Abstract
Using a bioinformatics analysis of public Arabidopsis (Arabidopsis thaliana) microarray data, we propose here a novel regulatory program, combining transcriptional and posttranslational controls, which participate in modulating fluxes of amino acid metabolism in response to abiotic stresses. The program includes the following two components: (1) the terminal enzyme of the module, responsible for the first catabolic step of the amino acid, whose level is stimulated or repressed in response to stress cues, just-in-time when the cues arrive, principally via transcriptional regulation of its gene; and (2) the initiator enzyme of the module, whose activity is principally modulated via posttranslational allosteric feedback inhibition in response to changes in the level of the amino acid, just-in-case when it occurs in response to alteration in its catabolism or sequestration into different intracellular compartments. Our proposed regulatory program is based on bioinformatics dissection of the response of all biosynthetic and catabolic genes of seven different pathways, involved in the metabolism of 11 amino acids, to eight different abiotic stresses, as judged from modulations of their mRNA levels. Our results imply that the transcription of the catabolic genes is principally more sensitive than that of the biosynthetic genes to fluctuations in stress-associated signals. Notably, the only exception to this program is the metabolic pathway of Pro, an amino acid that distinctively accumulates to significantly high levels under abiotic stresses. Examples of the biological significance of our proposed regulatory program are discussed.
Plant cells possess multiple highly regulated metabolic networks that play central regulatory roles in their growth. Among the various metabolic networks, those leading to the synthesis of amino acids have gained considerable interest not only because the amino acids are vital for the synthesis of proteins but also because amino acids serve as precursors for a large array of metabolites with multiple functions in plant growth and response to various stresses. Just to mention a few examples, the aromatic amino acids are used as precursors for numerous metabolites, such as hormones, cell wall components, and a large group of multiple functional secondary metabolites (Radwanski and Last, 1995; Wittstock and Halkier, 2002; Pichersky et al., 2006; Tempone et al., 2007); Met is a precursor for the synthesis of the hormone ethylene, polyamines, cellular energy glucosinolates, and also provides a methyl group to DNA methylation, chlorophyll biosynthesis, cell wall biosynthesis, and to a large number of secondary metabolites (Amir et al., 2002; Wittstock and Halkier, 2002; Rebeille et al., 2006; Goyer et al., 2007); Thr conversion into Gly is important for seed development (Jander et al., 2004; Joshi et al., 2006); Ile catabolism leads to the production of cellular energy (Mooney et al., 2002); and Pro catabolism is important for the recovery of plants from various abiotic stresses (Rontein et al., 2002).
Biochemical studies of plant amino acid metabolism have so far been mostly devoted to the biosynthesis of the amino acids. These studies showed that the biosynthesis of plant amino acids is largely regulated by end product feedback inhibition loops in which specific enzymes in a given amino acid biosynthesis pathway (called allosteric enzymes) are feedback inhibited by the amino acids that they synthesize (Galili, 1995; Radwanski and Last, 1995). Eliminations of feedback inhibition traits in specific allosteric/biosynthetic enzymes generally resulted in increased levels of the corresponding amino acids (Widholm, 1972; Galili, 1995, 2002; Radwanski and Last, 1995; Li and Last, 1996), leading to the notion that the biosynthetic/allosteric enzymes represent major regulatory factors determining the rate of the fluxes in metabolic pathways of amino acids. Consequently, the potential roles of downstream enzymes that convert amino acids into other metabolites (defined as catabolic enzymes of the amino acids) in the regulation of fluxes of amino acid metabolism under specific physiological conditions have been largely ignored. Nevertheless, direct experimental evidence (Dixon and Paiva, 1995; Moulin et al., 2000; Galili et al., 2001; Galili, 2002; Mikkelsen et al., 2003; Stepansky and Galili, 2003; Jander et al., 2004) and results of publicly available microarray results (H. Less and G. Galili, unpublished data) support such regulatory roles by showing that the transcript level of many of the catabolic enzymes of the amino acids is highly regulated by developmental, metabolic, and environmental cues.
In this report, we used a bioinformatics approach, based on publicly available microarray data from the model plant Arabidopsis (Arabidopsis thaliana), to dissect the potential function of catabolic enzymes of amino acids in regulating fluxes of amino acid metabolism upon the exposure of plants to various abiotic stresses. Our reasons for selecting abiotic stresses were as follows: (1) various abiotic stresses stimulate the expression of a number of genes encoding catabolic enzymes of amino acids; and (2) the experiments employing abiotic stresses are most detailed in the publicly available microarray results by having multiple time points during the stresses, enabling efficient data mining. Our results show that genes encoding catabolic enzymes of amino acids are principally much more sensitive and respond faster to abiotic stresses than genes encoding biosynthetic (allosteric and nonallosteric) enzymes and hence play major regulatory roles in amino acid metabolism upon exposure to these stresses. Yet, the spatial and temporal response patterns of genes encoding catabolic and biosynthetic enzymes are distinct for different metabolic pathways in response to different stress conditions. Our results also propose that catabolic enzymes contribute to changes of fluxes via different branches of amino acid metabolic pathways upon exposure to stress conditions, apparently to allow timely metabolic adjustments.
RESULTS
Selection of Metabolic Pathways of Amino Acids and Their Associated Genes
To discover regulatory principles of amino acid metabolism, we focused on genes encoding biosynthetic enzymes as well as enzymes catalyzing the first catabolic steps of the amino acids. Our definition of catabolism included the breakdown of the amino acid into carbon, nitrogen, and energy-associated molecules and the utilization of the amino acids for the synthesis of other special metabolites, such as secondary metabolites.
In few special cases, a given amino acid is converted into another amino acid by a second pathway, and the first enzyme of this second pathway can be defined either as a catabolic enzyme of the first pathway or as a biosynthetic enzyme of the second pathway (see below). For simplicity, we defined such an enzyme as a catabolic enzyme because it uses an amino acid as a substrate with only one exception of Thr deaminase, the first enzyme converting Thr into Ile. Thr deaminase was subjectively defined as a biosynthetic enzyme because it is an allosteric enzyme that is feedback inhibited by Ile (see below). Since amino acid metabolism is largely regulated by feedback inhibition of biosynthetic/allosteric enzymes as well as by enzymes responsible for the first catabolic steps of the amino acids, we focused only on amino acid metabolic pathways in which the genes encoding these enzymes have been identified. Hence, our research covered 11 of the 20 amino acids. We did not include in this analysis the following amino acids: (1) Gln, Glu, Asp, and Asn, which are the central regulators of carbon/nitrogen metabolism, interacting with multiple metabolic networks; (2) Ala, which is synthesized by one enzymatic step from pyruvate; (3) His, for which no catabolic gene has been identified; (4) Gly and Ser, whose metabolism occurs by several different pathways, one of which is strongly associated with photorespiration; and (5) Cys, which is used as a main sulfur donor in plants and as such is used for multiple catabolic processes catalyzed by multiple catabolic enzymes (see The Arabidopsis Information Resource [TAIR] and ARACYC). The 11 selected amino acids were grouped into four distinct metabolic networks (Fig. 1) whose enzymes and the genes encoding them are detailed in Table I.
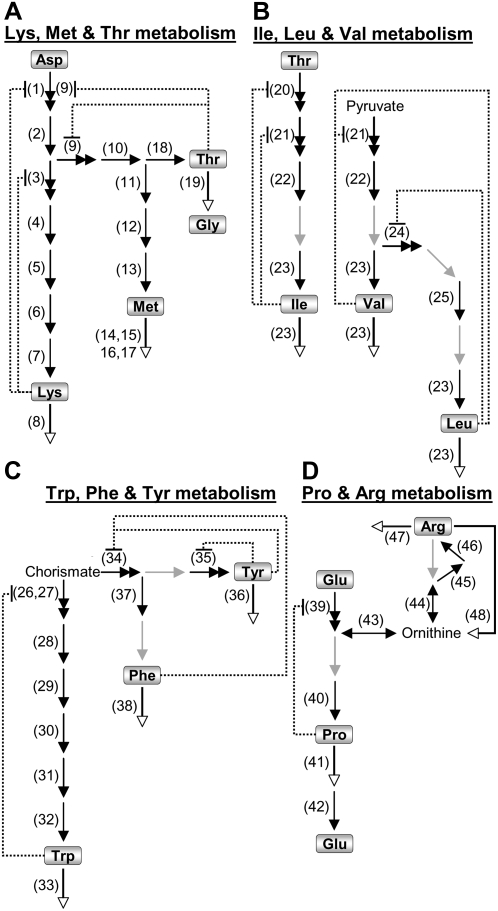
Figure 1.
Schematic representation of the four different metabolite networks analyzed in this report. The positions of the different amino acids in the different networks are marked with boxes. Biosynthetic/allosteric, biosynthetic/nonallosteric, and catabolic enzymatic steps are indicated by double-headed, black, and white-headed arrows, respectively, while enzymatic steps with no known genes are indicated by gray arrows. Numbers near each arrow refer to enzyme names provided in Table I. A, The network of Lys, Met, and Thr metabolism, composed of three pathways (from Asp to Lys, from Asp to Met, and from Asp to Thr). B, The network of Ile, Leu, and Val metabolism, composed of three pathways (from Thr to Ile, from pyruvate to Val, and from pyruvate to Leu), which were analyzed together (as one pathway). C, The network of Trp, Phe, and Tyr metabolism, composed of tree pathways (from chorismate to Trp, from chorismate to Phe, and from chorismate to Tyr), from which the pathways leading to Phe and Tyr were analyzed together (as one pathway). D, The network of Pro and Arg metabolism, composed of two pathways (from Glu and Orn to Pro and from Glu and Orn to Arg), which were analyzed together (as one pathway). The reasons for the definition and joining of the different pathways are explained in the text. Dotted lines ending with a bar sign represent feedback inhibition loops.
Table I.
A list and additional relevant information of all genes analyzed in the present study
| Pathway | Enzyme | Stepa | Symbol | ATGb | Probesetc | Biosyntheticd | Catabolice |
|---|---|---|---|---|---|---|---|
| Lys metabolism | Monofunctional Asp kinase | 1 | AK | AT3G02020 | 258977_s_at | Yes (Lys) | – |
| AK | AT5G14060 | 258977_s_at | Yes (Lys) | – | |||
| AK1 | AT5G13280 | 250291_at | Yes (Lys) | – | |||
| Asp-semialdehyde dehydrogenase | 2 | ASD | AT1G14810 | 262841_at | Yes | – | |
| Dihydrodipicolinate synthase | 3 | DHDPS1 | AT3G60880 | 251392_at | Yes (Lys) | – | |
| DHDPS2 | AT2G45440 | 245145_at | Yes (Lys) | – | |||
| Dihydrodipicolinate reductase | 4 | DHDPR | AT2G44040 | 267237_s_at | Yes | – | |
| DHDPR | AT3G59890 | 267237_s_at | Yes | – | |||
| DHDPR | AT5G52100 | 248402_at | Yes | – | |||
| l,l-Diaminopimelate aminotransferase | 5 | AGD2 | AT4G33680 | 253308_at | Yes | – | |
| Diaminopimelate epimerase | 6 | DAPE | AT3G53580 | 251948_at | Yes | – | |
| Diaminopimelate decarboxylase | 7 | DAPD | AT3G14390 | 258365_s_at | Yes | – | |
| DAPD | AT5G11880 | 258365_s_at | Yes | – | |||
| Lys-ketoglutarate reductase/saccharopine dehydrogenase | 8 | LKR/SDH | AT4G33150 | 253373_at | – | Lys | |
| Met metabolism | Asp kinase/homo-Ser dehydrogenase | 9 | AK/HSDH1 | AT1G31230 | 263696_at | Yes (Thr) | – |
| AK/HSDH2 | AT4G19710 | 254535_at | Yes (Thr) | – | |||
| Asp-semialdehyde dehydrogenase | 2 | ASD | AT1G14810 | 262841_at | Yes | – | |
| Homo-Ser kinase | 10 | HSK | AT2G17265 | 264855_at | Yes | – | |
| Cystathionine γ-synthase | 11 | CGS1 | AT3G01120 | 259279_at | Yes | – | |
| CGS | AT1G33320 | 256531_at | Yes | – | |||
| Cystathionine β-lyase | 12 | CBL | AT3G57050 | 251666_at | Yes | – | |
| Met synthase | 13 | MS1 | AT5G17920 | 259343_s_at | Yes | – | |
| MS2 | AT3G03780 | 259343_s_at | Yes | – | |||
| MS3 | AT5G20980 | 246185_at | Yes | – | |||
| S-Adenosyl-Met synthetase | 14 | SAMS1 | AT1G02500 | 260913_at | – | Met | |
| SAMS4 | AT2G36880 | 263838_at | – | Met | |||
| SAMS3 | AT3G17390 | 258415_at | – | Met | |||
| SAMS2 | AT4G01850 | 255552_at | – | Met | |||
| Met γ-lyase | 15 | MGL | AT1G64660 | 261957_at | – | Met | |
| Methylthioalkylmalate synthase | 16 | MAM1 | AT5G23010 | 249866_at | – | Met | |
| MAM-L | AT5G23020 | 249867_at | – | Met | |||
| Met-oxo-acid transaminase | 17 | BCAT4 | AT3G19710 | 257021_at | – | Met | |
| Thr metabolism | Asp kinase/homo-Ser dehydrogenase | 9 | AK/HSDH1 | AT1G31230 | 263696_at | Yes (Thr) | – |
| AK/HSDH2 | AT4G19710 | 254535_at | Yes (Thr) | – | |||
| Asp-semialdehyde dehydrogenase | 2 | ASD | AT1G14810 | 262841_at | Yes | – | |
| Homo-Ser kinase | 10 | HSK | AT2G17265 | 264855_at | Yes | – | |
| Thr synthase | 18 | TS | AT1G72810 | 262380_at | Yes | – | |
| TS | AT4G29840 | 253700_at | Yes | – | |||
| Thr aldolase | 19 | THA1 | AT1G08630 | 264777_at | – | Thr | |
| THA2 | AT3G04520 | 258599_at | – | Thr | |||
| Ile, Leu, and Val metabolism | Thr deaminase | 20 | TD | AT3G10050 | 258884_at | Yes (Ile) | Thr |
| Acetolactate synthase | 21 | AHASS1 | AT2G31810 | 263460_at | Yes (Ile,Leu,Val) | – | |
| AHASS2 | AT5G16290 | 250111_at | Yes (Ile,Leu,Val) | – | |||
| AHAS | AT3G48560 | 252325_at | Yes (Ile,Leu,Val) | – | |||
| Ketol acid reductoisomerase | 22 | KARI | AT3G58610 | 251536_at | Yes | – | |
| Branched-chain amino acid aminotransferase | 23 | BCAT1 | AT1G10060 | 264525_at | – | Ile,Leu,Val | |
| BCAT2 | AT1G10070 | 264524_at | – | Ile,Leu,Val | |||
| BCAT3 | AT3G49680 | 252274_at | – | Ile,Leu,Val | |||
| BCAT5 | AT5G65780 | 247158_at | – | Ile,Leu,Val | |||
| BCAT6 | AT1G50110 | 261636_at | – | Ile,Leu,Val | |||
| BCAT7 | AT1G50090 | 261690_at | – | Ile,Leu,Val | |||
| Isopropylmalate synthase | 24 | IPMS1 | AT1G18500 | 261668_at | Yes (Leu) | – | |
| IPMS2 | AT1G74040 | 260384_at | Yes (Leu) | – | |||
| 3-Isopropylmalate dehydrogenase | 25 | IMD1 | AT5G14200 | 263706_s_at | Yes | – | |
| IMD2 | AT1G80560 | 260285_at | Yes | – | |||
| IMD3 | AT1G31180 | 263706_s_at | Yes | – | |||
| Trp metabolism | Anthranilate synthase β | 26 | ASB | AT1G24807 | 247864_s_at | Yes (Trp) | – |
| ASB | AT1G24909 | 247864_s_at | Yes (Trp) | – | |||
| ASB | AT1G25083 | 247864_s_at | Yes (Trp) | – | |||
| ASB | AT1G25155 | 247864_s_at | Yes (Trp) | – | |||
| ASB | AT1G25220 | 247864_s_at | Yes (Trp) | – | |||
| ASB | AT5G57890 | 247864_s_at | Yes (Trp) | – | |||
| Anthranilate synthase α | 27 | ASA1 | AT5G05730 | 250738_at | Yes (Trp) | – | |
| ASA2 | AT2G29690 | 266671_at | Yes (Trp) | – | |||
| ASA | AT3G55870 | 251716_at | Yes (Trp) | – | |||
| Anthranilate phosphoribosyltransferase | 28 | TRP | AT1G70570 | 260311_at | Yes | – | |
| TRP1 | AT5G17990 | 250014_at | Yes | – | |||
| Phosphoribosylanthranilate isomerase | 29 | PAI1 | AT1G07780 | 259770_s_at | Yes | – | |
| PAI2 | AT5G05590 | 259770_s_at | Yes | – | |||
| PAI3 | AT1G29410 | 259770_s_at | Yes | – | |||
| Indole-3-glycerol phosphate synthase | 30 | IGPS | AT2G04400 | 263807_at | Yes | – | |
| IGPS | AT5G48220 | 248688_at | Yes | – | |||
| Trp synthase α | 31 | TSA2 | AT3G54640 | 251847_at | Yes | – | |
| TSA | AT4G02610 | 255487_at | Yes | – | |||
| Trp synthase β | 32 | TSB1 | AT5G54810 | 253898_s_at | Yes | – | |
| TSB2 | AT4G27070 | 253898_s_at | Yes | – | |||
| TSB | AT5G38530 | 249515_at | Yes | – | |||
| Cytochrome P450 | 33 | CYP79B3 | AT2G22330 | 264052_at | – | Trp | |
| CYP79B2 | AT4G39950 | 252827_at | – | Trp | |||
| Phe and Tyr metabolism | Chorismate mutase | 34 | CM1 | AT3G29200 | 257746_at | Yes (Tyr, Phe) | – |
| CM2 | AT5G10870 | 250407_at | Yes | – | |||
| CM3 | AT1G69370 | 260360_at | Yes (Tyr, Phe) | – | |||
| Arogenate dehydrogenase | 35 | AAT1 | AT5G34930 | 255859_at | Yes (Tyr) | – | |
| AAT2 | AT1G15710 | 259486_at | Yes (Tyr) | – | |||
| Tyr aminotransferase | 36 | TAT3 | AT2G24850 | 263539_at | – | Tyr | |
| TAT | AT5G53970 | 248207_at | – | Tyr | |||
| Arogenate dehydratase | 37 | PD1 | AT2G27820 | 266257_at | Yes | – | |
| PD | AT1G08250 | 261758_at | Yes | – | |||
| PD | AT1G11790 | 262825_at | Yes | – | |||
| PD | AT3G07630 | 259254_at | Yes | – | |||
| PD | AT3G44720 | 252652_at | Yes | – | |||
| PD | AT5G22630 | 249910_at | Yes | – | |||
| Phe-Ala ammonia lyase | 38 | PAL1 | AT2G37040 | 263845_at | – | Phe | |
| PAL2 | AT3G53260 | 251984_at | – | Phe | |||
| PAL3 | AT5G04230 | 245690_at | – | Phe | |||
| Pro and Arg metabolism | Δ-1-Pyrroline-5-carboxylate synthetase | 39 | P5CS1 | AT2G39800 | 251775_s_at | Yes (Pro) | – |
| P5CS2 | AT3G55610 | 251775_s_at | Yes (Pro) | – | |||
| Pyrroline-5-carboxylate reductase | 40 | P5CR | AT5G14800 | 246594_at | Yes | – | |
| Pro dehydrogenase | 41 | ERD5 | AT3G30775 | 257315_at | – | Pro | |
| PRODH | AT5G38710 | 249527_at | – | Pro | |||
| 1-Pyrroline-5-carboxylate dehydrogenase | 42 | P5CDH | AT5G62530 | 247436_at | Yes | – | |
| Orn δ-aminotransferase | 43 | δ-OAT | AT5G46180 | 248879_at | Yes | – | |
| Orn carbamoyltransferase complex | 44 | OCT | AT1G75330 | 261122_at | Yes | – | |
| Arginosuccinate synthase | 45 | ARS | AT4G24830 | 254134_at | Yes | – | |
| Argininosuccinate lyase | 46 | ARL | AT5G10920 | 250403_at | Yes | – | |
| Arg decarboxylase | 47 | ADC1 | AT2G16500 | 263241_at | – | Arg | |
| ADC2 | AT4G34710 | 253203_at | – | Arg | |||
| Arg | 48 | ARGAH1 | AT4G08900 | 255065_s_at | – | Arg | |
| ARGAH2 | AT4G08870 | 255065_s_at | – | Arg |
Step numbers represent enzymatic steps described in Figure 1.
ATG represents the common locus number using TAIR nomenclature.
Probeset represents specific identifiers according to Affymetrix AtH1 microarray.
Biosynthetic enzymes. For biosynthetic/allosteric enzymes, the inhibiting amino acid is indicated.
Catabolic enzymes. The amino acid substrate of each catabolic enzyme is indicated.
The first network, called the Asp family network (Fig. 1A), includes the amino acids Lys, Thr, and Met, whose synthesis initiates from Asp, and in which one pathway leads to Lys metabolism (enzymatic steps 1–8), a second pathway leads to Thr synthesis and its further catabolism into Gly (enzymatic steps, 2, 9, 10, 18, and 19), and the third pathway leads to Met metabolism (enzymatic steps 2 and 9–17). Thr also leads to Ile biosynthesis and therefore could also be included in this network, but because Ile biosynthesis is also highly coordinated with Leu and Val biosynthesis, we subjectively decided to include Ile in the second network that leads to the metabolism of these three amino acids, which are also called branched-chain amino acids. This second network (Fig. 1B) includes three pathways: (1) from Thr to Ile (enzymatic steps 20–23); (2) from pyruvate to Val (enzymatic steps 21–23); and (3) from pyruvate to Leu (enzymatic steps 21–25), but in this study we treated this network as one pathway, since most of its enzymatic steps (steps 21–23) are common to either two or three of these pathways. Another enzyme that could belong to both the Asp family and branched-chain amino acid networks (Fig. 1, A and B) is Met γ-lyase, which catabolizes Met into a number of products, including Ile (Rebeille et al., 2006). However, we subjectively localized it to the Asp family network (Fig. 1A, step 15) because its other catabolic products, methanethiol and S-methyl-Cys, are generally considered to be catabolic products of Met (Rebeille et al., 2006; Goyer et al., 2007). The third network (Fig. 1C) includes the aromatic amino acids, whose synthesis initiates from chorismate. This network includes three pathways: one leading to Trp metabolism (enzymatic steps 26–33), and the second and third leading to Phe and Tyr metabolism. Since the Phe and Tyr pathways together contain only a few enzymatic steps (enzymes 34–38), one of which is common to both pathways, we treated them as a single pathway. The fourth network (Fig. 1D) includes the amino acids Pro and Arg, whose metabolic pathways are linked to each other and therefore were considered a single pathway in this study (enzymatic steps 39–48). In all of the metabolic pathways, we included genes whose functional annotation has been identified, based on a combinatorial analysis of information from TAIR (http://www.arabidopsis.org/), ARACYC (http://www.arabidopsis.org/biocyc/index.jsp), and literature review.
Selection of Abiotic Stresses as Desirable Cues to Elucidate Regulatory Principles of Amino Acid Metabolism
Our selection of abiotic stresses as cues to elucidate regulatory principles of amino acid metabolism was for two major reasons. First, adaptation of plants to stress conditions requires an extensive shift in metabolism, including metabolic networks associated with amino acids (Dixon and Paiva, 1995; Zhao and Last, 1996; Ouwerkerk et al., 1999; Moulin et al., 2000; Galili et al., 2001; Amir et al., 2002; Galili, 2002; Mikkelsen et al., 2003; Stepansky and Galili, 2003). Second, our bioinformatics approach was based on publicly available Arabidopsis microarray results (Nottingham Arabidopsis Stock Centre [NASC]; http://affymetrix.arabidopsis.info/), and hence the quality of data mining was dependent on the quality and reliability of the data. The publicly available NASC database contains a group of eight detailed microarray experiments, analyzing the early response of Arabidopsis roots and shoots to various abiotic stresses (Kilian et al., 2007). In contrast to other microarray studies, this study used Arabidopsis plants of an identical genotype grown side-by-side under identical conditions, which were subjected to various stresses and which were measured at high resolution.
General Responses of Genes Encoding Biosynthetic and Catabolic Enzymes of Amino Acids to Abiotic Stresses
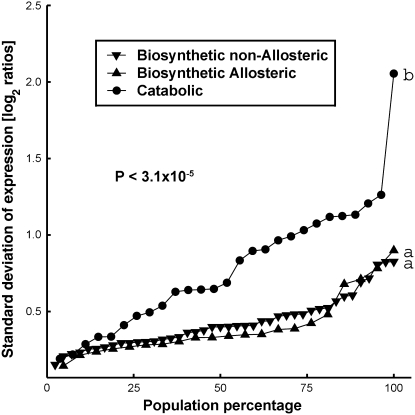
In order to elucidate the general behavior of the biosynthetic and catabolic genes belonging to all of the metabolic pathways shown in Figure 1, we divided the entire set of these genes into three groups: (1) genes encoding biosynthetic/allosteric enzymes, which are feedback inhibited by the amino acids; (2) genes encoding biosynthetic/nonallosteric enzymes; and (3) genes encoding enzymes responsible for the first catabolic steps of the amino acids (Table I). Our choice of placing the amino acids in a central position (biosynthesis before and catabolism after) was for the following reasons: (1) the amino acids are the metabolites that feedback inhibit the allosteric/biosynthetic enzymes; and (2) the amino acids are in the junction between metabolic pathways and protein synthesis and turnover. In cases in which a given enzymatic step is catalyzed by a number of isozymes (Table I), the expression level of each isozymic gene was analyzed separately. We then elucidated the relative fluctuation of mRNA levels of individual genes belonging to each group in response to the entire set of the abiotic stress conditions in both shoots and roots, by measuring their sd of expression (log2 ratios of treatments versus controls). As shown in Figure 2, the fluctuation of expression of the entire set of catabolic genes in response to the entire set of abiotic stresses exhibited a considerably different distribution of values compared with those of the combined sets of the biosynthetic/allosteric and the biosynthetic/nonallosteric genes, which showed no significant difference between them. This difference was highly significant using the Kolmogorov-Smirnov statistical test (P < 3.1 × 10−5). The different behavior between the catabolic and biosynthetic genes was not due to differences in the mean expression levels (absolute intensity) of the genes, which were statistically indistinguishable in the three groups (Supplemental Fig. S1).
Figure 2.
Distribution of the degree of fluctuations of mRNA levels of the entire sets of biosynthetic/nonallosteric (inverted triangles), biosynthetic/allosteric (triangles), and enzymes catalyzing the first catabolic step of the amino acids (circles) genes of the four metabolic networks described in Figure 1 in response to eight different abiotic stresses (both in shoots and roots). The degree of fluctuation of the mRNA levels was estimated using sd of expression (log2 ratio of treatments versus controls), and the values in each group were sorted along the x axis. Different letters (a and b at the right of each curve) represent statistically significantly different groups (P < 3.1 × 10−5).
We next wished to test whether the higher fluctuation of expression of the catabolic genes, compared with the biosynthetic/allosteric and biosynthetic/nonallosteric genes, is a general characteristic of all metabolic pathways or is specific to individual metabolic pathways. To address this, we compared the sd of expression of the individual genes belonging to each of the three enzyme groups, in each of the different metabolic pathways (Fig. 1), across the entire set of the abiotic stress conditions. As shown in Supplemental Figure S2, the biosynthetic/allosteric enzymes and the catabolic enzymes in the metabolic pathways are encoded by several isozymic genes, besides the pathways of Lys, Pro, and Arg, which possess respectively only a single catabolic or biosynthetic/allosteric gene (red and blue histograms, respectively). The biosynthetic/nonallosteric enzymes of the different pathways were generally, but not entirely, encoded by single genes. Notably, the gene with the highest fluctuation of expression in each of the metabolic pathways encodes a catabolic enzyme (Supplemental Fig. S2, red histograms), while the fluctuation of expression of the entire set of genes showed pathway-specific patterns. The metabolic pathways of Ile and Val and of Leu, Met, and Lys had significantly higher fluctuations of expression of the catabolic genes (red histograms) compared with the biosynthetic genes (blue and black histograms). The metabolic pathways of Pro and Arg, Phe and Tyr, and Trp and Thr also had some biosynthetic genes with relatively high fluctuation of expression, but these were as a whole lower than those of the catabolic genes (compare the black plus blue histograms with the red histograms).
It was also interesting to examine the molecular basis responsible for the higher sd of expression of the catabolic compared with the biosynthetic genes. To address this, we selected the catabolic genes and the biosynthetic/allosteric genes (as representatives of genes with high and low sd of expression, respectively) belonging to six representative metabolic pathways and analyzed their actual expression (log2 ratio of treatments versus controls) in response to the eight different abiotic stresses both in shoots and in roots. The entire results are presented in Supplemental Figures S3 (for shoots) and S4 (for roots). The higher sd of the mRNA levels of the catabolic genes, compared with the biosynthetic genes (Fig. 2; Supplemental Fig. S2), generally reflected two important characteristics: (1) a higher fluctuation of mRNA levels of the catabolic than the biosynthetic/allosteric genes around the zero expression level, either in pathways that do not respond to the stress or in pathways that respond to the stress, but at earlier time points before the stress effects occurred (Supplemental Figs. S3 and S4). This indicates that the catabolic genes are generally more sensitive to varying growth conditions that contribute to the “noise” of the experiments; and (2) in cases in which a given pathway responded to a given stress, the mRNA levels of the catabolic genes seemed to be stimulated much more frequently than the biosynthetic/allosteric genes (Supplemental Figs. S3 and S4).
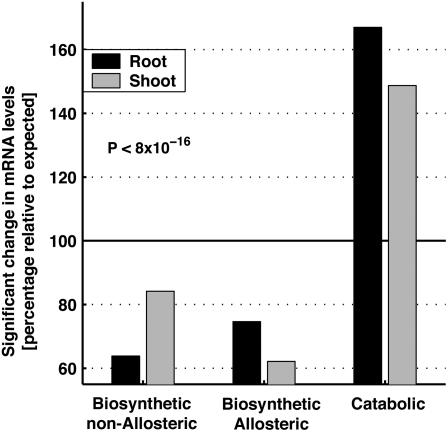
Next, we wished to test more accurately whether the catabolic genes generally respond more frequently than the biosynthetic/allosteric and the biosynthetic/nonallosteric genes to the entire set of abiotic stresses. To address this question, we examined the actual numbers of genes from each of the three groups that had a significant response (2-fold increase or decrease in mRNA level; P < 0.01) at any time point in any stress condition and compared these numbers with the expected numbers assuming no preferential responses of genes from each of the three groups to the different stresses (see “Materials and Methods” for more details and justification of the calculation). As shown in Figure 3, in both roots and shoots, the proportion of cases with a significant response was higher than expected (above 100%) in the group of the catabolic genes and lower then expected (below 100%) in both groups of the biosynthetic/allosteric and biosynthetic/nonallosteric genes. This deviation from the expected values was highly significant (P < 8 × 10−16) using a χ2 test.
Figure 3.
Comparison of the relative number of cases in which mRNA levels of genes belonging to each of the groups of the biosynthetic/nonallosteric enzymes, biosynthetic/allosteric enzymes, and the enzymes catalyzing the first catabolic step of the amino acids, derived from the four metabolic networks described in Figure 1, in response to the eight different abiotic stresses. Results are presented for roots and shoots separately. Each histogram represents the actual number of cases in which a statistically significant response was observed relative to the expected number of cases assuming no preferential response of each of the groups (see “Materials and Methods” for details). The hypothesis of no preferential response was rejected using a χ2 test (P < 8 × 10−16).
Specialized Response Patterns of the Different Metabolic Pathways to the Eight Different Stress Conditions
Since different metabolic pathways of amino acids fulfill different functions in plants, we tested further how the general principals described above (Figs. 2 and 3) are implemented with respect to the response of specific metabolic pathways to specific stress conditions. To address this, we analyzed the responses of the genes encoding each of the biosynthetic and catabolic steps in each of the metabolic pathways located within the four different metabolic networks (Fig. 1) to each of the eight different abiotic stresses. In order to view the results in a metabolic perspective rather than in an individual gene perspective, in each of the microarray chips we combined the mRNA signals of all genes encoding isozymes that catalyze the same enzymatic step in a given metabolic pathway (called “combined isozymic mRNA signals” below). Despite its obvious limitations, this is the only available way to estimate total mRNA levels contributing to individual enzymatic steps based on mRNA microarray results. The entire results are provided separately for shoots and roots in Supplemental Figures S5 and S6, respectively. To minimize the impact of experimental errors, we defined in this analysis that a reliable response of a given metabolic pathway to a given stress condition requires a minimum of a statistically significant (P < 0.05) 2-fold increase or 2-fold decrease in the combined isozymic mRNA signals of at least one of the biosynthetic or catabolic enzymatic steps of this pathway in at least two consecutive time points of the stress treatment, compared with the control. This statistical threshold fits our analysis better than false discovery rate controlling methods, because our aim was to elucidate general behaviors of amino acid metabolic modules under various abiotic stress conditions rather than elucidating specific behaviors of individual modules under particular stress conditions. The specific cases obeying this rule are marked by asterisks in Supplemental Figures S5 and S6. In total, 45 of the 112 cases (shoots plus roots) satisfied our rule, while the rest were considered cases in which a given metabolic pathway does not respond to a given stress cue. Notably, while based on our requirements none of the metabolic pathways responded to oxidative stress, neither in shoots nor in roots, at least one or more metabolic pathway responded to all other stresses, either in shoots and/or in roots. In addition, the different metabolic pathways responded differentially to the various stresses, as expected from the fact that they serve different physiological functions. Notably, the metabolic pathway of Pro and Arg generally behaved quite differently from the other metabolic pathways, apparently because Pro is a special amino acid accumulating to very high levels under various abiotic stresses (Rontein et al., 2002). Therefore, we will describe the Pro and Arg module separately from the other pathways.
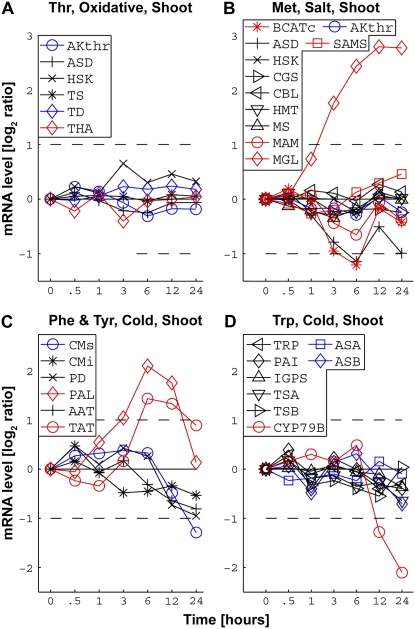
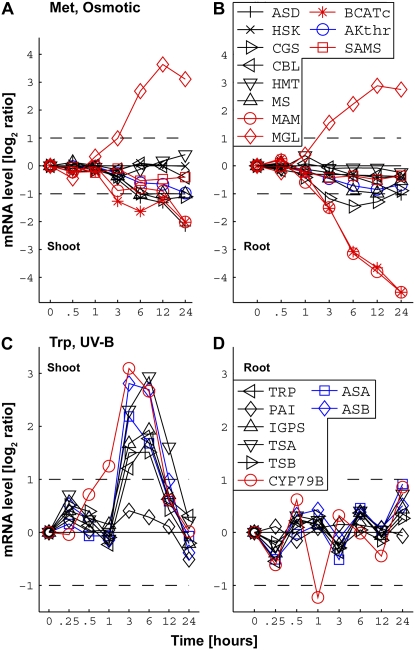
Analyzing the data for all metabolic pathways besides the Pro and Arg pathway (Supplemental Figs. S5 and S6), we could identify two principally different patterns, which accounted for the majority of the responses. The most frequent pattern signifies a situation in which the entire pathway, namely, the combined isozymic mRNA signals of all of the biosynthetic and catabolic enzymatic steps of the pathway, does not respond to a given stress cue (Fig. 4A, depicting the response of shoot Thr metabolism to oxidative stress). The second most frequent pattern signifies pathways in which only the combined isozymic mRNA signals of the catabolic steps (one or more catabolic steps) respond extensively to the stress cue, while the combined isozymic mRNA signals of the entire biosynthetic steps either do not respond or are only slightly down-regulated by the stress cue. This pattern could be divided into three main subpatterns. The first subpattern includes pathways in which the combined isozymic mRNA signals of one or more of the catabolic steps (each catabolic step leads to a different metabolic direction) are stimulated by the stress cue. This subpattern is depicted by the response of the Met pathway in shoots to salt stress via up-regulating only one of its four different catabolic enzymatic steps (Fig. 4B) and by the response of the Phe and Tyr pathway in shoots to cold stress via up-regulating its two different catabolic steps (Fig. 4C). The second subpattern includes a repression of the combined isozymic mRNA signals of the catabolic step by the stress cue (Fig. 4D, depicting the response of the Trp pathway to cold stress). The third subpattern includes both up-regulation and repression of the combined isozymic mRNA signals of different types of catabolic steps (Fig. 5, A and B, depicting the response of Met metabolism in roots and shoots to osmotic stress). In this specific case of the response of the Met pathway to osmotic stress, the combined isozymic mRNA signals of one of its biosynthetic steps (cystathionine γ-synthase) were also repressed in the roots, but the extent of their repression was significantly lower than that of the catabolic steps (Fig. 5B).
Figure 4.
The responses of combined isozymic mRNA levels of genes encoding isozymes of exemplary metabolic pathways to representative stress conditions in shoots. The combined isozymic mRNA levels of genes encoding biosynthetic/allosteric enzymes, biosynthetic/nonallosteric enzymes, and enzymes catalyzing the first catabolic steps of the amino acids are indicated by blue, black, and red lines, respectively. The names of the enzymes encoded by the different isozymic genes, as provided in Table I, are given in the boxes of each panel. A, Response of the Thr pathway to oxidative stress. B, Response of the Met pathway to salt stress. C, Response of the Phe and Tyr pathway to cold stress. D, Response of the Trp pathway to cold stress. The changes in mRNA levels are given in log2 ratios of treatments versus controls (y axes on the left). Only values greater than 2-fold increased or 2-fold decreased in mRNA levels (broken horizontal lines) were considered to be significant changes.
Figure 5.
Comparative responses between shoots and roots of combined isozymic mRNA levels of genes encoding isozymes of exemplary metabolic pathways to representative stress conditions. The combined isozymic mRNA levels of genes encoding biosynthetic/allosteric enzymes, biosynthetic/nonallosteric enzymes, and enzymes catalyzing the first catabolic steps of the amino acids are indicated by blue, black, and red lines, respectively. The names of the enzymes encoded by the different isozymic genes, as provided in Table I, are given in the boxes in B (for A and B) and in D (for C and D). A and B, Comparative responses of the Met pathway to osmotic stress in shoots and roots. C and D, Comparative responses of the Trp pathway to UV-B stress in shoots and roots. The changes in mRNA levels are given in log2 ratios of treatments versus controls (y axes on the left). Only values greater than 2-fold increased or 2-fold decreased in mRNA levels (broken horizontal lines) were considered to be significant changes.
Although the combined isozymic mRNA signals of the biosynthetic steps (allosteric and nonallosteric) of all of the metabolic pathways generally showed either no effect or slight down-regulation in response to the various stresses (Supplemental Figs. S5 and S6), the response of the Trp pathway to UV-B stress in shoots exhibited an exceptional situation in which the combined isozymic mRNA signals of both the catabolic step and many of the biosynthetic steps were stimulated by this stress cue (Fig. 5C). This expression pattern is in general agreement with previous reports using wet experimental research (Zhao and Last, 1996; Ouwerkerk et al., 1999; Glawischnig et al., 2004).
Our study also exposed several distinct spatial expression patterns of specific metabolic pathways in shoots and roots as well as distinct temporal expression patterns of closely associated metabolic pathways in either the shoots or the roots. In many cases, the behavior of a given metabolic pathway to a given stress cue was similar in the roots and shoots, even though it seemed that the general response was slightly lower in roots than in shoots (Supplemental Figs. S5 and S6). In some cases, the response of a given metabolic pathway to a given stress was observed only in shoots (Fig. 5, C and D, depicting the response of the Trp pathway to UV-B stress in shoots and roots, respectively). In other cases, the opposite was observed. The repression of the combined isozymic mRNA signals of the catabolic steps of the Met pathway in response to osmotic stress, and the stimulation of the combined isozymic mRNA signals of one of the catabolic steps of the Phe and Tyr pathway in response to salt stress, were much stronger in the roots than in the shoots (Fig. 5, A with B, for the Met pathway; Supplemental Figs. S5 and S6 for the Phe and Tyr pathway).
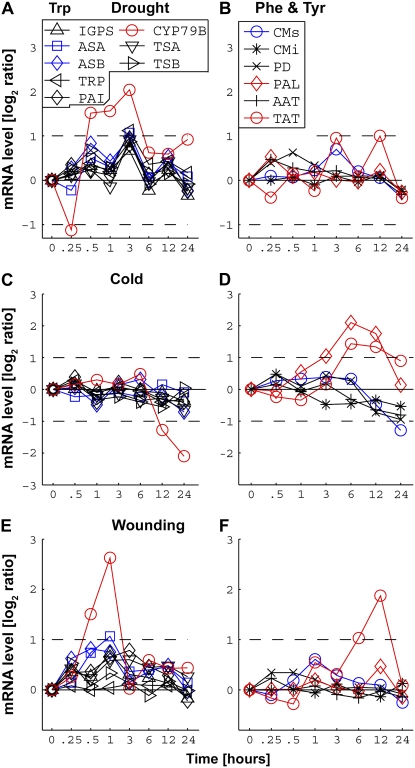
We also observed interesting interactions between the closely associated pathways of Trp and Phe and Tyr metabolism, which are synthesized from the common metabolite chorismate (Fig. 1). Exposure to drought stress stimulated the catabolism of Trp, but not Phe and Tyr, in the shoots (Fig. 6, A and B), while exposure to cold stress stimulated the catabolism of Phe and Tyr, and Trp catabolism was repressed in the shoots (Fig. 6, C and D). Finally, as shown in Figure 6, E and F, wounding stimulated the catabolism of both Trp and Tyr in the shoots, but the stimulation of Trp catabolism occurred earlier (between 0.5 and 1 h after wounding) than Tyr catabolism (between 6 and 12 h after wounding). A similar response of these two metabolic pathways was also observed upon exposure of the shoots to UV-B (Supplemental Fig. S5).
Figure 6.
Comparative responses of the combined isozymic mRNA levels of genes encoding isozymes of the closely related Trp (A, C, and E) and Phe and Tyr (B, D, and F) pathways to various stress conditions. A and B, Drought stress. C and D, Cold stress. E and F, Wounding stress. The combined isozymic mRNA levels of genes encoding biosynthetic/allosteric enzymes, biosynthetic/nonallosteric enzymes, and enzymes catalyzing the first catabolic steps of the amino acids are indicated by blue, black, and red lines, respectively. The names of the enzymes encoded by the different isozymic genes, as provided in Table I, are given in the boxes in A (for A, C, and E) and in B (for B, D, and F). The changes in mRNA levels are given in log2 ratios of treatments versus controls (y axes on the left). Only values greater than 2-fold increased or 2-fold decreased in mRNA levels (broken horizontal lines) were considered to be significant changes.
Not only the majority of the responses of the various metabolic pathways to the various stress cues were principally associated with stimulation or repression of their catabolic genes, but also the catabolic genes responded relatively fast to the stress cues. In the majority of the cases, pronounced stimulations or repressions of the catabolic genes were observed in the range of 0.5 to 3 h following the initiation of the stress, while in only a few cases did the responses occur after 6 to 12 h (Figs. 4–6; Supplemental Figs. S5 and S6).
Differential Response of the Pro and Arg Pathway to Osmotic, Drought, and Salt Stresses
Our analysis revealed that the Pro and Arg pathway has a distinct principal response to the different stress cues than the other metabolic pathways, which was particularly manifested in shoots and to a lesser extent in roots, exposed to osmotic, salt, and to a lesser extent also drought stresses. This response was manifested by an increase in the combined isozymic mRNA signals of the biosynthetic/allosteric step of Pro biosynthesis, namely, Δ-1-pyrroline-5-carboxylate synthetase (Supplemental Fig. S5). These results support published research showing significant accumulation of the osmolite amino acid Pro during drought, osmotic, and salt stresses as a part of the adaptation machineries of plants to these stresses (Rontein et al., 2002).
The Relative Contribution of Protein Breakdown to the Pool of Amino Acids during Abiotic Stresses
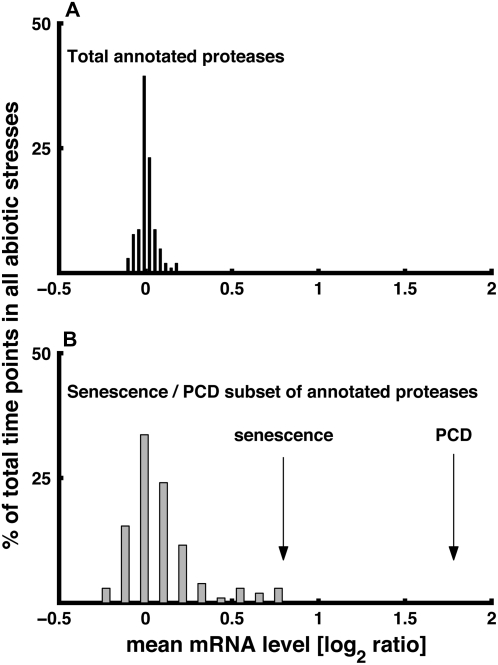
The pools of amino acids during abiotic stresses may be derived not only from their de novo synthesis but also from protein breakdown. To test the relative contribution of protein degradation to the pool of amino acids during abiotic stresses, we grouped all Arabidopsis genes annotated as proteases by TAIR (http://www.arabidopsis.org; Supplemental Table S2). Then, the mean mRNA levels (presented as log2 ratio of treatment versus control) of all of these genes were calculated for each time point in each of the eight stress conditions. Due to the relatively low number of total time points (104 time points), the distribution of the mean mRNA levels was estimated using 10 bins ranging from lowest to highest mean mRNA levels. As shown in Figure 7A, the majority of the time points possessed mean expression levels around zero. Detailed inspection (data not shown) indicated that this is because only a very small subset of the total annotated proteases operate during abiotic stresses. Thus, we decided next to focus only on annotated proteases that are up-regulated (log2 of treatment versus control > 1) during senescence or programmed cell death (PCD) in which there is massive protein degradation. Only approximately 18% of the total annotated proteases showed pronounced stimulation of mRNA levels under these conditions (data not shown) and were grouped as “senescence/PCD annotated proteases.” This subset of annotated proteases is indicated in Supplemental Table S2. The mean mRNA levels of these proteases were approximately 0.8 and approximately 1.8 (log2 ratio of treatment versus control) in the senescence and PCD treatments, respectively (Fig. 7B, values indicated by arrows). In contrast, the mean mRNA levels of these proteases at all time points of all abiotic stresses were distributed around zero, with only a minor proportion reaching the level observed during senescence (histogram in Fig. 7B).
Figure 7.
The distribution of the mean expression levels (log2 ratio of treatment versus control) of genes encoding total annotated proteases (A) and the subset of proteases that are induced in senescence and PCD (B) in individual time points of the entire set of abiotic stresses. The distribution of the mean mRNA levels was estimated using 10 bins ranging from lowest to highest mean mRNA levels. Arrows in B show the mean expression levels of the genes encoding the senescence/PCD subset of proteases in response to senescence and PCD (Buchanan-Wollaston et al., 2005).
It was also interesting to test the mean mRNA levels of the senescence/PCD annotated proteases in the different time points of each of the eight different abiotic stresses individually, both in shoots and in roots. As shown in Supplemental Figure S7, the mean mRNA levels of these proteases were hardly changed, either in roots or in shoots, under all stress conditions, besides the late stages of osmotic stress, in which these mean mRNA levels increased to comparable levels to those observed during senescence. Since some of the stresses also stimulate the conversion of aromatic amino acids into secondary metabolites (Supplemental Figs. S5 and S6), it is likely that protein breakdown is not absolutely required to generate the pools of aromatic amino acids needed for these secondary metabolites.
DISCUSSION
Regulatory Transcriptional Principles of Amino Acid Metabolism in Response to Abiotic Stresses
In this report, we used a bioinformatics approach to analyze the response of genes encoding enzymes in pathways controlling the metabolism of 11 of the 20 protein amino acids to eight different abiotic stresses in the model plant Arabidopsis, using data on mRNA levels derived from publicly available microarray experiments. Since microarray experiments do not allow differentiating between control mechanisms of transcription and mRNA stability, we include in our discussion both types of mechanisms under the term “transcriptional control.” Our results revealed several distinct regulatory transcriptional principles, which characterized all of the tested metabolic pathways, besides that of Pro and Arg metabolism, as follows: (1) genes encoding the catabolic enzymes of the amino acids are generally significantly more sensitive in terms of changes in their mRNA levels to environmental and stress-associated signals than genes encoding the biosynthetic/allosteric and biosynthetic/nonallosteric enzymes (this was evident by both higher “noise” fluctuations and higher magnitudes of responses to the stress cues); (2) in most cases in which there was a transcriptional response of a metabolic pathway to a given stress cue, this response was associated with major changes (stimulation or repression) in mRNA levels of catabolic genes, while the biosynthetic/allosteric and biosynthetic/nonallosteric genes mostly did not respond or in some cases were slightly reduced; (3) the transcriptional responses of the catabolic genes to the stress cues could be quite rapid, with significant stimulation of mRNA levels occurring in many cases between 0.5 and 3 h after the initiation of the stress; and (4) except for cases in which genes of metabolic pathways were entirely not responding in term of their mRNA levels to given stress cues, there was never a case in which catabolic genes of all of the pathways responded in terms of their mRNA levels to a given stress cue in the same manner, implying the occurrence of pathway-specific transcriptional behaviors. Notably, the metabolic pathway of Pro and Arg behaved differently from the other pathways, particularly in its transcriptional response to osmotic and salt stresses and to a lesser extent also to drought stress, which was associated with an induction of the mRNA level of biosynthetic genes encoding Δ-1-pyrroline-5-carboxylate synthetase. These results are in strong agreement with previously published molecular studies (Yoshiba et al., 1995; Strizhov et al., 1997). They also explain the unique function of this pathway to accumulate the osmolite amino acid Pro in response to osmotic, salt, and drought stresses, as a part of the machinery responsible for the adjustment of plants to these stresses (Rontein et al., 2002).
The Principal Transcriptional Regulatory Roles of the Catabolic Genes Are Manifested with Different Spatial and Temporal Response Patterns of the Different Metabolic Pathways to Different Stress Cues
Our results showed that the regulatory roles of the catabolic genes in response to the stress cues were manifested with variable, pathway-specific, spatial, and temporal transcriptional behaviors that include the following. (1) Differential response patterns between roots and shoots. This is exemplified by the occurrence of the relatively strong transcriptional response of the Trp pathway to UV-B in shoots, while no response is observed in roots (Fig. 5, C and D), an expected response taking into account that only aboveground tissues are naturally directly exposed to UV-B stress. (2) Variable, pathway-specific temporal response to a stress cue. This is exemplified by the temporal pattern of transcriptional coordination of the different catabolic steps in the Trp and Phe and Tyr metabolic pathways, whose three aromatic amino acids are synthesized from the common metabolite chorismate (Fig. 1C). Upon wounding of shoots, the mRNA level of the catabolic gene of Trp was transiently stimulated between 0.5 and 1 h, while the mRNA level of the catabolic step of Tyr was transiently stimulated between 6 and 24 h (Fig. 6, E and F, respectively). This temporal transcriptional pattern may lead to a temporal coordination of the flux from chorismate to secondary metabolites derived from Trp and Tyr. (3) The transcriptional response of the metabolic pathway of Trp in shoots to UV-B radiation signified a unique pattern, which included stimulation of the expression of genes encoding both the catabolic step and a number of the biosynthetic steps of this module (Fig. 5C). Yet, even in this unique situation, the combined mRNA signals of the catabolic step were stimulated faster than those of the biosynthetic/allosteric and biosynthetic/nonallosteric steps (Fig. 5C), supporting our design principle of a primary role of the catabolic steps in regulating flux via the pathway in response to a given stress cue.
A Hypothesis for a Two-Component Module Regulating Plant Amino Acid Metabolism in Response to Abiotic Stresses via Combined Transcriptional and Posttranslational Controls
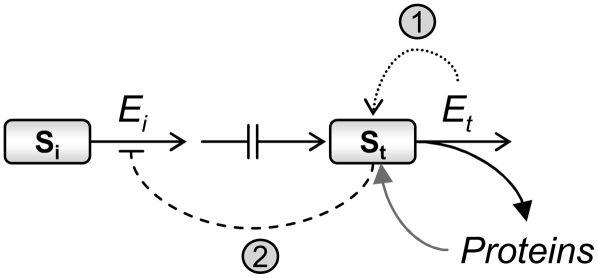
Our results, when taken together with extensivebiochemical studies on amino acid biosynthesis, imply that amino acid biosynthesis is predominantly regulated by posttranslational allosteric feedback inhibition loops, while amino acid catabolism is principally regulated at the transcriptional level. Based on these results, we wish to propose a hypothesis that integrates these two components into a principal program regulating plant amino acid metabolism in response to abiotic stresses. This hypothesis is schematically illustrated in Figure 8. A key regulatory factor in this program is the terminal enzyme of the metabolic module, Et, which is responsible for the first catabolic step of the amino acid. The level of this enzyme can be stimulated or repressed in a significant manner in response to arriving stress cues via a compound transcriptional regulation of its gene(s). A second regulatory component in this program is the biosynthetic/allosteric enzyme, which is the first enzyme of the module and therefore is defined as the initiator biosynthetic enzyme, Ei. The gene(s) encoding Ei as well as the other biosynthetic enzymes of the module are generally expressed at relatively basal levels, which can slightly fluctuate by diurnal and metabolic controls (Osuna et al., 2007), and may apparently enable sufficient flux through the module. Yet, Ei activity is allosterically inhibited by rising levels of the substrate of Et, namely, the amino acid itself (St). A single metabolic module may contain several different Et enzymes, leading to different metabolic directions, each of which may contain several isozymes. Hence, the level of the amino acid (St) is apparently determined by the combined modulation (repression or stimulation) of the entire set of the catabolic genes. Stated simply, the dynamics of the principal regulatory program occurs as follows. Combined stimulation or repression of expression of the gene(s) encoding all Et enzymes in a module in response to a stress cue causes the following consecutive steps: (1) stimulation or repression of Et activity(s); (2) reduction or increase in the level of amino acid, namely St (Fig. 8, curved dotted arrow); and (3) elevation or reduction of Ei activity upon an increase or a reduction in its allosteric feedback inhibition (Fig. 8, curved dashed line), resulting in an acceleration or slowdown of the flux into and throughout the metabolic module. This renders the catabolic Et activities as dominant factors controlling the flux into and through the metabolic module in response to abiotic stresses. Only in specific cases in which the levels of the biosynthetic (allosteric and nonallosteric) enzymes may not be sufficient to maintain adequate flux through the module in response to a stress cue, as was observed for the Trp pathway in response to UV-B stress (Fig. 5C), can expression of the gene(s) encoding the biosynthetic enzymes also be stimulated (Fig. 5C).
Figure 8.
Schematic representation of the proposed regulatory metabolic module. The regulatory steps are as follows. The major controller of the module is the gene encoding the catabolic enzyme Et, catalyzing the first catabolic step of the amino acid. Its upregulation stimulates the module by reducing the level of its substrate (St), namely, the amino acid (step 1; dotted curved arrow). Reduced levels of St stimulate the activity of allosteric biosynthetic Ei enzymes by reduction of its feedback inhibition by St (step 2; dashed curved line), stimulating the flux through the entire module. The broken line represents all biosynthetic/nonallosteric steps. The pool of the amino acid may also be determined by the extent of its incorporation into proteins (curved black arrow) and by the extent of protein breakdown (curved gray arrow). Our proposed module may also fit to actively growing (nonsenescence) tissues of plants grown under favorable (nonstress) conditions in which the catabolic enzymes are generally repressed, but the incorporation of the amino acids into proteins (black curved arrow) may transiently reduce the level of the amino acid (St) and as a consequence enhance the flux through the metabolic module by transiently reducing the feedback inhibition on the allosteric enzyme Ei.
Our hypothesis claiming that relatively basal levels of the biosynthetic (allosteric and nonallosteric) enzymes are generally sufficient to maintain efficient flux through the modules is supported by published evidence showing that removal by mutagenesis of the allosteric feedback inhibition traits of Ei in the two different modules of Lys and Trp metabolism generally results in overproduction of the substrates of Et, namely, the amino acids Lys and Trp (Galili, 1995; Li and Last, 1996). Yet, our current hypothesis still does not take into account other potential regulatory factors, such as posttranslational regulation of metabolic enzymes as well as intracellular compartmentation of enzymes and amino acids for which there is still limited published evidence. For instance, while the allosteric/biosynthetic enzymes are likely to respond strictly to levels of amino acids inside the organelle where they are localized (mostly plastids), amino acids can be sequestered inside vacuoles and by that become regulatorily inert, causing problems of interpreting metabolic fluxes from analyses of the steady-state levels of total pools of amino acids. Indeed, different studies show variable results with respect to the effects of different abiotic stresses on the levels of total pools of amino acids. Heat and drought stresses were shown to cause either increases or decreases in the total pool of several amino acids (Rizhsky et al., 2004), while cold stress caused an increase in the total pool of a number of amino acids (Kaplan et al., 2004; Usadel et al., 2008). The reason for the increased pool size of some amino acids in response to some abiotic stresses is yet unknown, and it is possible that this results from their reduced incorporation into proteins. However, our hypothesis assumes that the accumulating amino acids cannot reside in the plastid, as they would inhibit the allosteric biosynthetic enzymes. Lastly, not only amino acids but also metabolic enzymes may be localized in different compartments. At least some of the catabolic enzymes, such as LKR/SDH (Kemper et al., 1999; Zhu et al., 2000), are localized in the cytosol, and the operation of our hypothetic regulatory module assumes that amino acid transport is not a limiting factor under stress conditions. The regulatory significance of posttranslational regulation of amino acid metabolic enzymes and intracellular transport and sequestration of amino acids await future studies.
The pool of an individual amino acid is not only determined by the operation of our proposed two-component module but also by its incorporation into proteins and by protein breakdown (Fig. 8, curved arrows). Yet, although the potential contribution of posttranscriptional and posttranslational controls to protease activity was not addressed in this report, our transcriptional results (Fig. 7) imply that the contribution of protein breakdown to the pool of amino acids during the relatively early response to abiotic stresses is generally more limited than that occurring in senescence and PCD, in which there is massive protein degradation. This observation is also interesting taking into account that the aromatic amino acids lead to the synthesis of a large number of secondary metabolites. Thus, our results also imply that during the relatively early response to abiotic stresses, the pool of aromatic amino acids used for the production of secondary metabolism is apparently largely derived from primary metabolism via our proposed metabolic module rather than from extensive breakdown of proteins. Notably, only in the case of osmotic stress do the total mRNA levels of these proteases accumulate to a similar intensity to that occurring during senescence (Supplemental Fig. S7). Moreover, osmotic stress in shoots was also characterized as the single condition in which the mRNA levels of most catabolic genes were stimulated (Supplemental Fig. S3). Thus, our results suggest that osmotic stress is distinct from all other abiotic stresses with respect to the operation of amino acid metabolism. The physiological consequences of this interesting phenomenon await future studies.
Several differently designed programs of metabolism have been elucidated in different organisms using a variety of systems biology approaches. In bacteria, the stimulation of amino acid biosynthesis in response to its limiting level occurs in the following principally temporal pattern. The gene encoding the first enzyme of the biosynthetic pathway responds first, while there is a progressive time difference in the response of the genes encoding the consecutive enzymes of the same biosynthetic pathway (Zaslaver et al., 2004). This designed principle, named “just-in-time” (Zaslaver et al., 2004), is an energy-saving program that is apparently important for rapidly dividing organisms, but it requires highly accurate and stringent conservation of promoters during evolution. In contrast, human stem cells not only use a just-in-time program but also maintain a basal expression level of a large number of genes even in the absence of any cue, so that their gene products will be available “just-in-case” when they are needed (Golan-Mashiach et al., 2004; Domany, 2005). This strategy is energetically uneconomical but requires minimal transcriptional control, enabling the cells to have a diverse response to arriving cues, which is critical for their differentiation (Domany, 2005). The regulatory metabolic program suggested by our bioinformatics approach offers a novel strategy that combines the just-in-time (amino acid catabolic genes) and just-in-case (amino acid biosynthetic genes) strategies as well as transcriptional (Et) and posttranslational (feedback inhibition of Ei) controls into a single module.
How efficiently can our proposed two regulatory components (allosteric biosynthetic enzymes and catabolic enzymes of the amino acid) regulate the flux through amino acid metabolic modules? A possible answer to this question can be exemplified in the following published evidence: seeds of an Arabidopsis knockout mutant in the AtLKR/SDH gene of Lys catabolism (Et) accumulate only 2-fold higher Lys levels than wild-type seeds, and a comparable level of Lys also accumulates in transgenic Arabidopsis plants expressing a bacterial Lys-insensitive DHDPS (Ei of the Lys metabolic module; Zhu and Galili, 2003). Yet, combining the AtLKR/SDH knockout and the bacterial Lys-insensitive DHDPS traits by crossing these two plants results in a considerable synergistic increase in Lys level (Zhu and Galili, 2003). These results imply the following: (1) when the gene encoding LKR/SDH (Et) is shut down, a 2-fold increase in Lys level significantly inhibits the activity of DHDPS (Ei); and (2) when the allosteric feedback inhibition trait of DHDPS is released, the flux toward Lys is dramatically stimulated, but most of the overproduced Lys is further catabolized by LKR/SDH, whose activity is generally upregulated in seeds and can also be further induced by increased Lys levels (Karchi et al., 1994; Zhu and Galili, 2003).
The expected metabolic significance of our proposed two principal regulatory components, particularly the general localization of the allosteric/biosynthetic enzymes in branch points of metabolic pathways (Fig. 1), is that they apparently allow rapid and pronounced shifts in fluxes through specific metabolic pathways needed to achieve specific metabolic and physiological status for optimal adjustment of the plants to different stresses. In fact, our suggested principal regulatory components appear to be optimal for amino acid metabolism, because they apparently also regulate the interactions between the synthesis of amino acids and their incorporation into proteins, occurring most extensively under nonstress conditions. Consumption of the amino acid by its incorporation into proteins should analogously reduce the level of the amino acid and stimulate flux into the pathway by the same principles suggested in Figure 8.
Suitability of Bioinformatics Analysis of Microarray Results for Elucidating Regulatory Metabolic Principles
Our proposed principal regulatory program of amino acid metabolism is based on bioinformatics analysis of Affymetrix microarrays. The microarray technology is a well-established method to accurately monitor mRNA levels and has been widely used to elucidate regulatory principles of metabolism, even though it is naturally restricted to transcriptional and posttranscriptional regulation (Ihmels et al., 2004). In addition, our proposed principal regulatory program is also supported by published information concerning genes encoding individual enzymes belonging to the metabolic modules analyzed in this report: (1) the stimulation of mRNA level of the LKR/SDH gene of Lys catabolism by various abiotic stress conditions is supported by published evidence that salt or osmotic stress stimulates the levels of LKR/SDH mRNA, protein, and enzymatic activity (Moulin et al., 2000; Stepansky and Galili, 2003); (2) expression of a bacterial dihydrodipicolinate enzyme of Lys biosynthesis coupled with a knockout of the single LKR/SDH gene of Lys catabolism causes a dramatic synergistic increase in Lys level in Arabidopsis seeds, compared with seeds of plants expressing each of these traits alone, implying that the pathway of Lys metabolism has a high flux potential and is coordinately regulated by the allosteric/biosynthetic DHPS enzyme and the first catabolic enzyme of Lys, namely, LKR/SDH (Zhu and Galili, 2003); and (3) even though the steady-state level of Phe in plants is generally very low, stimulation of the expression of genes encoding its catabolic enzyme Phe-Ala ammonia lyase generally causes significant increases in the levels of many Phe-derived secondary metabolites (Shadle et al., 2003). This implies that the stimulated expression of Phe-Ala ammonia lyase enhances the flux through the allosteric chorismate mutase enzyme of Phe biosynthesis, even though the expression of its gene has not been transgenically enhanced.
CONCLUSION
Our study, covering metabolic pathways of 11 amino acids, proposes a hypothesis for a novel regulatory program controlling plant amino acid metabolism in response to abiotic stresses. In this program, the first catabolic enzyme of the amino acid responds to an external cue by transcriptional control, while the allosteric biosynthetic enzyme responds posttranslationally to resulting changes in the level of the amino acid. Our defined metabolic modules, which initiate by the allosteric/biosynthetic enzymes and terminate by the first catabolic enzymes of the amino acids, are generally located in branch points of metabolic networks and may apparently enable efficient metabolic redirections in response to external cues. Such metabolic modules may enable the redirection of fluxes of primary metabolism as well as the conversion of primary metabolism into secondary metabolism. Our results also imply that the balance of fluxes through the different metabolic modules is likely under specific spatial and temporal controls. Notably, the only exception to our proposed program is the pathway of Pro metabolism, an amino acid that distinctively accumulates to significantly high levels under abiotic stresses. The gene encoding the allosteric biosynthetic enzyme of this pathway is transcriptionally stimulated in response to a number of abiotic stresses. Finally, our hypothesis is based solely on transcriptional programs and posttranslational feedback inhibition loops of allosteric enzymes. It does not incorporate the potential influence of posttranslational controls of other biosynthetic enzymes and intracellular sequestration of amino acids, for which there is still very little published information. The influence of these factors on our proposed hypothesis awaits future studies.
MATERIALS AND METHODS
Data Source
Genes encoding biosynthetic/allosteric, biosynthetic/nonallosteric, and enzymes catalyzing the first catabolic steps of the amino acids in Arabidopsis (Arabidopsis thaliana) were identified and collected using the ARACYC database (http://www.arabidopsis.org/biocyc/index.jsp) supplemented with extensive literature confirmation to avoid false annotations. Genes encoding proteases were collected based on their annotation in TAIR (http://www.arabidopsis.org). Expression data were obtained from the NASC (http://affymetrix.arabidopsis.info/AffyWatch.html), which contains hundreds of publicly available expression profiles. In this study, we focused on well-documented experiments of abiotic stresses from the AtGenExpress project. In these data, each stress experiment contains several time points with replicates and, in total, 272 microarrays. For the evaluation of protease activities during senescence and PCD, two additional experiments from the AffyWatch database were used (NASCARRAYS-52 and NASCARRAYS-30, respectively; Buchanan-Wollaston et al., 2005).
Gene Expression Data Analysis
For gene expression raw data analysis, we used the RMA algorithm implemented under the R programming environment (http://www.bioconductor.org), which is currently the gold standard technique for high-density microarray data analysis. The mean of all replicates was used as the estimated expression level of each gene in each time point. The RMA results of all genes used in this study, derived from all microarray chips used in this study, are provided in Supplemental Table S1 in a CSV (comma separator) format.
Combination of Isozymic mRNA Signals
In order to estimate the transcript level of each enzymatic step, we summed the mRNA signals of genes encoding all isozymes of a given enzymatic step in each microarray separately (in a linear scale) and transformed these values back to a logarithmic scale as an estimator for the expression level of each enzymatic step. These were named combined isozymic mRNA signals.
Statistical Analysis
A Kolmogorov-Smirnov goodness-of-fit hypothesis test was used to estimate the difference between the sd of expression. The sd of the mRNA levels of the two groups of genes encoding biosynthetic/allosteric and biosynthetic/nonallosteric enzymes were not significantly different from each other (P = 0.25); therefore, their data were joined and tested against the group of the catabolic genes. This test was also used to examine the differences in the absolute expression levels of the genes in the different groups. A χ2 test was used to estimate the significance of the data described in Figure 3. First, we calculated the number of actual statistically significant changes in mRNA level (treatment versus control; P < 0.01; absolute log2 ratio > 1) for each gene in each tissue (roots or shoots) in each time point of each of the stresses. Based on this number, we next calculated the expected number of statistically significant changes in each of the three groups of genes, based on the relative number of genes in each group (roots and shoots separately), assuming no preferential response of the genes of each group to each of the stresses. Although it is common to use various methods to control the false discovery rate when analyzing microarray data, we preferred not to use false discovery rate, since the χ2 test is much more valid with large numbers of observations and we focused on general tendencies rather than on specific genes.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The three groups of genes possess no significant difference in their absolute expression levels.
Supplemental Figure S2. The degree of fluctuation of mRNA levels of individual genes.
Supplemental Figure S3. Response of shoot mRNA levels of biosynthetic/allosteric and catabolic genes to abiotic stresses.
Supplemental Figure S4. Response of root mRNA levels of biosynthetic/allosteric and catabolic genes to abiotic stresses.
Supplemental Figure S5. Responses of shoot combined isozymic mRNA levels of all genes to abiotic stresses.
Supplemental Figure S6. Responses of root combined isozymic mRNA levels of all genes to abiotic stresses.
Supplemental Figure S7. Responses of annotated protease mRNA levels to abiotic stresses.
Supplemental Table S1. Raw data of mRNA expression levels.
Supplemental Table S2. List of annotated proteases.
Supplementary Material
Acknowledgments
We thank Dr. Yitzhak Pilpel and Dr. Eitan Rubin for critical reading of the manuscript and helpful comments. G.G. is an incumbent of the Bronfman Chair of Plant Science at the Weizmann Institute of Science.
This work was supported by grants from the United States–Israel Binational Agricultural Research and Development Fund (grant no. IS–3331–02) and by the Israel Science Foundation (grant no. 764/07).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gad Galili (gad.galili@weizmann.ac.il).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Amir R, Hacham Y, Galili G (2002) Cystathionine gamma-synthase and threonine synthase operate in concert to regulate carbon flow towards methionine in plants. Trends Plant Sci 7 153–156 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJ (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42 567–585 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domany E (2005) Analysis of DNA-chip and antigen-chip data: studies of cancer, stem cells and autoimmune diseases. Comput Phys Commun 169 183–187 [Google Scholar]
- Galili G (1995) Regulation of lysine and threonine synthesis. Plant Cell 7 899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G (2002) New insights into the regulation and functional significance of lysine metabolism in plants. Annu Rev Plant Biol 7 153–156 [DOI] [PubMed] [Google Scholar]
- Galili G, Tang G, Zhu X, Gakiere B (2001) Lysine catabolism: a stress and development super-regulated metabolic pathway. Curr Opin Plant Biol 4 261–266 [DOI] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA (2004) Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci USA 101 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan-Mashiach M, Dazard JE, Gerecht-Nir S, Amariglio N, Fisher T, Jacob-Hirsch J, Bielorai B, Osenberg S, Barad O, Getz G, et al (2004) Design principle of gene expression used by human stem cells: implication for pluripotency. FASEB J 18 147–149 [DOI] [PubMed] [Google Scholar]
- Goyer A, Collakova E, Shachar-Hill Y, Hanson AD (2007) Functional characterization of a methionine gamma-lyase in Arabidopsis and its implication in an alternative to the reverse trans-sulfuration pathway. Plant Cell Physiol 48 232–242 [DOI] [PubMed] [Google Scholar]
- Ihmels J, Levy R, Barkai N (2004) Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat Biotechnol 22 86–92 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Joshi V, Fraga M, Rugg A, Yu SX, Li LL, Last RL (2004) Application of a high-throughput HPLC-MS/MS assay to Arabidopsis mutant screening: evidence that threonine aldolase plays a role in seed nutritional quality. Plant J 39 465–475 [DOI] [PubMed] [Google Scholar]
- Joshi V, Laubengayer KM, Schauer N, Fernie AR, Jander G (2006) Two Arabidopsis threonine aldolases are nonredundant and compete with threonine deaminase for a common substrate pool. Plant Cell 18 3564–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchi H, Shaul O, Galili G (1994) Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proc Natl Acad Sci USA 91 2577–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper EL, Neto GC, Papes F, Moraes KCM, Leite A, Arruda P (1999) The role of Opaque2 in the control of lysine-degrading activities in developing maize endosperm. Plant Cell 11 1981–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50 347–363 [DOI] [PubMed] [Google Scholar]
- Li J, Last RL (1996) The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble tryptophan. Plant Physiol 110 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA (2003) Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol 131 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney BP, Miernyk JA, Randall DD (2002) The complex fate of alpha-ketoacids. Annu Rev Plant Biol 53 357–375 [DOI] [PubMed] [Google Scholar]
- Moulin M, Deleu C, Larher F (2000) L-Lysine catabolism is osmo-regulated at the level of lysine-ketoglutarate reductase and saccharopine dehydrogenase in rapeseed leaf discs. Plant Physiol Biochem 38 577–585 [DOI] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, Gibon Y, Bläsing OE, Höhne M, Günter M, Kamlage B, Trethewey R, Scheible WR, Stitt M (2007) Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J 49 463–491 [DOI] [PubMed] [Google Scholar]
- Ouwerkerk PBF, Hallard D, Verpoorte R, Memelink J (1999) Identification of UV-B light-responsive regions in the promoter of the tryptophan decarboxylase gene from Catharanthus roseus. Plant Mol Biol 41 491–503 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Noel JP, Dudareva N (2006) Biosynthesis of plant volatiles: nature's diversity and ingenuity. Science 311 808–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwanski ER, Last RL (1995) Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell 7 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeille F, Jabrin S, Bligny R, Loizeau K, Gambonnet B, Van Wilder V, Douce R, Ravanel S (2006) Methionine catabolism in Arabidopsis cells is initiated by a gamma-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc Natl Acad Sci USA 103 15687–15692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang HJ, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide: the response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontein D, Basset G, Hanson AD (2002) Metabolic engineering of osmoprotectant accumulation in plants. Metab Eng 4 49–56 [DOI] [PubMed] [Google Scholar]
- Shadle GL, Wesley SV, Korth KL, Chen F, Lamb C, Dixon RA (2003) Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of L-phenylalanine ammonia-lyase. Phytochemistry 64 153–161 [DOI] [PubMed] [Google Scholar]
- Stepansky A, Galili G (2003) Synthesis of the Arabidopsis bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase enzyme of lysine catabolism is concertedly regulated by metabolic and stress-associated signals. Plant Physiol 133 1407–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strizhov N, Abrahám E, Okrész L, Blickling S, Zilberstein A, Schell J, Koncz C, Szabados L (1997) Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J 12 557–569 [DOI] [PubMed] [Google Scholar]
- Tempone AG, Sartorelli P, Mady C, Fernandes F (2007) Phenylpropanoids as naturally occurring antioxidants: from plant defense to human health. Cell Mol Biol 15 15–25 [PubMed] [Google Scholar]
- Usadel B, Blasing OE, Gibon Y, Poree F, Hohne M, Gunter M, Trethewey R, Kamlage B, Poorter H, Stitt M (2008) Multilevel genomic analysis of the response of transcripts, enzyme activities and metabolites in Arabidopsis rosettes to a progressive decrease of temperature in the non-freezing range. Plant Cell Environ 31 518–547 [DOI] [PubMed] [Google Scholar]
- Widholm JM (1972) Cultured Nicotiana tabacum cells with an altered anthranilate synthetase which is less sensitive to feedback inhibition. Biochim Biophys Acta 261 52–58 [DOI] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7 263–270 [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K (1995) Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J 7 751–760 [DOI] [PubMed] [Google Scholar]
- Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette MG, Alon U (2004) Just-in-time transcription program in metabolic pathways. Nat Genet 36 486–491 [DOI] [PubMed] [Google Scholar]
- Zhao JM, Last RL (1996) Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis. Plant Cell 8 2235–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Galili G (2003) Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also trans regulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 15 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Tang G, Galili G (2000) Characterization of the two saccharopine dehydrogenase isozymes of lysine catabolism encoded by the single composite AtLKR/SDH locus of Arabidopsis. Plant Physiol 124 1363–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.