Abstract
Tobacco (Nicotiana tabacum) is a member of the Solanaceae, one of the agronomically most important groups of flowering plants. We have performed an in silico analysis of 1.15 million gene-space sequence reads from the tobacco nuclear genome and report the detailed analysis of more than 2,500 tobacco transcription factors (TFs). The tobacco genome contains at least one member of each of the 64 well-characterized TF families identified in sequenced vascular plant genomes, indicating that evolution of the Solanaceae was not associated with the gain or loss of TF families. However, we found notable differences between tobacco and non-Solanaceae species in TF family size and evidence for both tobacco- and Solanaceae-specific subfamily expansions. Compared with TF families from sequenced plant genomes, tobacco has a higher proportion of ERF/AP2, C2H2 zinc finger, homeodomain, GRF, TCP, zinc finger homeodomain, BES, and STERILE APETALA (SAP) genes and novel subfamilies of BES, C2H2 zinc finger, SAP, and NAC genes. The novel NAC subfamily, termed TNACS, appears restricted to the Solanaceae, as they are absent from currently sequenced plant genomes but present in tomato (Solanum lycopersicum), pepper (Capsicum annuum), and potato (Solanum tuberosum). They constitute approximately 25% of NAC genes in tobacco. Based on our phylogenetic studies, we predict that many of the more than 50 tobacco group IX ERF genes are involved in jasmonate responses. Consistent with this, over two-thirds of group IX ERF genes tested showed increased mRNA levels following jasmonate treatment. Our data are a major resource for the Solanaceae and fill a void in studies of TF families across the plant kingdom.
The Solanaceae (nightshade family) is one of the largest and most important families of flowering plants, with over 3,000 species. Many of its members are important crop plants, such as tomato (Solanum lycopersicum), potato (Solanum tuberosum), aubergine (Solanum melongena), and chili pepper (Capsicum annuum). Others are prized for their medicinal, poisonous, or psychotropic effects and are the source of drugs such as atropine, scopolamine, and hyoscyamine (Oksman-Caldentey, 2007). As a result, the Solanaceae has been the focus of considerable research, including genome sequencing projects for both tomato and potato (Mueller et al., 2005; Mullins et al., 2006).
Regulation of gene expression at the level of transcription is a major control point in many biological processes, and plant genomes devote approximately 7% of their coding sequence to transcription factors (TFs; Udvardi et al., 2007). In plants, changes in transcription rates are seen as the plant grows and develops and also as the plant is required to respond to changes in the environment. A plant's ability to respond appropriately to these cues may ultimately influence its chances of survival and affect yields in crop plants. Transcriptional regulation can determine a number of agronomically important traits; therefore, studies of TFs form a major focus in plant biology (Richardt et al., 2007; Udvardi et al., 2007).
The first genome-wide analysis of TFs in any plant species was reported soon after the first plant genome sequence, that of Arabidopsis (Arabidopsis thaliana), was completed (Riechmann et al., 2000). This analysis revealed that Arabidopsis contains at least 29 families of TFs, with 16 families appearing to have no counterpart in animals. However, this first analysis was incomplete due to the discovery of additional TF families and refinements in bioinformatic analysis used to find the genes. The Database of Arabidopsis Transcription Factors (DATF; Guo et al., 2005) currently lists 64 families of TFs as present in Arabidopsis. All 64 of these families are also present in poplar (Populus spp.; Zhu et al., 2007), while the monocot rice (Oryza sativa) contains only 63 families (Gao et al., 2006), missing the STERILE APETALA (SAP) family that is represented by only a single gene in both of the dicot species. Recent large-scale analyses of plant TFs have produced databases for TF studies across the entire plant kingdom (Richardt et al., 2007; Udvardi et al., 2007). These are excellent resources, but they are largely restricted to sequences already present in public databases and contain no comprehensive analysis of a member of the Solanaceae.
Tobacco (Nicotiana tabacum) has been a model plant for decades and is one of the most studied higher plant species. Many resources are already available to facilitate functional genomics in tobacco, including transformation and regeneration systems, reduced complexity cell culture lines, and extensive mutant and breeding collections (Geelen and Inze, 2001). However, no large data set of TFs is available for either tobacco or any member of the Solanaceae. This constitutes a major void in both comparative plant genomics and Solanaceae-based research into gene regulatory mechanisms.
Given its large genome size of approximately 4.5 Gb (Zonneveld et al., 2005), complete sequencing of the tobacco genome would be difficult. Fortunately, several methods are now available that can provide sequence information for the majority of genes in a species without the need to sequence and assemble the entire genome. One of these is methylation filtration (MF), which preferentially clones the hypomethylated fraction of the genome, effectively reducing the size of the genome to be sequenced. MF has already been applied successfully in maize (Zea mays), sorghum (Sorghum bicolor), and cowpea (Vigna unguiculata; Rabinowicz et al., 1999; Palmer et al., 2003; Whitelaw et al., 2003; Chen et al., 2007). The development of MF followed studies of genome architecture that revealed that repetitive elements tend to form clusters within plant genomes that become heavily methylated (hypermethylated), leaving stretches of less-methylated (hypomethylated), low-copy gene-rich space scattered in islands throughout the genome (Bennetzen et al., 1994; Bedell et al., 2005).
The Tobacco Genome Initiative (TGI; http://www.tobaccogenome.org/) has generated over one million gene-space sequence reads (GSRs) from MF tobacco genomic DNA libraries prepared from ‘Hicks Broadleaf’, a highly inbred and fairly homozygous cultivar. Based on predictions of enrichment gained by MF (Gadani et al., 2003), the approximately 1.2 Gb of gene space sequenced to date sequence tags more than 90% of the open reading frames in the tobacco genome (C. Opperman, personal communication). The availability of these GSRs facilitates genome-wide analysis, large-scale functional genomics, and gene discovery.
Here, we have carried out an in silico analysis of approximately 1.15 million gene GSRs from the tobacco genome and report the detailed analysis of approximately 2,500 TFs present in the tobacco genome. This is currently the largest data set of TF genes from any member of the Solanaceae, and our analysis of the TF genes revealed novel subfamilies that are absent from currently sequenced plant genomes. We have established a publicly available Web site, TOBFAC: The Database of Tobacco Transcription Factors (http://compsysbio.achs.virginia.edu/tobfac/), to facilitate access to our findings and as a knowledge base of tobacco TFs for use by the broader plant community (Rushton et al., 2008).
RESULTS AND DISCUSSION
TFs in the Tobacco Genome
To identify genes encoding TFs present in the tobacco genome, a data set of 1,159,022 GSRs with an average read length of approximately 600 bp was downloaded from the TGI (http://www.tobaccogenome.org/). Based on a predicted 10-fold enrichment resulting from MF (Gadani et al., 2003), the approximately 1.2 Gb of sequence contained in this data set provides at a minimum 1.0× to 1.4× coverage of the gene space and should sequence tag 90% of the open reading frames in the tobacco genome (C. Opperman, personal communication). In fact, this estimate is likely the lower end of coverage, since BLAST searches with a sample of 56 randomly chosen published tobacco genes, including 10 non-TFs (e.g. PUTRESCINE N-METHYLTRANSFERASE [NtPMT1], PYRUVATE DEHYDROGENASE E1α, and NtpolI-like1), showed that 94.6% of the genes (53 of 56) were tagged in the 1.15-million GSR data set. Therefore, we anticipated that sequences encoding the majority of the TF genes are present.
We developed a procedure for the identification of GSRs encoding TFs that proved to be well suited for gene discovery from large GSR data sets (Supplemental Fig. S1; see “Materials and Methods”). This approach consisted of searching the GSR data set by tBLASTn using conserved protein domains from 64 plant TF families previously identified in vascular plants (Guo et al., 2005), followed by the assembly of contigs and manual curation to remove false positives. Using this approach, we found 2,882 tobacco GSR assemblies (contigs and singletons) representing a minimum of 2,513 TF genes (Table I). The actual number of genes in our data set is between 2,513 (the minimum number of genes) and 2,882 (the total number of sequences). This value is greater than the predicted number of TFs present in Arabidopsis (1,922) and rice (O. sativa indica, 2,025, O. sativa japonica, 2,384; Guo et al., 2005, 2006) and equivalent to that reported in poplar (2,576; Zhu et al., 2007). N. tabacum is considered to be an ancient allotetraploid (2n = 4x = 48) resulting from an interspecific hybridization event between Nicotiana sylvestris and Nicotiana tomentosiformis (Goodspeed, 1954; Murad et al., 2002). As a result of its polyploid nature, it is difficult to predict the total number of tobacco TFs, but this number is likely to be over 3,000. Each predicted tobacco TF gene was given an arbitrary number. Supplemental Table S1 lists each predicted gene, together with the accessions numbers of all of the individual GSRs that are components of this gene. These data allowed the first analysis of TF genes from a member of the Solanaceae using a large data set.
Table I.
Size distribution of the TF families found in tobacco
The table shows the minimum number of members identified in each of the 64 families of TFs found in the tobacco GSR data set. Each family is identified by its abbreviated name, and the estimated minimum number of gene family members is given in parentheses.
| ABI3-VP1 (76) | Alfin (9) | AP2/ERF (35/239) | ARF (12) | ARID (8) |
|---|---|---|---|---|
| AS2 (75) | AUX-IAA (35) | BBR-BPC (3) | BES1 (19) | bHLH (190) |
| bZIP (75) | C2C2-CO-like (40) | C2C2-Dof (46) | C2C2-GATA (28) | C2C2-YABBY (10) |
| C2H2 (161) | C3H (69) | CAMTA (6) | CCAAT-DR1 (3) | CCAAT-HAP2 (12) |
| CCAAT-HAP3 (15) | CCAAT-HAP5 (6) | CPP (3) | E2F-DP (6) | EIL (6) |
| FHA (12) | GARP-ARR-B (11) | GARP-G2-like (64) | GeBP (15) | GIF (4) |
| GRAS (45) | GRF (23) | HB (129) | HMG (9) | HRT-like (2) |
| HSF (34) | JUMONJI (18) | LFY (2) | LIM (22) | LUG (4) |
| MADS (119) | MBF1 (5) | MYB (194) | MYB-related (56) | NAC (152) |
| Nin-like (11) | NZZ (2) | PcG (20) | PHD (59) | PLATZ (15) |
| S1Fa-like (1) | SAP (4) | SBP (27) | SRS (12) | TAZ (8) |
| TCP (43) | TULP (13) | Trihelix (40) | ULT (4) | VOZ (2) |
| Whirly (2) | WRKY (93) | ZF-HD (38) | ZIM (13) |
At least one gene family member was identified from each of the 64 TF families examined in this study. The minimum number of genes identified in each family is shown in Table I. The largest gene family is the ERF family, which has at least 239 members. There are also at least 35 of the related AP2 genes. Among the other large TF families in tobacco are the R2R3MYB, bHLH, C2H2 zinc finger, NAC, homeodomain, MADS box, WRKY, bZIP, and AS2 gene families. At the other end of the spectrum are the LEAFY, HRT-like, NOZZLE, VOZ, and whirly families, which have only two members each, and the S1Fa-like family, which is represented by only a single gene in our data set.
Having identified members of the TF families, we then examined these families relative to what has been reported for the size and complexity of the homologous families in the genomes of Arabidopsis, rice, and poplar. Fifty-one of the 64 identified TF families in tobacco were of similar size to the families present in the three other plant species. Thirteen families, however, appeared to be significantly different in size when comparing our tobacco data set with the TFs from the three complete plant genomes (Supplemental Table S2). Tobacco appears to have proportionally more ERF/AP2, C2H2 zinc finger, homeodomain, GRF, TCP, zinc finger homeodomain, BES, and SAP genes. With some families (e.g. BES and SAP) this increase is associated with the acquisition of a novel subfamily (see below). Other TFs, including the ARF, CCAAT HAP5, CPP, ZIM, and PcG gene families, may be underrepresented in tobacco. Together, our data suggest that the evolution of the Solanaceae was not associated with the wholesale gain or loss of TF families; rather, Solanaceae-specific, or even tobacco-specific, expansion of TF subfamilies occurred. This suggestion is consistent with the results of an earlier study that compared 37 gene families in rice and Arabidopsis and found several lineage-specific TF subfamilies but no lineage-specific families (Xiong et al., 2005). It is also in accordance with a recent study of land plant TFs that indicated that all angiosperm TF families were already present in the earliest land plants (Richardt et al., 2007). While we have found evidence for both Solanaceae-specific and tobacco-specific expansion of TF subfamilies, we have found no evidence for the loss of TF families.
Similarities between Tobacco TFs and Those from Sequenced Plant Genomes
To study the TF gene families in more detail, phylogenetic trees were constructed with MEGA version 4 (Tamura et al., 2007) using the neighbor-joining method. In each case, the conserved domains were used to construct the phylogenetic trees. Two phylogenetic trees were produced for each TF family, one containing only the tobacco TF family members and the other containing the tobacco sequences and sequences of selected members of the Arabidopsis TF family as markers for specific subgroups. We used the Arabidopsis domains because they are the most complete and well-characterized data set. The latter phylogenies enable direct comparisons to be made between family members from the two species and allow the rapid identification of any novel lineages within the tobacco gene family. Differences represent components of potentially novel gene regulatory mechanisms, and we predict that some of these TFs regulate Solanaceae- or tobacco-specific processes.
We analyzed eight of the largest TF families in tobacco, the ERF, R2R3MYB, bHLH, NAC, homeodomain, MADS box, WRKY, and bZIP families. These account for over one-third of the total number of tobacco TFs. Our analyses showed that seven of these families are similar in composition to other vascular plants, the one notable exception being the NAC family. Despite the broad overall similarity of these seven families, each provides insights into the evolution, subfamily distribution, and possible functional relatedness of the gene family members. All trees were produced by the neighbor-joining method using MEGA version 4 (Tamura et al., 2007), and similar trees were produced using other methods.
The ERF Family
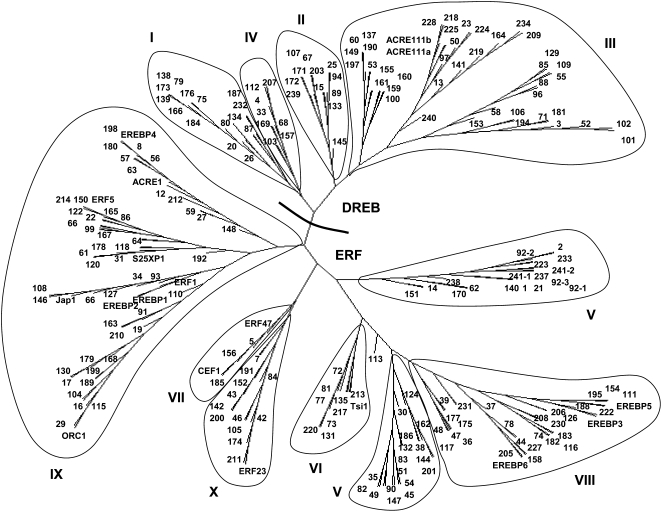
The ERF family of TFs was first discovered in tobacco (Ohme-Takagi and Shinshi, 1995) and is a large multigene family whose members play important roles in biotic and abiotic stress responses as well in the regulation of plant growth and development (McGrath et al., 2005; Nakano et al., 2006). The ERFs are part of the larger AP2/ERF superfamily that is defined by the presence of the AP2 domain. Our searches revealed 241 tobacco sequences that contain complete or partial ERF domains. Of these, 10 contain only the N-terminal part of the domain and seven contain only the C-terminal portion. Based on their amino acid sequences and phylogenetic positions, a maximum of two N-terminal fragments could potentially correspond to C-terminal fragments; therefore, the minimum number of ERF genes in tobacco is 239. A minimum of 35 AP2 genes are also present (Table I), and the 274 tobacco AP2/ERF genes form the largest single family of tobacco TFs. Phylogenetic analysis of the tobacco ERF gene family shows that it is broadly similar to other ERF gene families from both monocot and dicot plants (Fig. 1). The gene family forms major clades that correspond to subgroups I to X that have been found in both rice and Arabidopsis (McGrath et al., 2005; Nakano et al., 2006). There is, however, one major difference between the tobacco ERF family and that from Arabidopsis: the group V genes from tobacco form two separate clades. We suggest that this may be a common feature of ERF gene families, as we have observed that the group V genes from cowpea also form two clades in a similar way (Timko et al., 2008). Additionally, the group V genes from rice are also not monophyletic (Nakano et al., 2006). Overall, the tobacco ERF gene family confirms the similarity of the ERF gene families from both monocot and dicot plants and also suggests a modification to the classification of group V ERF genes.
Figure 1.
Tobacco ERF genes. Shown is an unrooted phylogenetic tree of the ERF domains constructed using the neighbor-joining method. Each gene is designated by an arbitrary number, and published genes are represented by their published names. Roman numerals indicate previously defined subgroups, and a line separates the ERF and DREB subfamilies. Group V is found as two separate clades.
The WRKY Family
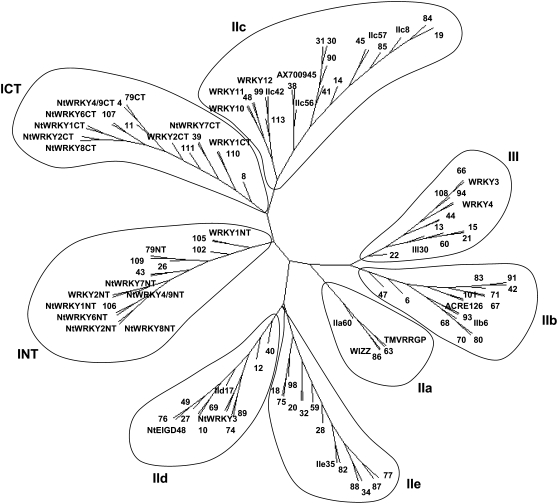
The WRKY TFs are one of the largest groups of transcriptional regulators in plants (Rushton et al., 1995, 1996; Eulgem et al., 2000; Ulker and Somssich, 2004; Zhang and Wang, 2005), mediating responses not only to biotic stresses (Rushton et al., 1996) but also to abiotic stresses, such as wounding, drought, and cold adaptation (Eulgem et al., 2000; Ulker and Somssich, 2004). WRKY genes are classified into groups I, IIa, IIb, IIc, IId, IIe, and III based on their primary amino acid sequence and the structure of their zinc finger motifs (Eulgem et al., 2000; Zhang and Wang, 2005). A total of 131 full or partial WRKY domains, representing a minimum of 93 WRKY genes, were identified and used in our phylogenetic analysis. Included in this number are 18 published tobacco WRKY sequences whose domain-coding regions were not found in our GSR data set. Fifteen of these genes were in fact tagged in the GSRs but only contained sequence information outside of the conserved domain used in our analysis.
The composition of the WRKY gene family in tobacco is similar to that from other dicot plants, as it consists of the same seven groups of WRKY genes (I, IIa, IIb, IIc, IId, IIe, and III; Fig. 2). The relative number of members in each group is also similar. This contrasts with the WRKY gene family from rice, in which the number of genes in group III is greatly expanded (Zhang and Wang, 2005), making up approximately one-third of all WRKY genes. This is not seen in the tobacco gene family (Fig. 2), and it appears that amplification of this group is associated with the evolution of monocots. The initial classification of WRKY genes into groups I, IIa, IIb, IIc, IId, IIe, and III was based only partly on phylogeny and relied also on the number and structures of the WRKY domains (Eulgem et al., 2000). Recently, phylogenetic analysis led Zhang and Wang (2005) to propose that group II WRKY genes are not monophyletic but instead form three distinct clades: IIa + IIb, IIc, and IId + IIe. The tobacco WRKY gene family provides new data to test this and suggests that this new classification is correct. Group II WRKY genes are not monophyletic, and IIa + IIb, IIc, and IId + IIe are to be found in different areas of the phylogenetic tree (Fig. 2).
Figure 2.
Tobacco WRKY genes. Shown is an unrooted phylogenetic tree of the WRKY domains constructed using the neighbor-joining method. Each gene is designated by an arbitrary number, and published genes are represented by their published names. Arabidopsis domains are included for identification purposes and indicated by the name of their group. Group I WRKY genes normally have two WRKY domains, and both were used individually to construct the phylogenetic tree. INT and ICT denote the N-terminal and C-terminal domains from these group I genes.
The Homeodomain Family
Homeodomain proteins were initially discovered during the study of homeotic mutants in Drosophila (Gehring, 1987) and play important roles in developmental regulation in all eukaryotic lineages. Tobacco genes were isolated by searches with the homeodomains from each of the major subgroups of plant homeodomain proteins (Chan et al., 1998). We found that this approach was crucial, as the different subgroups of tobacco homeodomain genes are dissimilar. BLAST searches with the homeodomains from one group often failed to identify genes from other groups, even when using a very high e-value cutoff. To illustrate this, searches with the homeodomain from glabra2 resulted in 74 hits, whereas searches with that from knotted1 yielded only 15 hits and none of these were present in the data set obtained with glabra2. The combined results from all searches revealed a minimum of 129 homeodomain genes in tobacco. The tobacco homeodomain gene family is similar to the plant homeodomain families in other higher plants (Chan et al., 1998; Haecker et al., 2004; Hake et al., 2004; Prigge et al., 2005) and can be divided into nine major subgroups, the knotted class 1, knotted class 2, PHD finger, Bell, glabra, HDZip class 1, HDZip class 2, HDZip class 3, and WOX classes (Supplemental Fig. S2). This phylogenetic similarity should aid the identification of functional homologs in tobacco of characterized homeodomain genes from other higher plants.
The MYB Family
The MYB family is the largest family of TFs in many plant species (Qu and Zhu, 2006), and our data suggest that it is the second largest in tobacco (Table I). MYB TFs are defined by up to three imperfect repeats of the MYB DNA-binding domain (Ogata et al., 1992). The largest MYB subfamily is the R2R3MYB family, and these appear to be predominantly involved in plant-specific processes such as the regulation of plant secondary metabolism and the identity and fate of plant cells (Kranz et al., 1998; Stracke et al., 2001). We identified 232 R2R3MYB sequences, and this represents a minimum number of 194 R2R3MYB genes in tobacco. Tobacco also contains a minimum of 56 MYB-related genes (Table I); therefore, the MYB family in tobacco contains at least 250 genes. The phylogenetic tree of the R2R3MYB genes can be divided into numerous small clades (Supplemental Fig. S3). This is similar to Arabidopsis, in which at least 23 R2R3MYB subgroups have been defined (Stracke et al., 2001). As there is no clear cross-species nomenclature, we have given the tobacco clades arbitrary numbers. Previous analysis of the Arabidopsis R2R3MYB family concluded that there are clear examples of functional conservation between related members of the R2R3MYB family across species (Stracke et al., 2001). This potential conservation has enabled us to use the phylogenetic tree to identify tobacco genes that are similar to the key regulators GLABROUS1, WEREWOLF, TRANSPARENT TESTA, ALTERED TRYPTOPHAN REGULATION1, and GAMYB.
The bZIP Family
A total of 75 bZIP genes were identified in the tobacco GSR data set. These genes form 10 clades, corresponding to groups A to E, G to I, M, and S previously defined in Arabidopsis (Jakoby et al., 2002; Supplemental Fig. S4). In this regard, the tobacco bZIP gene family is broadly similar to that found in Arabidopsis. However, tobacco may be missing homologs of the Arabidopsis group F bZIP genes, as none were identified in the GSR data set. The absence of group F genes in tobacco could indicate that this group is part of a species- or family-specific expansion.
The MADS Box Family
Tobacco contains at least 119 MADS box genes (Supplemental Fig. S5). Members of the MADS box family are known to be predominantly involved in developmental processes (Parenicova et al., 2003) and are found in animal as well as plant species. In plants, the gene family has greatly expanded, and this appears to have been instrumental in shaping the evolution of the true flower approximately 120 to 150 million years ago (Rijpkema et al., 2007). The MADS box genes can be divided into five subgroups (MIKC, Mα, Mβ, Mγ, and Mδ) based on the phylogenetic relationships of the MADS box domain (Parenicova et al., 2003). The MADS box genes in tobacco are found in these same five subgroups, and it appears that the MADS box gene family is similar in structure and composition across higher plants.
The bHLH Family
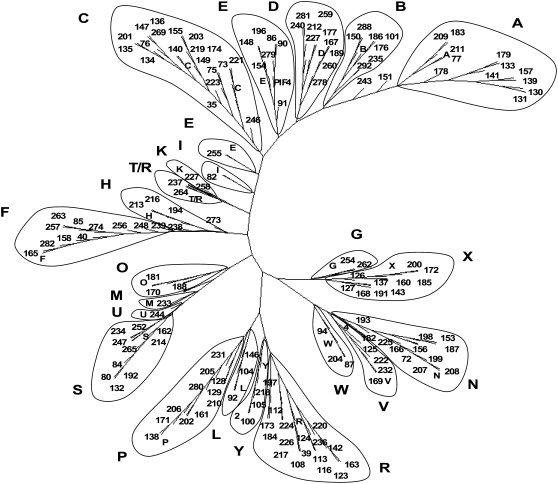
Of all the similarities we observed between tobacco TF families and those from sequenced plant genomes, the bHLH family is the most striking. bHLHs form one of the largest families of TFs in plants (Bailey et al., 2003) and regulate numerous processes. Analysis of the bHLH family is technically more difficult than for most of the other TF families (Bailey et al., 2003; Heim et al., 2003; Toledo-Ortiz et al., 2003); therefore, we performed a total of 30 independent searches using a representative bHLH domain from each of the 23 subfamilies described by Heim et al. (2003). This resulted in the identification of 192 bHLH sequences representing a minimum of 190 bHLH genes. Figure 3 shows a phylogenetic tree of the tobacco bHLH domains together with a marker domain (shown by the letter of the subfamily) for each of the 23 subfamilies. There is clear evidence suggesting that tobacco contains members of all 23 bHLH subfamilies that are present in Arabidopsis. Even smaller subfamilies, such as groups I, M, and U, appear to have at least one tobacco member. This supports the detailed classification of Arabidopsis bHLH genes (Bailey et al., 2003) and suggests conservation of the composition of the bHLH gene family among vascular plants. The phylogenetic tree identifies possible tobacco homologs of key regulators of light responses, such as the PIF/PILs (subgroup E), and jasmonate responses, such as AtMYC2/jin1/jai1 (subgroup N). Recently, it was shown that MYC2, a key transcriptional activator of jasmonate responses in Arabidopsis, interacts with the JAZ family of transcriptional repressors (Chini et al., 2007; Thines et al., 2007). The JAZ proteins are members of the ZIM family of TFs and interact with another central regulator of JA signaling, the F box protein COI1 (Chini et al., 2007; Thines et al., 2007). We have found at least 13 ZIMs in tobacco (Table I), and there are apparent homologs of the complete COI1/JAZ1/MYC2 jasmonate-inducible signaling cascade in tobacco (data not shown).
Figure 3.
Tobacco bHLH genes. Shown is an unrooted phylogenetic tree of the bHLH domains constructed using the neighbor-joining method. Each tobacco gene identified in the GSR data set is designated by an arbitrary number. Large letters indicate previously defined subfamilies. To identify subfamilies, the bHLH domain from one representative gene of each subfamily was included and is indicated by the small letter within the phylogenetic tree. The Arabidopsis gene groups used were as follows: A, AtbHLH087 (At3g21330); B, AtbHLH139 (At5g43175); C, AtbHLH44 (At1g18400) and AtbHLH31 (At1g59640); D, AtbHLH130 (At2g42280); E, AtbHLH26 (At1g02340) and AtbHLH73 (At5g67110); F, AtbHLH103 (At4g21340); G, AtbHLH135 (At1g74500); H, AtbHLH150 (At3g05800); I, AtbHLH142 (At5g64340); K, AtbHLH12 (At4g00480); L, AtbHLH102 (At1g69010); M, AtbHLH34 (At3g23210); N, AtbHLH6 (At1g32640); O, AtbHLH95 (At1g49770); P, AtbHLH96 (At1g72210); R, AtbHLH22 (At4g21330); S, AtbHLH20 (At2g22770); T/R, AtbHLH1 (At5g41315); U, AtbHLH41 (At5g56960); V, AtbHLH30 (At1g68810); W, AtbHLH117 (At3g22100); X, AtbHLH55 (NM_101125.3); and Y, AtbHLH91 (At2g31210). The bHLH domain from PIF4 (At2g43010) is also included for comparison.
Differences between Tobacco TFs and Those from Sequenced Plant Genomes
We found a number of notable differences in the composition of several TF families in tobacco compared with those in poplar, Arabidopsis, and rice. This includes a number of novel TF subfamilies that may be components of regulatory circuits specific to tobacco or the Solanaceae.
The NAC Family
A major difference is found in the NAC gene family, one of the largest families of plant-specific TFs (Guo et al., 2005; Olsen et al., 2005). NACs have been implicated in regulating diverse processes, including flower development, reproduction, defense against insect pests and pathogens, abiotic stress responses, and responses to hormones (Olsen et al., 2005). NAC TFs are defined by the presence of the NAC domain, a conserved DNA-binding domain that appears to have no known close structural homologs (Aida et al., 1997; Ernst et al., 2004).
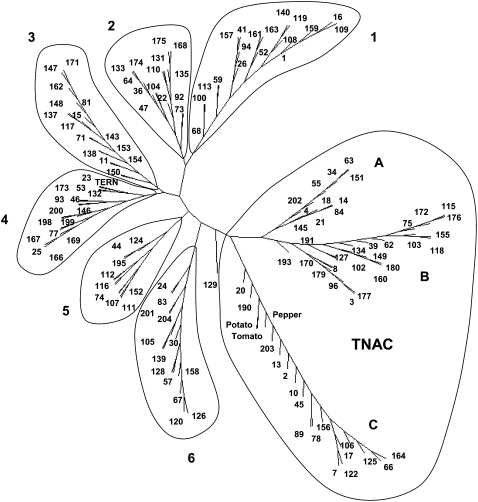
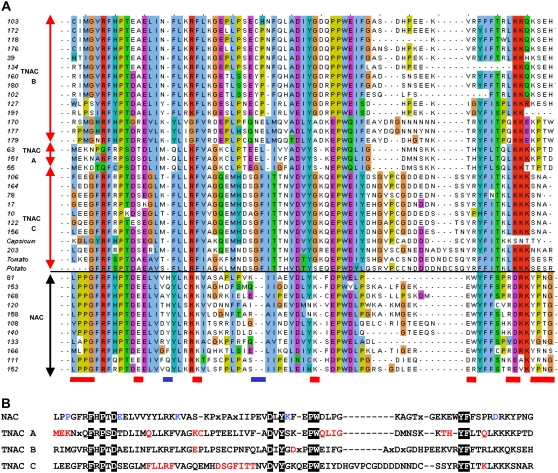
We found 203 complete or partial NAC domains in tobacco and a minimum number of 152 NAC genes. Previous phylogenetic analysis of NAC TFs has been limited. The most comprehensive study of NACs is by Ooka et al. (2003), who divided the rice and Arabidopsis NAC family into two major subgroups and numerous minor groups. Figure 4 shows the phylogenetic relationship of members of the tobacco NAC gene family. We identified seven major subfamilies, six of which are present in tobacco and other plant species and a seventh subfamily that contains the largest number of tobacco NAC genes and appears unique to the Solanaceae. This subfamily, termed TNACS, represents not only a novel subgroup of NAC genes but also a major difference between tobacco and all sequenced plant genomes. There are approximately 50 TNAC genes, and they account for approximately one-quarter of all NAC genes in tobacco. The TNAC genes can be further subdivided into three major clades (A, B, and C), with members in each clade having clearly different primary amino acid sequences in their NAC domains. The differences among the NAC domain sequences in the TNAC genes (subdomains A–C) and how they differ from the NAC domain consensus derived from the other six groups of tobacco NACs are illustrated in Figure 5. The TNACs lack the LPPG motif at the N-terminal end of the NAC domain, and the conserved D/EEE motif found in other NACs is replaced by D/ExE in TNACS. In addition, the TNACs are missing the YPNG motif (underlined in red in Fig. 5), and there are two areas where additional amino acids are present in either the NACs or TNACs (underlined in blue in Fig. 5). Differences between TNACS and NACs are also found in the D and E subdomains (data not shown).
Figure 4.
Tobacco NAC genes. Shown is an unrooted phylogenetic tree of the NAC domains constructed using the neighbor-joining method. Each tobacco gene identified in the GSR data set is designated by an arbitrary number. Six clades (1–6) are found in tobacco and other plant species, and three clades, designated TNAC A to TNAC C, are found in tobacco and other Solanaceae species. EST sequences from TNAC genes of potato (CV505554), tomato (BI422367), and pepper (U204177) are included in TNAC clade C. The published tobacco NAC gene TERN (AB021178) is indicated.
Figure 5.
Amino acid sequence comparisons of the N-terminal half of tobacco NAC domains. A, Amino acid sequences of the N-terminal half of tobacco NAC domains from the TNAC A, B, and C clades and other NAC genes. Areas of significant difference are indicated by red boxes at the bottom of the alignment, and missing/additional amino acids are indicated by blue boxes. Three EST sequences from other Solanaceae TNACs are also included: tomato (BI422367), potato (CV505554), and pepper (U204177). B, Consensus sequences of NAC and the three TNAC clades. Blue letters show amino acids that are conserved in NAC domains but absent from TNAC domains. Red letters show amino acids that are characteristic of TNAC clades. Amino acids that are conserved in the consensus sequence for all four are highlighted in black boxes.
We sought to determine the distribution of TNACs across the plant kingdom. Extensive BLAST searches of the National Center for Biotechnology Information databases (all GenBank + EMBL + DDBJ + PDB sequences, Plant Genomes EST sequences, Genome Survey Sequence, Unfinished High Throughput Genomic Sequences) and the Sol Genomics Network (http://www.sgn.cornell.edu/) revealed that there are no published TNAC genes and that they are completely absent from all currently sequenced plant genomes. A total of just six TNAC ESTs, one from tomato (BI422367), one from potato (CV505554), one from pepper (U204177), and three from tobacco (AM845922, BP137257, and AF211685), were found in the databases, together with one tomato TNAC gene (SL_MboI0012J01 3) from a Genome Survey Sequence and one predicted TNAC gene on tomato chromosome 2 (C02SLm0076E07). Like tobacco, the crop plants tomato, potato, and pepper are also members of the Solanaceae, and this large group of novel NAC genes appears to be a previously unknown feature of transcriptional regulation in the Solanaceae.
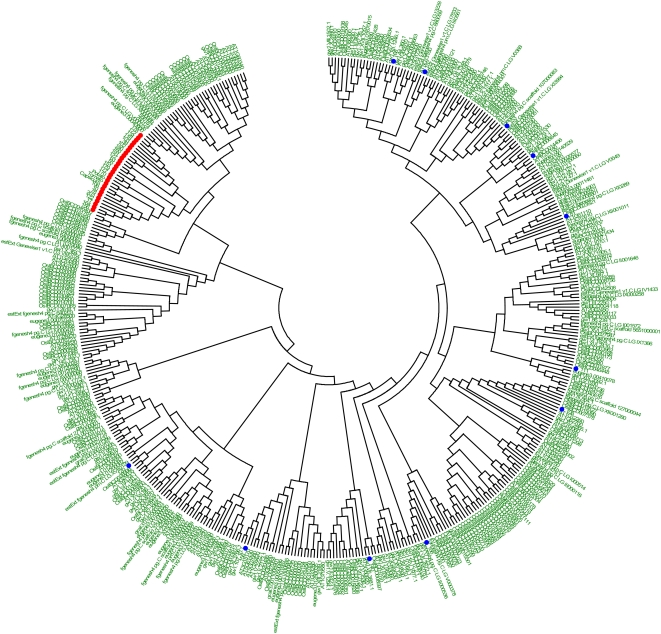
To demonstrate the Solanaceae-specific nature of the TNAC subfamily, we constructed a phylogenetic tree containing the complete NAC gene families from Arabidopsis, poplar, and rice (indica), together with the tobacco NAC genes shown in Figure 5. The A to C portions of the NAC domains were used for its construction. Figure 6 shows this combined phylogeny of over 450 NAC genes. The non-TNAC genes from tobacco (blue dots) are widely scattered across the phylogenetic tree, illustrating that they belong to NAC subfamilies that are present in different plant species. In stark contrast, the TNAC genes (red dots) clearly form a separate clade, and this demonstrates that TNACs are absent from the other three plant genomes and are likely to be specific to the Solanaceae. Due to the limited previous phylogenetic information on the NAC gene family, our analysis of over 450 NAC genes should be of broad use for studies of NAC genes.
Figure 6.
Phylogenetic analysis of the complete NAC gene families from Arabidopsis, poplar, and rice together with NAC and TNAC genes from tobacco. TNAC genes are highlighted with red dots and tobacco NAC genes are highlighted with blue dots. The phylogeny contains over 450 genes and shows that the TNACs form a separate clade. All rice, poplar, and Arabidopsis NAC genes are indicated by their accession numbers.
The TNAC ESTs from tobacco come from both the A and C clades. The A clade tobacco EST AF211685 comes from cell suspension cultures harvested 30 min after treatment with the Avr9 peptide from the fungus Cladosporium fulvum. The other tobacco A clade EST (AM845922) appears to come from the TNAC gene NtNAC84 and was isolated from cold-treated whole plants. This suggests that some A clade genes may play roles in stress responses in tobacco. We have performed expression studies on a small number of representative TNAC genes and found that members of all three clades are expressed in tobacco. Some TNAC genes, such as the B clade gene NtNAC176 and the C clade gene NtNAC156, appear to be expressed in most plant tissues, whereas the A clade gene NtNAC151 is mostly expressed in roots and young leaves (data not shown). We suggest that TNACs are components of regulatory circuits, some of which are specific to tobacco or the Solanaceae.
The BES Family
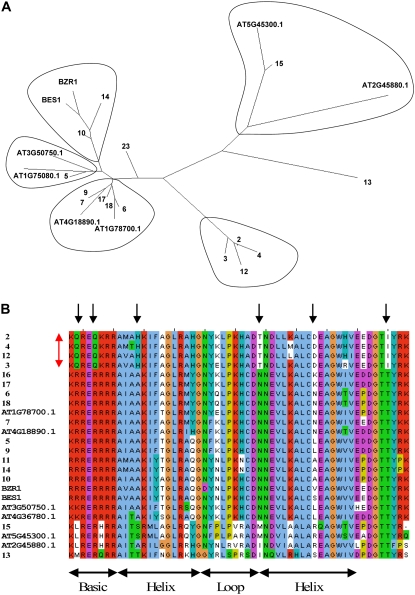
The prototype of the BES family of TFs is BES1 (Yin et al., 2002, 2005), a TF that binds to, and activates, brassinosteroid-regulated gene promoters. Tobacco contains at least 19 BES-like TFs distributed in five main clades (Fig. 7A). Four clades have members with homologs in other plant species. Among the best characterized BES genes are the Arabidopsis genes BES1 and BZR1, which play roles in stem elongation and senescence (Yin et al., 2002, 2005). Based on sequence homology, NtBES10 and NtBES14 are likely to be their functional homologs in tobacco (Fig. 7). The fifth clade is composed of four tobacco BES-like genes that are significantly different from members of the other four clades. Figure 7B shows a comparison of the conserved N-terminal regions of the tobacco BES genes and their Arabidopsis counterparts. There are at least six amino acid differences in the conserved domain that are a hallmark of the tobacco-specific group. This N-terminal domain contains a bipartite nuclear localization signal, together with a highly conserved bHLH-like DNA-binding domain (Yin et al., 2005) that is responsible not only for heterodimer formation with bHLH TFs but also for direct binding to E box elements (CANNTG). The differences between the novel tobacco subgroup of BES TFs and other BESs are found within both the basic region and the two helices (Fig. 7B). This raises intriguing questions regarding the interacting partners of these tobacco BES genes, their DNA binding preferences, and their roles in planta. Database searches of the National Center for Biotechnology Information databases (all GenBank + EMBL + DDBJ + PDB sequences, Plant Genomes EST sequences, Genome Survey Sequence, Unfinished High Throughput Genomic Sequences) and the Sol Genomics Network (http://www.sgn.cornell.edu/) failed to find any members of this BES group in other plants, and it is possible that these BES-like genes are specific to tobacco and related Nicotiana species.
Figure 7.
Phylogenetic analysis of tobacco and Arabidopsis BES genes. A, Unrooted phylogenetic tree of tobacco and Arabidopsis BES genes constructed using the neighbor-joining method. Each tobacco gene is designated by an arbitrary number, and five clades are circled. Arabidopsis BES genes are indicated by accession numbers, except for BES1 (At1g19350) and BZR1 (At1g75080), which are indicated by their published names. A small number of tobacco BES sequences that do not contain complete domains (e.g. NtBES1, NtBES8, and NtBES19) are omitted from the analysis. B, Multiple sequence alignment of the bHLH-like domains from the tobacco and Arabidopsis BES genes. The alignment was produced with ClustalW and displayed using Jalview. The bHLH domain is indicated, and the black arrows show amino acids that are conserved within the tobacco-specific clade. The red arrow indicates the members of the tobacco-specific clade.
The SAP Family
A recessive mutation in Arabidopsis SAP causes severe aberrations in inflorescence and flower and ovule development (Byzova et al., 1999). Together with the organ identity gene AGAMOUS, SAP is required for the maintenance of floral identity, acting in a manner similar to AP1. Both Arabidopsis and poplar have just a single SAP gene, and it is the only family among the 64 TF families that appears to be absent in rice (Gao et al., 2006). Tobacco has at least four SAP genes (Fig. 8). Although similar to the Arabidopsis and poplar genes, the tobacco SAP genes are more similar to each other than they are to the genes from the other two species. Whether expansion of the tobacco SAP gene family led to redundant, partially redundant, or distinct functions for the SAP genes remains to be experimentally determined. In the near future, genome sequences for tomato and potato will be available, and this information should help clarify whether expansion in the SAP gene family is a general feature of the Solanaceae.
Figure 8.
Phylogenetic analysis of SAP genes from tobacco, Arabidopsis, and poplar. Shown is a phylogenetic tree of SAP genes from tobacco, Arabidopsis (At5g35770), and poplar (fgenesh4_pg.C_LG_XIV001059) constructed using the neighbor-joining method. Numbers indicate bootstrap values from 1,000 replications.
The C2H2 Family
The C2H2 zinc finger family appears to be overrepresented in the tobacco genome compared with other plants (Supplemental Table S2). This is due almost entirely to the presence of 23 unusual gene-coding sequences in the tobacco GSR data set that contain tandem zinc fingers (up to nine repeats), mostly of the C-x2-C-x12-H-x3-H type (data not shown). None of these GSRs have corresponding homologs in currently available EST sequence data. Based on BLAST homology, the domains in these GSRs may be related to the pfam domain DUF1644 found in a number of hypothetical plant proteins of unknown function. A lack of clear homology makes it is difficult to determine an exact number for these genes, as they are not particularly similar to known plant proteins. The tandem zinc fingers are most similar to transcriptional repressors like Kruppel (Hanna-Rose et al., 1997), but their roles in plants remain unknown.
Group IX ERF Genes Regulate Jasmonate Responses in Tobacco
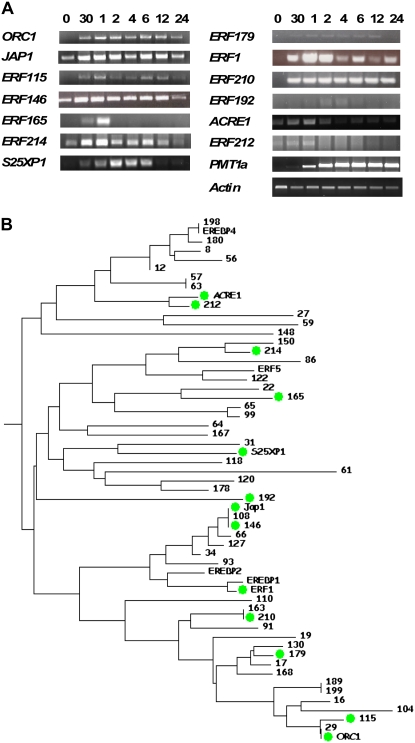
Group IX ERF genes have been implicated in regulating defense responses mediated by methyl jasmonate (MeJA) in a number of plant species, including Arabidopsis (McGrath et al., 2005), Catharanthus roseus (van der Fits and Memelink, 2000), and tobacco (Goossens et al., 2003; De Sutter et al., 2005). In C. roseus, the group IX ERF ORCA3 has been shown to be a master regulator of primary and secondary metabolism during jasmonate responses (van der Fits and Memelink, 2000, 2001). In tobacco, the MeJA-responsive group IX ERFs NtORC1 and NtJAP1 are able to positively regulate the jasmonate-inducible gene PMT, which plays a key role in secondary metabolism (De Sutter et al., 2005). These results suggest that group IX ERFs are among the key regulators of jasmonate responses in plants. Therefore, we used our phylogenetic analysis of the tobacco ERF gene family to determine whether we could use phylogenetic position to reveal potential function. We predicted that phylogenetically similar genes may have similar functions and specifically that many group IX ERF genes play roles in jasmonate responses in tobacco. This translational biology approach led us to predict that many tobacco group IX genes would be inducible by MeJA. To test this, we treated tobacco ‘Bright Yellow-2’ (BY-2) cells with MeJA and used semiquantitative reverse transcription (RT)-PCR together with gene-specific primers (Supplemental Table S3) to determine whether 19 representative tobacco group IX ERF genes are indeed inducible by MeJA. Figure 9A shows that the BY-2 cells responded well to MeJA, as judged by induction of the PMT1a gene that is up-regulated as part of a reprogramming of secondary metabolism in tobacco. Of the 19 genes studied, 13 were inducible by MeJA and the remaining six yielded no product. The inducible genes are found across the whole of the group IX phylogenetic tree (Fig. 9B).
Figure 9.
Analysis of MeJA induction of tobacco group IX ERF genes. A, Results of RT-PCR analysis of the expression levels of 13 ERF genes in tobacco BY-2 cells following treatment with 50 μm MeJA. Numbers represent the time of treatment in hours (0, 1–24) or minutes (30). The NtPMT1a gene, a well-documented MeJA-induced transcript in tobacco, was included as a positive control, and actin was used as a non-MeJA-induced negative control. PCR products were separated by electrophoresis on 2% agarose gels. Experiments were replicated at least three times, and representative data are shown. B, Phylogenetic tree of group IX tobacco ERF genes. Genes that are inducible by MeJA in BY-2 cells are indicated by green circles. [See online article for color version of this figure.]
The induction of the ERF genes followed several different kinetic patterns, ranging from rapid and sustained induction (NtERF210, NtERF179, and ORC1) to very rapid and transient induction (NtERF165 and ACRE1), suggesting that several different mechanisms of gene activation are involved. The maximum level of mRNA was after 1 to 2 h of treatment for most of the ERF genes, with the maximum NtPMT1a mRNA level being reached later, approximately 4 h after treatment. One of the most notable patterns of mRNA accumulation was found with NtERF210. NtERF210 mRNA was undetectable in the absence of MeJA treatment but then showed an extremely rapid induction, with maximum levels being reached within 30 min of hormone treatment. This maximum level was then sustained throughout the subsequent 24-h period. In addition to the striking pattern of mRNA accumulation, our phylogenetic analysis of the tobacco group IX ERF genes revealed that NtERF210 is found in a small clade (containing NtERF163, NtERF210, and NtERF91) that contains the genes that are the most similar to ORCA3, the key regulator of jasmonate responses in C. roseus. This is a validation of the use of phylogenies as an aid for functional prediction, as it not only suggests that many tobacco group IX ERF genes are indeed involved in jasmonate responses but also identifies NtERF210 as a potential key regulator of secondary metabolism during jasmonate responses in tobacco.
CONCLUSION AND PERSPECTIVES
Reduced representation sequence data sets, such as the MF data set used here, are useful resources for plant species in which whole genome sequence projects are lacking. They deliver sequences for the majority of genes and are invaluable tools for functional genomics and systems biology. However, they do lack information on some genes. Our discovery of members from 64 TF gene families in tobacco, including those that consist of only one or two genes in other plants, suggests that a large percentage of genes have been tagged in the tobacco gene space data set. Due to the incomplete nature of gene space sequences, some TF contigs in our data set contain partial DNA binding domains, normally due to the presence of introns. For these reasons, it is only possible to give a minimum number of TF genes that are present in tobacco based on the largest number of independent sequences that contained a certain portion of the conserved domain. The actual number of tobacco TFs in our data set is between 2,513 (the minimum number of genes) and 2,882 (the total number of contigs). Given the allopolyploid nature of tobacco, it is probable that the actual number of TF genes in tobacco is over 3,000.
Seven of the nine largest TF gene families in tobacco are very similar in size and complexity to their counterparts in Arabidopsis, rice, and poplar (Guo et al., 2005; Gao et al., 2006; Zhu et al., 2007). This allowed us to identify possible functional homologs in tobacco of genes characterized in other plant species. This translational biology approach has identified ERF, WRKY, homeodomain, MADS box, bHLH, bZIP, and R2R3MYB genes from tobacco that are prime candidates as regulators of many plant processes, including disease resistance, abiotic stress responses, light signaling, flowering time, secondary metabolism, senescence, organ identity, germination, and the circadian clock.
Our data constitute the largest currently available data set of TF sequences from any member of the Solanaceae and contain, to our knowledge, the first studies of over 40 tobacco TF families. To facilitate subsequent comparison and analysis, we constructed an expandable knowledge base called TOBFAC: The Database of Tobacco Transcription Factors (http://compsysbio.achs.virginia.edu/tobfac/). TOBFAC provides access to the data presented here and integrates it with available EST data, published reports, sequences of tobacco TFs, and a large quantity of other data concerning TFs in tobacco. TOBFAC provides a major resource for the study of gene expression in tobacco and the Solanaceae and helps to fill a void in studies of TF families across the plant kingdom (Rushton et al., 2008).
MATERIALS AND METHODS
Identification of Tobacco TFs
A data set of 1,159,022 GSRs was downloaded from the TGI (http://www.tobaccogenome.org/) to the Cowpea Genespace/Genomics Knowledge Base (Chen et al., 2007). To identify GSRs that encode TFs, searches were performed using the amino acid sequences of the conserved domains from members of each of 64 TF families previously identified in vascular plants that are listed in the DATF (Guo et al., 2005). Normally, the only similarity between all members of a TF multigene family is the DNA-binding domain, and with most gene families the searches were performed with this domain. The exact amino acid sequences used are listed in Supplemental Table S4. For most families, 5 to 10 independent searches were performed. These searches included the most variant versions of the conserved domain, so that all major subfamilies and divergent members of the tobacco (Nicotiana tabacum) gene families were found. In most cases, the domains were chosen based on multiple sequence alignments found at the DATF (http://datf.cbi.pku.edu.cn/). As the sequence length and the conservation rate among the DNA-binding domains of different families vary greatly, searches were designed to enable isolation of all possible gene family members. This approach was combined with a high cutoff e value of 10 in order to rigorously ensure that all possible gene family members were identified. This high e-value cutoff was an important part of the identification strategy and was designed to ensure that all GSRs that encode any part of a conserved domain were found. More rigorous cutoff values resulted in a smaller data set that lacked many GSRs that contained short exons, only small amounts of coding region, or coded for divergent members of the TF families.
For each family of TFs, all GSRs identified in the searches were combined into one data set and assembled into contigs using the Phrap program on the Cowpea Genespace/Genomics Knowledge Base (Chen et al., 2007), with the default settings for pairwise alignments and assembly except for the minimum overlap match, which was set at 30 bp. These parameters give positive alignment scores when regions of two sequences that are 70% or more identical are extended. In almost all cases in which we were able to assemble contigs, the alignments were 90% identical or greater. The average contig length was approximately 1,000 bp, with each contig containing on average between three and four independent GSRs per contig. The shortest contig was approximately 250 bp and the longest was approximately 2.8 kb. Each predicted gene was then individually manually curated by BLASTx searches against the nonredundant protein database housed at http://www.ncbi.nlm.nih.gov/. This served two purposes. First, all sequences were verified as coding for at least part of a conserved domain (i.e. all false positives that did not encode any part of the conserved domain from the gene family were discarded at this point). Second, the searches identified whether the sequence contained all of the conserved domain or only part of it. If the hit was only partial, the searches also identified which part of the conserved domain was present. This information was used to determine the minimum number of members in each TF gene family by calculating the number of independent sequences that contained any certain portion of the domain (e.g. amino acids 20–30). For larger gene families, the genes were first assigned to known subfamilies and then the minimum number of genes was calculated for each subfamily.
The data sources for the Arabidopsis (Arabidopsis thaliana), poplar (Populus spp.), and rice (Oryza sativa) data sets were the following. The Arabidopsis TFs were from version 2 of DATF that was updated in July 2006. It is based on the Arabidopsis Sequence of The Arabidopsis Information Resource version 6 (20051108). The rice and poplar TFs were taken from the Plant Transcription Factor Databases (http://planttfdb.cbi.pku.edu.cn/) using the version that was updated on November 20, 2007.
Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 (Tamura et al., 2007). For each analysis, introns were excised and the amino acid sequences of the DNA-binding domains were used to construct multiple sequence alignments using Clustal. Multiple sequence alignments were manually adjusted to optimize the alignments. Short partial domains were discarded, as they cannot be used to construct acceptable phylogenies. Phylogenetic trees were produced by the neighbor-joining method (settings: gaps/missing, pairwise deletion; model, amino number of differences; substitutions to include, all; pattern among lineages, same; rates among sites, uniform). Statistical support for the nodes in the phylogenetic trees (bootstrap values from a minimum of 1,000 trials) were obtained for each tree and are available on the TOBFAC Web site. We chose to present the overview phylogenies of the larger gene families as radial trees using PhyloDraw (Choi et al., 2000), as these provide a simple and clear overview of each family. The tree files are available for download at TOBFAC and can be displayed in other formats to show exact phylogenetic similarities and to identify each gene. The NAC domain phylogenetic tree was produced using the complete NAC gene families from Arabidopsis, poplar, and rice as found at the Plant Transcription Factor Databases (http://planttfdb.cbi.pku.edu.cn/) and the tobacco NAC genes that are shown in Figures 4 and 5. In each case, the amino acid sequences of the A, B, and C domains of the NAC domains were used. Less than 10 fragmentary NAC domains were excluded, as their amino acid sequences are uncertain and they could not be aligned correctly.
Preparation of RNA from BY-2 Cells and RT-PCR Analysis of MeJA Responsiveness
Tobacco BY-2 cell suspension cultures were grown in Murashige and Skoog medium containing 3% (w/v) Suc and 0.2 mg/L 2,4-dichlorophenyoxyacetic acid, pH 5.8, and subcultured in fresh Murashige and Skoog medium every 7 d (Nagata et al., 1992; Goossens et al., 2003). For MeJA treatment, BY-2 cells were subcultured and grown for 1 d, after which the suspensions were treated with 50 μm MeJA. Cells were collected by filtration at the times indicated, and 1 to 2 g of frozen cells were used to prepare RNA by the method developed by the RIKEN TAB project (http://mrg.psc.riken.go.jp/strc/Protocols.htm).
Total RNA (5 μg) was used for cDNA synthesis using the ThermoScript RT-PCR system (Invitrogen) according to the manufacturer's instructions. Each PCR contained 0.2 μg of cDNA and the gene-specific primers listed in Supplemental Table S3. To ensure that each product represented the mRNA level from a single gene, each gene was compared at the DNA level with its closest neighbor in the phylogeny, and two primers were designed for each gene that were specific for this ERF member. The NtPMT1a gene, a well-documented MeJA-induced transcript in tobacco (Nagata et al., 1992; Goossens et al., 2003), was included as a positive control, and actin was used as a non-MeJA-induced control. PCR products were separated by electrophoresis on 2% agarose gels. Experiments were replicated at least three times, and representative data are shown.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Flow diagram for the identification of GSRs encoding TFs in tobacco.
Supplemental Figure S2. Tobacco homeodomain gene family.
Supplemental Figure S3. Tobacco R2R3MYB gene family.
Supplemental Figure S4. Tobacco bZIP gene family.
Supplemental Figure S5. Tobacco MADS box gene family.
Supplemental Table S1. Comprehensive list of predicted TF genes in TOBFAC and the GenBank accession numbers of their constituent GSRs.
Supplemental Table S2. Comparison of predicted TF gene family sizes in tobacco with those from sequenced plant genomes.
Supplemental Table S3. Gene-specific primers used in RT-PCR analysis of MeJA-induced gene expression.
Supplemental Table S4. Comprehensive list of amino acid sequences of the TF domains used in BLAST homology searches.
Supplementary Material
Acknowledgments
We thank Jennifer Murphy for her help with the RT-PCR experiments and the preparation of Figure 6. We also thank Alain Goossens and Sofie Tilleman for providing the tobacco BY-2 cell culture line, Charles Opperman, Steve Lommel, and Mark Burke for their assistance in accessing the TGI data and helpful discussions in our analysis of the GSRs, and Karam Singh for his critical reading and commentary on the manuscript.
This work was supported by funding from Philip Morris USA.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Michael P. Timko (mpt9g@virginia.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Werber M, Weisshaar B (2003) Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15 2497–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell JA, Budiman MA, Nunberg A, Citek RW, Robbins D, Jones J, Flick E, Rohlfing T, Fries J, Bradford K, et al (2005) Sorghum genome sequencing by methylation filtration. PLoS Biol 3 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Schrick K, Springer PS, Brown WE, Sanmiguel P (1994) Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37 565–576 [DOI] [PubMed] [Google Scholar]
- Byzova MV, Franken J, Aarts MGM, de Almeida-Engler J, Engler G, Mariani C, Van Lookeren Campagne MM, Angenent GC (1999) Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev 13 1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RL, Gago GM, Palena CM, Gonzalez DH (1998) Homeoboxes in plant development. Biochim Biophys Acta 1442 1–19 [DOI] [PubMed] [Google Scholar]
- Chen X, Laudeman TW, Rushton PJ, Spraggins TA, Timko MP (2007) CGKB: an annotation knowledge base for cowpea (Vigna unguiculata L.) methylation filtered genomic genespace sequences. BMC Bioinformatics 8 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 666–671 [DOI] [PubMed] [Google Scholar]
- Choi JH, Jung HY, Kim HS, Cho HG (2000) PhyloDraw: a phylogenetic tree drawing system. Bioinformatics 16 1056–1058 [DOI] [PubMed] [Google Scholar]
- De Sutter V, Vanderhaeghen R, Tilleman S, Lammertyn F, Vanhoutte I, Karimi M, Inze D, Goossens A, Hilson P (2005) Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. Plant J 44 1065–1076 [DOI] [PubMed] [Google Scholar]
- Ernst HA, Olsen AN, Skriver K, Larsen S, Lo Leggio L (2004) Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 199–206 [DOI] [PubMed] [Google Scholar]
- Gadani F, Hayes A, Opperman CH, Lommel SA, Sosinski BR, Burke M, Hi L, Brierly R, Salstead A, Heer J (2003) Large scale genome sequencing and analysis of Nicotiana tabacum: the tobacco genome initiative. In 5émes Journées Scientifiques du Tabac de Bergerac. Institut du Tabac-Bergerac, Dordogne, France, pp 117–130
- Gao G, Zhong Y, Guo A, Zhu Q, Tang W, Zheng W, Gu X, Wei L, Luo J (2006) DRTF: a database of rice transcription factors. Bioinformatics 22 1286–1287 [DOI] [PubMed] [Google Scholar]
- Geelen DNV, Inze DG (2001) A bright future for the Bright Yellow-2 cell culture. Plant Physiol 127 1375–1379 [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ (1987) Homeotic genes, the homeobox, and the spatial-organization of the embryo. Harvey Lect 81 153–172 [PubMed] [Google Scholar]
- Goodspeed TH (1954) The Genus Nicotiana. Chronica Botanica, Waltham, MA
- Goossens A, Hakkinen ST, Laakso I, Seppanen-Laakso T, Biondi S, De Sutter V, Lammertyn F, Nuutila AM, Soderlund H, Zabeau M, et al (2003) A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc Natl Acad Sci USA 100 8595–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, He K, Liu D, Bai SN, Gu XC, Wei LP, Luo JC (2005) DATF: a database of Arabidopsis transcription factors. Bioinformatics 21 2568–2569 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131 657–668 [DOI] [PubMed] [Google Scholar]
- Hake S, Smith HMS, Holtan H, Magnani E, Mele G, Ramirez J (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20 125–151 [DOI] [PubMed] [Google Scholar]
- Hanna-Rose W, Licht JD, Hansen U (1997) Two evolutionarily conserved repression domains in the Drosophila Kruppel protein differ in activator specificity. Mol Cell Biol 17 4820–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20 735–747 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7 106–111 [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16 263–276 [DOI] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Tanksley SD, Giovannoni JJ, van Eck J, Stack S, Choi D, Kim BD, Chen MS, Cheng ZK, Li CY, et al (2005) The Tomato Sequencing Project, the first cornerstone of the International Solanaceae Project (SOL). Comp Funct Genomics 6 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins E, Milbourne D, Petti C, Doyle-Prestwich BM, Meade C (2006) Potato in the age of biotechnology. Trends Plant Sci 11 254–260 [DOI] [PubMed] [Google Scholar]
- Murad L, Lim KY, Christopodulou V, Matyasek R, Lichtenstein CP, Kovarik A, Leitch AR (2002) The origin of tobacco's T genome is traced to a particular lineage within Nicotiana tomentosiformis (Solanaceae). Am J Bot 89 921–928 [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cell line as the HeLa cell in the cell biology of higher plants. Int Rev Cytol 132 1–30 [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K, Hojo H, Aimoto S, Nakai T, Nakamura H, Sarai A, Ishii S, Nishimura Y (1992) Solution structure of a DNA-binding unit of Myb: a helix turn helix-related motif with conserved tryptophans forming a hydrophobic core. Proc Natl Acad Sci USA 89 6428–6432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA-binding proteins that interact with an ethylene-responsive element. Plant Cell 7 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksman-Caldentey KM (2007) Tropane and nicotine alkaloid biosynthesis: novel approaches towards biotechnological production of plant-derived pharmaceuticals. Curr Pharm Biotechnol 8 203–210 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Lo Leggio L, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10 79–87 [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10 239–247 [DOI] [PubMed] [Google Scholar]
- Palmer LE, Rabinowicz PD, O'Shaughnessy AL, Balija VS, Nascimento LU, Dike S, de la Bastide M, Martienssen RA, McCombie WR (2003) Maize genome sequencing by methylation filtrations. Science 302 2115–2117 [DOI] [PubMed] [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LJ, Zhu YX (2006) Transcription factor families in Arabidopsis: major progress and outstanding issues for future research. Commentary. Curr Opin Plant Biol 9 544–549 [DOI] [PubMed] [Google Scholar]
- Rabinowicz PD, Schutz K, Dedhia N, Yordan C, Parnell LD, Stein L, McCombie WR, Martienssen RA (1999) Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat Genet 23 305–308 [DOI] [PubMed] [Google Scholar]
- Richardt S, Lang D, Reski R, Frank W, Rensing SA (2007) PlanTAPDB, a phylogeny-based resource of plant transcription-associated proteins. Plant Physiol 143 1452–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rijpkema AS, Gerats T, Vandenbussche M (2007) Evolutionary complexity of MADS complexes. Curr Opin Plant Biol 10 32–38 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Bokowiec MT, Laudeman TW, Brannock JF, Chen X, Timko MP (2008) TOBFAC: the database of tobacco transcription factors. BMC Bioinformatics 9 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol Biol 29 691–702 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4 447–456 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448 661–665 [DOI] [PubMed] [Google Scholar]
- Timko MP, Rushton PJ, Laudeman TW, Bokowiec MT, Chipumuro E, Cheung F, Town CD, Chen X (2008) Sequencing and analysis of the gene-rich space of cowpea. BMC Genomics 9 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Kakar K, Wandrey M, Montanari O, Murray J, Andriankaja A, Zhang JY, Benedito V, Hofer JMI, Chueng F, et al (2007) Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol 144 538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7 491–498 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289 295–297 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J (2001) The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J 25 43–53 [DOI] [PubMed] [Google Scholar]
- Whitelaw CA, Barbazuk WB, Pertea G, Chan AP, Cheung F, Lee Y, Zheng L, van Heeringen S, Karamycheva S, Bennetzen JL, et al (2003) Enrichment of gene-coding sequences in maize by genome filtration. Science 302 2118–2120 [DOI] [PubMed] [Google Scholar]
- Xiong YQ, Liu TY, Tian CG, Sun SH, Li JY, Chen MS (2005) Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol Biol 59 191–203 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109 181–191 [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Wang LJ (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Guo AY, Gao G, Zhong YF, Xu M, Huang M, Luo J (2007) DPTF: a database of poplar transcription factors. Bioinformatics 23 1307–1308 [DOI] [PubMed] [Google Scholar]
- Zonneveld BJM, Leitch IJ, Bennett MD (2005) First nuclear DNA amounts in more than 300 angiosperms. Ann Bot (Lond) 96 229–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.