro transcription. For the second set of experiments, RNA amplification was done using the Ambion MessageAmp kit.
Microarray Hybridization and Data Analysis
The amplified antisense RNA samples were labeled with Cy3-NHS or Cy5-NHS fluorescent dyes (Amersham Biosciences) according to the manufacturer's instructions. Labeled targets were purified using RNeasy MiniElute kits (QIAGEN). Arabidopsis whole genome long oligonucleotide microarrays (http://cals.arizona.edu/microarray/deconvolutionver3.0.html) were employed for hybridization, following standard procedures in the Galbraith laboratory (http://cals.arizona.edu/microarray/methods.html), and as previously described (Zanetti et al., 2005). Microarrays were scanned using a GenePix 4200AL microarray scanner (Molecular Devices). Fluorescent intensity values were extracted using GenePix Pro 6.0 software. For each hybridized slide, the median signals of Cy3 and Cy5 were saved as two separate text files, and treated as two data sets. For the first set of experiments, a total of eight data sets were prepared from four hybridized slides. These data sets were analyzed using GeneSpring GX7.3 (Agilent Technologies). Data were not normalized before or after being imported into GeneSpring. In the first set of experiments, to test for significant differences between signals representing PCC and non-PCC nuclei, statistical analysis was carried out using a built-in one-way ANOVA test (GeneSpring). The list of genes with statistically significant changes (P < 0.05) was then filtered using another built-in tool, Volcano Plot (GeneSpring). The fold difference was set to 2 and the P-value cutoff to 0.01. The resulting gene list is reported in Supplemental Table S1. Because this approach was used for preliminary screening of genes, P values were not adjusted for multiple testing, but this approach was vindicated by the resulting PCC cell-specific expression patterns for the transgenics that were tested.
For the second set of experiments, a data set was prepared from one additional hybridized slide. For the combined data sets from the first and second sets of experiments, the data were submitted to analysis with the program Pointillist (Hwang et al., 2005a, 2005b). The median intensity values for all the data sets were expressed as ratios of PCC/non-PCC, normalized (quantile) and converted to log values. The variances were calculated and then combined to determine which transcripts had an FDR less than 0.05. Those that were considered significant by these criteria, and additionally had mean intensities at least 1.5-fold higher in the PCC nuclei than in other nuclei, were used to represent the transcripts enriched in the PCC. This list of transcripts was further analyzed using the BiNGO plug-in of the Cytoscape program (Maere et al., 2005) to determine which GO annotational headings were overrepresented among the NPCC transcripts as compared to the whole Arabidopsis genome. This determination was done for the Bioprocess, Molecular Function, and Cell headings; overrepresentation was only found within the first two headings.
Cluster analysis was performed on the log-transformed intensity values for the list of PCC-enriched genes, using the program Genesis (http://genome.tugraz.at/genesisclient/genesisclient_description.shtml). The resulting clusters of genes were further analyzed with Promomer (http://bbc.botany.utoronto.ca/ntools/cgi-bin/BAR_Promomer.cgi) for the presence of 5-mer motifs that are significantly more common in 95% or greater of the 1,000-bp upstream sequences for the gene clusters compared to the Arabidopsis genome in general. Promomer provides a P value for the comparison of cluster genes to the whole genome, and those 5 mers with a P value of 0.05 or smaller are given in Figure 4. Further analysis was done with the aid of the PLACE database (http://www.dna.affrc.go.jp/PLACE/signalscan.html; Prestridge, 1991; Higo et al., 1999).
Transgenic Analysis of Promoter Specificity
To determine the expression patterns of the candidate genes identified through microarray analysis, plasmid constructs were made comprising nuclear-localized versions of GFP coding sequences placed under control of the upstream regulatory sequences. Genomic sequences about 2 kb upstream and up to 24 bp downstream of translational start (ATG) were amplified using PfuUltra high-fidelity DNA polymerase (Stratagene). The primers and related information are listed in Supplemental Table S2. The amplified sequences were ligated into plasmid vectors pBIn1GFP or pBIn2GFP (provided by Dr. Ramin Yadegari, University of Arizona), which express fusion proteins of HTB2-GFP and HTB2-GFP-GFP, respectively. Plasmids carrying the above constructs were introduced into Agrobacterium tumefaciens strain GV3101. Arabidopsis Col-0 was transformed using the floral dip method (Clough and Bent, 1998). T1 seeds were harvested and screened on half-strength MS agar plates supplemented with 0.5% Suc, 0.6% agar, 35 mg L−1 kanamycin, and 25 mg L−1 cefotaximine. Kanamycin-resistant seedlings (approximately 10 d after germination) were transferred to fresh half-strength MS agar plates supplemented with 0.5% Suc, 1.2% agar, and 25 mg L−1 cefotaximine. The plates were kept vertical for 3 d to allow roots to grow on the surface of the medium. Roots were examined for GFP fluorescence using confocal microscopy. Confirmed transformants were transferred to soil.
Confocal Microscopy
Roots from T1 or T2 seedlings were counterstained with 10 μg mL−1 of propidium iodide (Sigma) for 2 min, and were placed on slides carrying a drop of water for observation. GFP fluorescence was imaged by confocal microscopy using a MRC 1024MP (Bio-Rad) confocal scanner attached to an Olympus BX-50 upright microscope, equipped with UPlanFl 4×/0.13, UPlanFl 10×/0.30, and UPlanApo 20×/0.70 objective lenses. LaserSharp2000 (Bio-Rad) was employed for image collection and color merging. For each construct, at least seven independent T1 seedlings were observed.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number DQ370422.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Organization of cell types within a primary root of wild-type Arabidopsis.
Supplemental Figure S2. Confocal analysis of a root expressing pSULTR2;1∷HTA6-GFP.
Supplemental Figure S3. Correlation of global transcript abundances between total and nuclear poly(A+) RNA pools.
Supplemental Figure S4. Confocal analysis of a root of a transgenic NPCC2 plant.
Supplemental Figure S5. Confocal analysis of a root of a transgenic NPCC4 plant.
Supplemental Figure S6. Confocal analysis of a root of a transgenic NPCC5 plant.
Supplemental Figure S7. Confocal analysis of a root of a transgenic NPCC6 plant.
Supplemental Figure S8. Confocal analysis of a root of a transgenic NPCC12 plant.
Supplemental Figure S9. Confocal analysis of a root of a transgenic NPCC1 plant.
Supplemental Figure S10. Confocal analysis of a root of a transgenic NPCC3 plant.
Supplemental Figure S11. Confocal analysis of a root of a transgenic NPCC7 plant.
Supplemental Figure S12. Confocal analysis of a root of a transgenic NPCC8 plant.
Supplemental Figure S13. Confocal analysis of a root of a transgenic NPCC9 plant.
Supplemental Figure S14. Confocal analysis of a root of a transgenic NPCC10 plant.
Supplemental Figure S15. Confocal analysis of a root of a transgenic NPCC11 plant.
Supplemental Table S1. Genes whose transcripts are up- or down-regulated within PCC nuclei (Experiment 1).
Supplemental Table S2. Sequence information for genes NPCC1 to NPCC12.
Supplemental Table S3. Genes whose transcripts are up- or down-regulated within PCC nuclei (Combined Experiments).
Supplemental Table S4. Comparison of gene lists resulting from this work and that of Brady et al. (2007).
Supplementary Material
Acknowledgments
We thank R. Yadegari (University of Arizona) for his support and helpful suggestions.
This work was supported by the National Science Foundation Plant Genome Research Program (grant DBI–0211857 to D.W.G.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: David W. Galbraith (galbraith@arizona.edu).
The online version of this article contains Web-only data.
Open Access article can be viewed online without a subscription.
References
- Barthelson RA, Lambert GM, Vanier C, Lynch RM, Galbraith DW (2007) Comparison of the contributions of the nuclear and cytoplasmic compartments to global gene expression in human cells. BMC Genomics 8 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum SF, Dubrovsky JG, Rost TL (2002) Apical organization and maturation of the cortex and vascular cylinder in Arabidopsis thaliana (Brassicaceae) roots. Am J Bot 89 908–920 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN (2005) Cell-type specific expression profiling in plants using fluorescent reporter lines, protoplasting, and cell sorting. Nat Methods 2 1–5 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Kock J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318 801–806 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Listead P, Poethig S, Roberts K, Scheres B (1993) Cellular organization of the Arabidopsis thaliana root. Development 119 71–84 [DOI] [PubMed] [Google Scholar]
- Elmayan T, Tepfer M (1995) Evaluation in tobacco of the organ specificity and strength of the rolD promoter, domain A of the 35S promoter and the 35S2 promoter. Transgenic Res 4 388–396 [DOI] [PubMed] [Google Scholar]
- Fontaine JX, Saladino F, Agrimonti C, Bedu M, Terce-Laforgue T, Tetu T, Hirel B, Restivo FM, Dubois F (2006) Control of the synthesis and subcellular targeting of the two GDH genes products in leaves and stems of Nicotiana plumbaginifolia and Arabidopsis thaliana. Plant Cell Physiol 47 410–418 [DOI] [PubMed] [Google Scholar]
- Fontaine V, Hartwell J, Jenkins GI, Nimmo HG (2002) Arabidopsis thaliana contains two phosphoenolpyruvate carboxylase kinase genes with different expression patterns. Plant Cell Environ 25 115–122 [Google Scholar]
- Galbraith DW (2007) Protoplast analysis using flow cytometry and sorting. In J Dolezel, J Greilhuber, J Suda, eds, Flow Cytometry with Plant Cells. Wiley-VCH, Weinheim, Germany, pp 231–250
- Galbraith DW, Birnbaum K (2006) Global studies of cell type-specific gene expression in plants. Annu Rev Plant Biol 57 451–475 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Elumalai R, Gong FC (2004) Integrative flow cytometric and microarray approaches for use in transcriptional profiling. Methods Mol Biol 263 259–279 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JR, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220 1049–1051 [DOI] [PubMed] [Google Scholar]
- Hafke JB, Furch ACU, Reitz MU, van Bel AJE (2007) Functional sieve element protoplasts. Plant Physiol 145 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell J, Gill A, Nimmo GA, Wilkins MB, Jenkins GI, Nimmo HG (1999) Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J 20 333–342 [DOI] [PubMed] [Google Scholar]
- Higo KY, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D, Rust AG, Ramsey S, Smith JJ, Leslie DM, Weston AD, de Atauri P, Aitchison JD, Hood L, Siegel AF, et al (2005. a) A data integration methodology for systems biology. Proc Natl Acad Sci USA 102 17296–17301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D, Smith JJ, Leslie DM, Weston AD, Rust AG, Ramsey S, de Atauri P, Siegel AF, Bolouri H, Aitchison JD, et al (2005. b) A data integration methodology for systems biology: experimental verification. Proc Natl Acad Sci USA 102 17302–17307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T, Kusano H, Kajigaya Y, Ichikawa M, Shimada H (2007) A rice dihydrosphingosine C4 hydroxylase (DSH1) gene, which is abundantly expressed in the stigmas, vascular cells and apical meristem, may be involved in fertility. Plant Cell Physiol 48 1108–1120 [DOI] [PubMed] [Google Scholar]
- Kichey T, Le Gouis J, Sangwan B, Hirel B, Dubois F (2005) Changes in the cellular and subcellular localization of glutamine synthetase and glutamate dehydrogenase during flag leaf senescence in wheat (Triticum aestivum L.). Plant Cell Physiol 46 964–974 [DOI] [PubMed] [Google Scholar]
- Kwart M, Hirner B, Hummel S, Frommer WB (1993) Differential expression of two amino acid transporters with differing substrate specificity in Arabidopsis thaliana. Plant J 4 993–1002 [DOI] [PubMed] [Google Scholar]
- Lappartient AG, Vidmar JJ, Leustek T, Glass A, Touraine B (1999) Inter-organ signaling in plants: regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. Plant J 18 1–89 [DOI] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Yao N, Song LT, Luo S, Lu H, Greenberg JT (2003) Ceramides modulate programmed cell death in plants. Genes Dev 17 2636–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde B, Moore C, Varani G (2007) RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21 3448–3449 [DOI] [PubMed] [Google Scholar]
- Mølhøj M, Verma R, Reiter WD (2003) The biosynthesis of the branched-chain sugar D-apiose in plants: functional cloning and characterization of a UDP-D-apiose/UDP-D-xylose synthase from Arabidopsis. Plant J 35 693–703 [DOI] [PubMed] [Google Scholar]
- Morales A, Lee H, Goni FM, Kolesnick R, Fernandez-Checa JC (2007) Sphingolipids and cell death. Apoptosis 12 923–939 [DOI] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlich A, von Orlow M, Rontein D, Hanson AD, Dörmann P (2007) Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria. Plant Physiol 144 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto S, Koch W, Tegeder M, Fischer WN, Biehl A, Leister D, Stierhof YD, Frommer WB (2004) Root phloem-specific expression of the plasma membrane amino acid proton co-transporter AAP3. J Exp Bot 55 2155–2168 [DOI] [PubMed] [Google Scholar]
- Okumoto S, Schmidt R, Tegeder M, Fischer WN, Rentsch D, Frommer WB, Koch W (2002) High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. J Biol Chem 277 45338–45346 [DOI] [PubMed] [Google Scholar]
- Prestridge DS (1991) SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci 7 203–206 [DOI] [PubMed] [Google Scholar]
- Scheres B, van den Toorn H, Heidstra R (2004) Root genomics: towards digital in situ hybridization. Genome Biol 5 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Sauer N (1996) The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot Acta 109 299–306 [Google Scholar]
- Tani M, Okino N, Mori K, Tanigawa T, Izu H, Ito M (2000) Molecular cloning of the full-length cDNA encoding mouse neutral ceramidase. A novel but highly conserved gene family of neutral/alkaline ceramidases. J Biol Chem 275 11229–11234 [DOI] [PubMed] [Google Scholar]
- Weir IE (2001) Analysis of apoptosis in plant cells. Methods Cell Biol 63 505–526 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Schmidt RJ (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J 17 209–214 [DOI] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong FC, Galbraith DW, Bailey-Serres J (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol 138 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CQ, Gong FC, Lambert GM, Galbraith DW (2005) Cell type-specific characterization of nuclear DNA contents within complex tissues and organs. Plant Methods 1 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.