Abstract
Catalytic antibodies have shown great promise for catalyzing a tremendously diverse set of natural and unnatural chemical transformations. However, few catalytic antibodies have efficiencies that approach those of natural enzymes. In principle, random mutagenesis procedures such as phage display could be used to improve the catalytic activities of existing antibodies; however, these studies have been hampered by difficulties in the recombinant expression of antibodies. Here, we have grafted the antigen binding loops from a murine-derived catalytic antibody, 17E8, onto a human antibody framework in an effort to overcome difficulties associated with recombinant expression and phage display of this antibody. “Humanized” 17E8 retained similar catalytic and hapten binding properties as the murine antibody while levels of functional Fab displayed on phage were 200-fold higher than for a murine variable region/human constant region chimeric Fab. This construct was used to prepare combinatorial libraries. Affinity panning of these resulted in the selection of variants with 2- to 8-fold improvements in binding affinity for a phosphonate transition-state analog. Surprisingly, none of the affinity-matured variants was more catalytically active than the parent antibody and some were significantly less active. By contrast, a weaker binding variant was identified with 2-fold greater catalytic activity and incorporation of a single substitution (Tyr-100aH → Asn) from this variant into the parent antibody led to a 5-fold increase in catalytic efficiency. Thus, phage display methods can be readily used to optimize binding of catalytic antibodies to transition-state analogs, and when used in conjunction with limited screening for catalysis can identify variants with higher catalytic efficiencies.
Since the generation of catalytic antibodies began a decade ago, an impressive array of antibodies have been produced that catalyze a wide range of distinct chemical transformations (1, 2). The standard approach to prepare a catalytic antibody involves immunizing animals with a stable transition-state analog for the reaction of interest. This approach relies on the assumption that a correlation exists between catalytic activity and transition-state analog affinity. In principal, these tailored catalysts could be enormously useful for both medical and industrial applications. Although some catalytic antibodies have been reported with efficiencies similar to that of corresponding natural enzymes (3), in general it has been difficult to obtain highly active catalysts using the standard approach. For this reason, an important challenge has been to find ways of boosting the catalytic activities of these antibodies. One approach to this problem is to attempt to improve existing antibody catalysts by using protein engineering techniques (4, 5). However, structure–activity relationships have been studied for only a limited number of catalytic antibodies, making it difficult to predict mutations that might enhance activity. A more promising approach may be through the use of random mutagenesis, provided suitable selection strategies can be devised which allow for the evolution of catalytic activity (6).
The catalytic efficiency, kcat/Km, for a catalytic antibody that binds to a hapten mimicking the reaction transition-state should approximate the ratio of the uncatalyzed rate constant over the inhibition constant of transition-state analog binding (kuncat/Ki) (7, 8). Indeed, a number of studies have demonstrated that within a series of related antibodies (9–11) or enzyme mutants (12), catalytic efficiency usually increases as a function of increased affinity for a transition-state analog. Given the anticipated relationship between activity and hapten binding, we reasoned that in vitro affinity maturation of a catalytic antibody might provide a general means of increasing the catalytic efficiency. Whereas the affinity maturation of antibodies occurs in vivo via the process of somatic hypermutation (13), in vitro optimization of antigen affinities has been achieved in selected cases using phage display methods (14–17). Increases in antigen binding affinity of up to 1,000-fold have been demonstrated, based on favorable mutations in the antigen binding loops of antibody molecules (17).
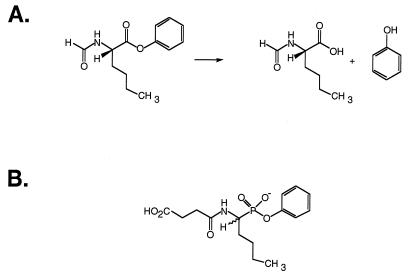
The catalytic antibody 17E8 catalyzes the hydrolysis of different amino acid phenyl esters. 17E8 was obtained by immunization with a norleucine phosphonate hapten that mimics the hydrolytic transition-state (18) (Fig. 1); thus we reasoned that improving the affinity of 17E8 for this same transition-state analog might improve the catalytic efficiency. This catalytic antibody seemed an ideal candidate for affinity optimization, as the three-dimensional structure of the hapten-bound complex is known (19), thus facilitating the design of phage display libraries. Furthermore, the hapten binding affinity (Ki = 500 nM) is not overly tight, suggesting that this could possibly be improved. Here we describe the use of mutagenesis and phage display to optimize the affinity of humanized 17E8 for a transition-state analog, and report both the hapten binding affinities and catalytic activities for several of the selected variants.
Figure 1.
(A) Ester hydrolysis reaction catalyzed by antibody 17E8 and (B) structure of the phosphonate transition-state analog used to elicit this antibody (18).
MATERIALS AND METHODS
Construction of Chimeric and Humanized 17E8 Phagemid Vector.
Antibody 17E8 binds the hapten phenyl[1-(1-N-succinylamino)pentyl]phosphonate and has been cloned (18). The VL and VH encoding sequences of 17E8 were subcloned into p4xH (20) to give plasmid pRS-1, which encodes a chimeric murine V-region/human C-region 17E8 Fab fragment. An EcoRI–ApaI fragment of this was subcloned into the phagemid vector pMB4-19 (21) to generate pMB8-1. This construct was similar to pRS-1, except that the 3′ end of the heavy chain gene is followed by an amber codon and then codons 249–406 of M13 gene III.
A humanized form of 17E8 (hu17E8) was generated by site-directed mutagenesis using a deoxyuridine-containing template of pMB4-19 (21). Six mutagenic oligonucleotides were used to substitute each of the 17E8 complementarity-determining regions (CDRs) onto a human VLκI–VHIII framework (22). The transplanted CDR sequences were chosen according to the sequence definition of Kabat (23), except for CDR-H1, which was extended to encompass both the sequence and structural (24) definition, viz VH residues 26–35. The final construct, pMB8-15, encoded a human VLκI–Cκ1 light chain and human VHIII–CH1γ1 heavy chain Fd-gene III fusion.
Expression and Purification of hu17E8.
Expression of soluble hu17E8 Fab and mutants thereof was performed in shake flasks as described (21). Periplasmic lysates were prepared from cell pellets by freezing for at least 2 h at −20°C, resuspending in 12.5 ml of 10 mM Tris (pH 7.6) containing 5 mM MgCl2 and 75 mM CaCl2, and shaking gently for 90 min at 4°C. Spheroplasts were removed by centrifugation (10,000 × g for 15 min), and Fab was purified by protein G affinity chromatography (Pharmacia Biotech). Purified Fab samples were characterized by electrospray mass spectrometry, and concentrations were determined according to absorbance at 280 nm (ɛ = 67,340 M−1⋅cm−1 for hu17E8) (25).
Construction of 17E8 Mutants and Randomized Library.
Mutants and the randomized hu17E8 library were constructed by site-directed mutagenesis according to the method of Kunkel et al. (26). On the basis of the 17E8-hapten crystallographic structure (19), three hu17E8 libraries were designed according to antibody residues that either directly contact or are in close proximity to the hapten: Gly-34L, Leu-46L, Gln-90L, Tyr-91L, Arg-96L (library 1); Lys-93H, Tyr-96H, Tyr-97H, Ser-100H, Val-100bH, Asp-101H (library 2); Tyr-36L, Leu-89L, Phe-98L, Val-37H, Trp-103H (library 3). For library construction, template vectors were first prepared by replacing the codons to be randomized with TAA stop triplets. Oligonucleotides were then used to randomly mutate target codons to NNS (N = G, A, T, C; S = G, C), except for codon 93H (library 2), which coded only for AAG (Lys) or AGG (Arg). Libraries were electroporated into Escherichia coli XL-1 Blue cells (Stratagene), and each library gave ≈108 individual transformants. The randomly mutated regions were sequenced for 10 clones from each starting library. Eighty percent of the library 1 clones, 80% from library 2, and 10% from library 3 contained random sequence (i.e. non-TAA) for all of the codons randomized, implying that >99% of all possible DNA sequences were represented in libraries 1 and 2, whereas ≈30% of all possible sequences were represented in library 3. Phagemid particles monovalently displaying the hu17E8 Fab fragments were propagated as described (21). Typically, final phagemid stocks contained ≈1014 pfu of phage/ml.
Selection of Hapten-Specific Phagemids.
For each library, microtiter plate wells were coated with a keyhole limpet hemocyanin conjugate of hapten (18) at a concentration of 10 μg/ml in PBS overnight at 4°C. After blocking with 6% skim milk in PBS and washing, ≈1012 pfu of phage in 0.6% skim milk were added to the wells. Plates were shaken gently for 2 h and washed, and then bound phage were eluted with 100 μl of 0.1 M glycine (pH 2.5) containing 0.05% Tween-20. Eluted phagemids were immediately neutralized with 25 μl of 1 M Tris⋅HCl (pH 8.0). An aliquot of this was used to titer the number of phage eluted, and the remainder used to repropagate the enriched library.
Binding and Activity Assays.
The hapten binding affinities of selected hu17E8 variants were measured directly as gene III fusion proteins on the surface of phagemid particles using a phage ELISA assay (27). Briefly, microtiter plates (Nunc; Maxisorb, 96 wells) were coated overnight at 4°C with 100 μl of 4 μg/ml keyhole limpet hemocyanin–hapten conjugate. Wells were blocked and washed, then serial dilutions of competing unconjugated hapten and a subsaturating concentration of hu17E8 phage were added in 100 μl of 1% skim milk in PBS. After 2 h the plates were washed, and the bound phagemid was labeled with anti-M13 polyclonal antibody-horseradish peroxidase conjugate (Pharmacia), and assayed. Binding affinities (EC50) were calculated as the concentration of competing hapten required to reduce maximal phagemid binding by 50%. Each binding assay was performed in duplicate, and standard errors in the EC50 determinations were ≤25%. The parent clone (hu17E8) was included in each binding experiment so that relative binding affinities [EC50 (hu17E8)/EC50 (mutant)] were always determined using data from a single experiment.
Steady state kinetic constants (kcat, Km) were determined for hydrolysis of the L-enantiomerically enriched phenyl ester of N-formylnorleucine by hu17E8 mutants. So that kinetic data could be directly compared with that of previous studies, all assays were performed at pH 8.7 as described (18). Background rates were subtracted to give the true antibody-catalyzed rate, and substrate concentrations were corrected to effective L-substrate concentrations, taking into account the 50% enantiomeric excess of the L-isomer (18). Km and kcat were determined by fitting the rate data to the Michaelis–Menten equation using the computer program enzymekinetics.
RESULTS AND DISCUSSION
Humanization of 17E8.
Previous attempts to express a soluble Fab fragment of antibody 17E8 in E. coli were not successful, although low levels of expression (≈50 μg purified Fab/L culture) were obtained for a chimeric murine V-region/human C-region Fab fragment. Initial efforts at monovalent display of the chimeric 17E8 Fab on phage produced only low levels of functional Fab, most likely due to poor expression of the Fab–gene III fusion. We were concerned that the low level of chimeric 17E8 Fab display would bias the panning of phage libraries for selection of mutants with enhanced expression rather than tighter hapten binding affinity. In an effort to increase functional Fab display on phage, we sought to engineer a fully humanized form of 17E8, given that several humanized Fabs have been shown to express at higher levels than their chimeric counterparts (28).
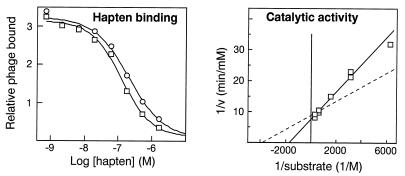
hu17E8 was generated by grafting the murine CDR sequences onto a human VLκ subgroup I and VH subgroup III variable framework (22). All framework residues were maintained as the human sequence. Comparison of humanized versus chimeric 17E8 Fab showed a 200-fold improvement in the level of functional Fab display. Similarly, expression of the soluble Fab was vastly improved and typical expression yields were 1.5 mg purified Fab/L culture. Most importantly, the hapten binding affinity and catalytic activity of hu17E8 were similar to that of the parent murine (or chimeric) forms of the antibody (Fig. 2). Thus, CDR grafting was sufficient to generate a humanized version of 17E8 with similar binding and catalytic properties to the parent murine antibody. This humanized form of 17E8 was used in all subsequent studies.
Figure 2.
Comparison of hapten binding and catalytic activity for hu17E8 Fab (squares) and chimeric or murine 17E8 (circles). Hapten binding affinities were determined by phage ELISA for humanized and chimeric 17E8 Fab: hu17E8, EC50 = 122 ± 13 nM; chimeric 17E8, EC50 = 197 ± 29 nM. Catalytic constants were determined for soluble hu17E8 Fab: kcat = 59 ± 1 min−1, Km = 580 ± 41 μM. The dotted line represents the calculated activity for the same concentration of murine 17E8 Fab, based on kinetic constants previously determined under identical assay conditions (18): kcat = 51 ± 0.3 min−1, Km = 259 ± 2 μM (kcat is half that reported in ref 18 to correct for the presence of two active sites in whole IgG).
hu17E8 Library Selectants and Analysis of Hapten Binding.
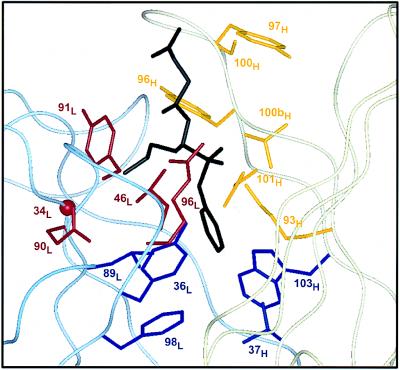
Three hu17E8 libraries were prepared by randomly mutating antibody residues in close proximity to the hapten binding site (Fig. 3). These libraries were subjected to multiple rounds of hapten binding selection (libraries 1 and 2, six rounds; library 3, five rounds) after which individual clones were selected and sequenced, and their hapten binding affinities determined by phage ELISA (Table 1).
Figure 3.
Design of hu17E8 libraries. Residues targeted for randomization are shown on the structure of 17E8 bound to the norleucine phosphonate hapten (19). The antibody light chain is represented by a pale blue ribbon, the heavy chain in pale yellow and bound hapten is black. Library 1 (red) targeted residues that help form the binding pocket of the substrate side chain; Gly-34L (CDR-L1), Leu-46L (FR-L2), Gln-90L, Tyr-91L, Arg-96L (CDR-L3). Library 2 (yellow) randomized heavy chain residues in CDR-H3 and also included a binary randomization of Lys-93H; Lys-93H (FR-H3), Tyr-96H, Tyr-97H, Ser-100H, Val-100bH, Asp-101H (CDR-H3). Library 3 (blue) randomized hydrophobic residues at the bottom of the substrate/hapten binding pocket; Tyr-36L (FR-L2), Leu-89L (CDR-L3), Phe-98L (FR-L4), Val-37H (FR-H2), Trp-103H (FR-H4). Omitted from these libraries were residues His-35H and Ser-95H; given their putative role in the catalytic mechanism of 17E8 (19), we chose not to randomize these positions. FR, framework region. All residues are numbered according to Kabat et al. (23).
Table 1.
Variants of hu17E8 and relative hapten binding affinities
| Clone | Residues randomized | EC50 hu17E8
|
|||||
|---|---|---|---|---|---|---|---|
| EC50 mutant | |||||||
| Library 1 | |||||||
| G34L | L46L | Q90L | Y91L | R96L | |||
| 1.1 | (7) | G | L | Q | Y | Y | 0.11 |
| 1.2 | G | L | G | Y | W | 0.27 | |
| 1.3 | G | L | Q | H | W | 0.26 | |
| 1.4 | G | L | Q | H | R | 0.05 | |
| 1.5 | G | A | Q | Y | R | ND | |
| 1.10* | G | L | Q | Y | W | 4.8 | |
| Library 2 | |||||||
| K93H | Y96H | Y97H | S100H | V100bH | D101H | ||
| 2.1 | K | Y | K | R | V | D | 4.0 |
| 2.2 | K | E | Y | H | G | K | 3.0 |
| 2.3 | K | F | I | H | R | E | 2.5 |
| 2.4 | K | E | Y | K | G | S | 2.0 |
| 2.5 | K | E | Y | G | G | N | 1.9 |
| 2.6 | K | E | Y | R | G | T | 1.2 |
| 2.7 | K | W | A | K | R | D | 0.75 |
| 2.8 | K | F | R | R | P | E | 0.20 |
| 2.9 | K | R | R | D | Q | P | ND |
| 2.10 | K | F | R | S | G | E | ND |
| 2.11 | K | S | K | I | R | D | ND |
| 2.12 | K | W | A | K | K | D | ND |
| 2.20* (2.1 R96LW) | 6.5 | ||||||
| 2.21* (2.3 R96LW) | 7.5 | ||||||
| Library 3 | |||||||
| Y36L | L89L | F98L | V37H | W103H | |||
| 3.1 | (6) | Y | I | W | I | W | 0.52 |
| 3.2 | Y | L | W | V | W | 0.16 | |
| 3.3 | Y | C | F | V | W | 0.11 | |
| 3.4 | R | D | M | R | Q | ND | |
Selection of non-wild-type residues is denoted by bold type. Numbers in parentheses indicate the number of times each mutant appeared among selected clones if greater than once. ND, not determined.
Clones 1.10 (Arg-96LTrp), 2.20 (Arg-96LTrp/Tyr-97HLys/Ser-100HArg), and 2.21 (Arg-96LTrp/Tyr-96HPhe/Tyr-97HIle/Ser-100HHis/Val-100bHArg/Asp-101HGlu) were generated by site-directed mutagenesis.
Analysis of the selectants from library 1 showed a strong preference for retention of the parent hu17E8 sequence at four of the five positions randomized. The exception was residue 96L, where arginine was replaced in most selectants by tyrosine (clone 1.1). To a lesser degree, tryptophan and arginine were also selected at 96L, although only in the context of additional substitutions. All of the library 1 selectants bound hapten more weakly than hu17E8, including the consensus Arg-96LTyr selectant (Table 1). However, we noted that while the Tyr-91LHis selectant (clone 1.4) bound hapten 20-fold less tightly, the double mutant Tyr-91LHis/Arg-96LTrp (clone 1.3) bound only 4-fold more weakly. This result suggested that the replacement of arginine with tryptophan at position 96L improves hapten affinity, despite the single mutant not appearing in the final pool of library 1 selectants. Accordingly, we prepared the Arg-96LTrp mutant of hu17E8 by site-directed mutagenesis (clone 1.10). Phage ELISA analysis indicated that this single substitution resulted in a 5-fold improvement in hapten binding relative to hu17E8.
By contrast with library 1, selectants from library 2 showed less conservation of the parent sequence. Most of the residues randomized were only partially consensed, with the exception of position 93H, which was retained exclusively as lysine. Residue 101H had a preference for acidic amino acids, presumably to maintain a buried salt bridge with Lys-93H (Fig. 4) (19). Other residues randomized in library 2 are largely solvent exposed; this may explain the lack of any strong consensus at those positions. Most of the selectants analyzed for hapten binding showed minor improvements relative to the parent antibody (Table 1), although no single substitution stood out as responsible for these gains. It is surprising that variant 2.2 bound hapten more tightly than hu17E8, given the electrostatic repulsion which would occur between Lys-101H and Lys-93H if these side chains were to pack as in the 17E8 structure (Fig. 4). Perhaps the CDR-H3 loop of variant 2.2 repacks in such a way that any deleterious electrostatic interaction is avoided or alternatively the pKa of either Lys-101H or Lys-93H may be perturbed such that the side chain is unprotonated.
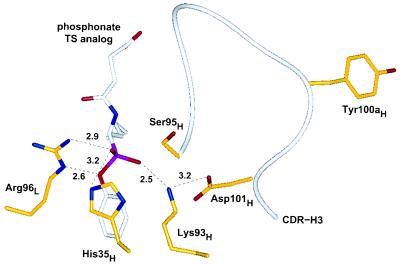
Figure 4.
Active site region of catalytic antibody 17E8 based upon the crystallographic structure of 17E8 bound to hapten (19). Key residues in proximity to the phosphonate moiety are depicted in addition to Tyr-100aH. Potential hydrogen bonds or ionic interactions are shown by dotted lines, along with distances in angstroms. All atoms of residue Tyr-100aH are at least 11 Å away from any hapten atom. Heteroatoms are shown in blue (nitrogen), red (oxygen), or pink (phosphorus), and the heavy chain CDR-H3 (residues 95H–102H) is shown by the white ribbon.
The sequences of mutants selected from library 3 were strongly consensed, with most clones containing the identical DNA sequence coding for three substitutions out of five positions randomized, viz Leu-89LIle/Phe-98LTrp/Val-37HIle (Table 1). In addition, this concensus clone also contained a spontaneous fourth substitution at position 100aH (Tyr → Asn) which was not programmed as part of the original randomization strategy. Although two of these substitutions (Phe-98LTrp/Val-37HIle) potentially reduce the size of the ligand binding cavity, this variant bound hapten only 2-fold weaker than hu17E8. As in library 1, it was surprising that the parent sequence was not selected in this library given the weaker hapten affinity of the consensus clone. This result was most likely due to the incomplete diversity of this library caused by inefficient mutagenesis when using multiple (four) oligonucleotides.
In addition to the variants analyzed from each of the three libraries, two additional variants were prepared by combining the Arg-96LTrp mutation with mutations from clones 2.1 and 2.3. These combination mutants (clones 2.20 and 2.21) bound hapten 6.5- and 7.5-fold tighter than hu17E8 according to comparison of the EC50 values (Table 1). Although the affinities of these combination mutants is higher than any of the individual clones, the binding improvements were less than that expected based on additivity of mutational effects (29).
Catalytic Activity of hu17E8 Selectants.
Soluble Fab was expressed for a number of the phage-selected variants and the catalytic activities of these were determined (Table 2). Surprisingly, all of the selectants with improved hapten binding were less active (lower kcat/Km) than hu17E8, and for some of these catalytic activity was reduced by over 20-fold. Thus increasing the affinity of the antibody for the transition-state analog did not lead to any improvement in catalysis in this system.
Table 2.
Catalytic and hapten binding properties of hu17E8 and variants
| Clone | kcat, min−1 | Km, μM | kcat/Km, M−1⋅min−1 |
kcat/Km (mutant)
|
EC50 (hu17E8)
|
|---|---|---|---|---|---|
| kcat/Km (hu17E8) | EC50 (mutant) | ||||
| hu17E8 | 59 | 580 | 1.0 × 105 | 1.0 | 1.0 |
| 1.1 | —* | —* | <5.0 × 103 | <0.05 | 0.11 |
| 1.4 | 36 | 2,700 | 1.3 × 104 | 0.13 | 0.050 |
| 1.10 | —* | —* | <5.0 × 103 | <0.05 | 4.8 |
| 2.1 | 34 | 930 | 3.7 × 104 | 0.37 | 4.0 |
| 2.2 | —* | —* | <5.0 × 103 | <0.05 | 3.0 |
| 2.3 | 96 | 2,400 | 4.0 × 104 | 0.40 | 2.5 |
| 2.5 | 7.1 | 800 | 9.0 × 103 | 0.09 | 1.9 |
| 3.1 | 110 | 530 | 2.0 × 105 | 2.0 | 0.52 |
| 3.2 | 46 | 1,600 | 3.0 × 104 | 0.3 | 0.16 |
| 2.21 | —* | —* | <5.0 × 103 | <0.05 | 7.5 |
Where catalytic activities were too weak to determine kcat and Km, kcat/Km was conservatively estimated as less than 5,000 M−1⋅min−1.
Decreases in catalytic activity (but not necessarily hapten binding) appeared to correlate with mutations to Arg-96L and Asp-101H, thus implicating that these two residues are important for catalysis. The catalytic activity of the Arg-96LTrp mutant (clone 1.10) was reduced by greater than 20-fold, even though this variant bound hapten 5-fold tighter than hu17E8. In the hapten-bound structure of 17E8 (19), the guanidinium group of Arg-96L forms a possible salt bridge with the pro-R phosphonyl oxygen and also hydrogen bonds to the aryl oxygen of the hapten (Fig. 4). These two interactions may play distinct roles in transition-state stabilization by helping to stabilize both the oxyanion intermediate formed during substrate hydrolysis and the incipient negative charge formed on the aryl leaving group. Interestingly, two other hydrolytic antibodies, 43C9 and 48G7, also contain arginine at position 96L, and in both cases mutagenesis studies have revealed that this residue is important for catalysis (30, 31).
Changes to Asp-101H in hu17E8 caused variable decreases in activity according to the nature of the substitution. Variants selected without an acidic residue at 101H were less active than those that had aspartate or glutamate at this position (Table 2). These results suggest that the Lys-93H–Asp-101H salt bridge is important for catalysis, although the ability to at least form a hydrogen bond, as in the case of the asparagine-containing variant 2.4, may compensate in part for the loss of an ionic interaction. As the side chain of Lys-93H forms potential salt bridges with both Asp-101H and the pro-S phosphonyl oxygen of the hapten (Fig. 4), a possible catalytic role for Asp-101H may be to raise the pKa of the buried ɛ-amino group of Lys-93H so that the protonated form of this residue can better stabilize the oxyanion intermediate formed during substrate hydrolysis.
Activity Improved Variant.
Although the consensus selectant from library 3 (clone 3.1) bound hapten less tightly than hu17E8, the catalytic activity of this variant was improved 2-fold due mostly to an increase in kcat. Clone 3.1 contained four substitutions relative to the parent sequence. Three of these changes were in buried residues which form the base of the ligand binding pocket. The fourth change (Tyr-100aH → Asn) was at a residue that resides in CDR-H3 and makes no direct contact with the hapten (Fig. 4). To examine the effect of this remote residue, we prepared a mutant of 3.1 by reinstalling tyrosine at position 100aH (clone 3.1y). The catalytic activity of this mutant was reduced 4-fold as was the hapten binding affinity (Table 3), suggesting that the Tyr-100aHAsn substitution does indeed contribute to the enhanced catalytic efficency of variant 3.1. We went on to replace residue 100aH with asparagine in both hu17E8 and variant 2.3 and in both cases this single amino acid substitution led to improvements in catalytic activities and hapten binding affinities (Table 3). Thus, even though residue 100aH is removed from the hapten binding site, it nevertheless influences both catalytic activity and hapten binding. This finding is similar to observations made by Schultz and coworkers (31) who found that the nine mutations which resulted in the in vivo maturation of hapten affinity and catalytic activity in antibody 48G7 were at residues that do not contact hapten. These results suggest that future efforts to optimize catalytic antibodies using phage display may benefit from a broader randomization strategy which is not limited to residues that interact directly with hapten.
Table 3.
Catalytic and hapten-binding properties of hu17E8 and mutants with tyrosine or asparagine at residue 100aH
| Clone | kcat, min−1 | Km, μM | kcat/Km, M−1⋅min−1 |
kcat/Km (mutant)
|
EC50 (hu17E8)
|
|---|---|---|---|---|---|
| kcat/Km (hu17E8) | EC50 (mutant) | ||||
| 3.1 | 110 | 530 | 2.0 × 105 | 2.0 | 0.52 |
| (Asn-100aH) | |||||
| 3.1y | 63 | 1,300 | 5.0 × 104 | 0.50 | 0.14 |
| (Tyr-100aH) | |||||
| hu17E8 | 59 | 580 | 1.0 × 105 | 1.0 | 1.0 |
| (Tyr-100aH) | |||||
| hu17E8n | 66 | 140 | 4.6 × 105 | 4.6 | 1.6 |
| (Asn-100aH) | |||||
| 2.3 | 96 | 2,400 | 4.0 × 104 | 0.40 | 2.5 |
| (Tyr-100aH) | |||||
| 2.3n | 200 | 2,500 | 8.0 × 104 | 0.80 | 4.0 |
| (Asn-100aH) |
Mutagenesis of the Serine–Histidine Dyad.
In characterizing the catalytic activities of phage-selected variants, we discovered that residues Arg-96L and Asp-101H both make a significant contribution to catalysis. To gain additional insights into the catalytic contribution of active site residues, we used mutagenesis to assess the role of Ser-95H and His-35H. These two residues are believed to be important for the activity of 17E8, based on crystallographic (19) and kinetic (18) data which suggests that these may participate in the hydrolytic mechanism in an analogous fashion to the serine and histidine in the catalytic triad of serine proteinases. Mutants of hu17E8 were prepared by separately replacing each of Ser-95H and His-35H with alanine and also by changing His-35H to glutamine. As expected, substitutions of His-35H resulted in decreases in catalytic activity (Table 4). Although the His-35HGln mutant was 12-fold less active than hu17E8, hapten binding was improved 2-fold. Once again, this reveals that replacement of key catalytic residues can result in significant departures from the kcat/Km versus kuncat/Ki relationship.
Table 4.
Catalytic properties of hu17E8 active-site mutants
| Mutation | kcat, min−1 | Km, μM | kcat/Km, M−1⋅min−1 |
kcat/Km (mutant)
|
EC50 (hu17E8)
|
|---|---|---|---|---|---|
| kcat/Km (hu17E8) | EC50 (mutant) | ||||
| hu17E8 | 59 | 580 | 1.0 × 105 | 1.0 | 1.0 |
| His-35HAla | —* | —* | <5.0 × 103 | <0.05 | ND |
| His-35HGln | 30 | 3,800 | 8.0 × 103 | 0.08 | 2.1 |
| Ser-95HAla | 44 | 210 | 2.1 × 105 | 2.1 | 1.4 |
ND = not determined.
Activity too weak to determine kcat and Km.
In contrast to substitutions of His-35H, replacement of Ser-95H with alanine yielded a fully active mutant (Table 4). In fact, the Ser-95HAla mutant exhibited a 2-fold improvement in kcat/Km. This result casts doubt on the notion that Ser-95H is involved in the formation of a covalent intermediate with substrate as a step in the catalytic mechanism of 17E8 (19). Although kinetic data (18) had suggested the formation of a covalent acyl-antibody intermediate, structural data in the context of the current mutagenesis results do not suggest any other antibody residue as a candidate nucleophile. Further work is required to better elucidate the mechanistic features of ester hydrolysis by 17E8. Our preparation of a humanized form of 17E8 which expresses well in E. coli should assist in these studies given the relative ease with which site-directed mutants of hu17E8 can be generated and their kinetic properties studied.
Conclusions and Perspectives.
Our selection of hu17E8 mutants with optimized hapten binding affinity did not lead to more active antibody catalysts. This breakdown in the relationship between binding of the transition-state analog and catalytic efficiency may be due to deficiencies in the ability of a phosphonate hapten to mimic the true hydrolytic transition-state. This result recapitulates the situation observed when monoclonal antibodies are raised in vivo against a stable transition-state analog;, that is, most of these do not possess catalytic activity (32). However, in vitro selection methods lend themselves to more elaborate selection strategies that may increase the chances of selecting improved antibody catalysts. For instance, phage displayed libraries could be sorted in the presence of competing substrate or product to select for variants that show high selectivity for binding the transition-state over the ground state, or variants could be selected based on the ability to covalently react with a mechanism-based inhibitor (33).
Phage display of catalytic antibodies provides a general methodology for optimizing transition-state analog binding and catalytic activity. By generating a humanized form of antibody 17E8 which expressed well in E. coli, we were able to display a library of randomized variants on phage and select those clones with improved binding for hapten. These studies have identified key residues that are important for the catalytic mechanism of 17E8 as well as a mutation that can increase the catalytic efficiency.
Acknowledgments
We thank the oligonucleotide synthesis group, James Bourell for mass spectrometric analysis and Michelle Arkin for her comments on the manuscript. T.S.S. acknowledges the National Institutes of Health for funding support (EM 50672).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: hu178, humanized form of 17E8; CDR, complementarity-determining region.
References
- 1.Hilvert D. Curr Opin Struct Biol. 1994;4:612–617. [Google Scholar]
- 2.Schultz P G, Lerner R A. Science. 1995;269:1835–1842. doi: 10.1126/science.7569920. [DOI] [PubMed] [Google Scholar]
- 3.Driggers E M, Schultz P G. Adv Prot Chem. 1996;49:261–287. doi: 10.1016/s0065-3233(08)60491-x. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin E, Schultz P G. Science. 1989;245:1104–1107. doi: 10.1126/science.2672338. [DOI] [PubMed] [Google Scholar]
- 5.Stewart J D, Krebs J F, Siuzdak G, Berdis A J, Smithrud D B, Benkovic S J. Proc Natl Acad Sci USA. 1994;91:7404–7409. doi: 10.1073/pnas.91.16.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kast P, Hilvert D. Pure Appl Chem. 1996;68:2017–2024. [Google Scholar]
- 7.Kraut J. Science. 1988;242:533–540. doi: 10.1126/science.3051385. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs J W. Bio/Technology. 1991;9:258–262. [Google Scholar]
- 9.Stewart J D, Benkovic S J. Nature (London) 1995;375:388–391. doi: 10.1038/375388a0. [DOI] [PubMed] [Google Scholar]
- 10.Angeles T S, Smith R G, Darsley M J, Sugasawara R, Sanchez R I, Kenten J, Schultz P G, Martin M T. Biochemistry. 1993;32:12128–12135. doi: 10.1021/bi00096a025. [DOI] [PubMed] [Google Scholar]
- 11.Fujii I, Tanaka F, Miyashita H, Tanimura R, Kinoshita K. J Am Chem Soc. 1995;117:6199–6209. [Google Scholar]
- 12.Phillips, M. A., Kaplan, A. P., Rutter, W. J. & Bartlett, P. A. (1992) Biochemistry 959–963. [DOI] [PubMed]
- 13.Kim S, Davis M, Sinn E, Patten P, Hood L. Cell. 1981;27:573–581. doi: 10.1016/0092-8674(81)90399-8. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins R E, Russell S J, Winter G. J Mol Biol. 1992;226:889–896. doi: 10.1016/0022-2836(92)90639-2. [DOI] [PubMed] [Google Scholar]
- 15.Jackson J R, Sathe G, Rosenberg M, Sweet R. J Immunol. 1995;154:3310–3319. [PubMed] [Google Scholar]
- 16.Yang W P, Green K, Pinz-Sweeney S, Briones A T, Burton D R, Barbas C F. J Mol Biol. 1995;254:392–403. doi: 10.1006/jmbi.1995.0626. [DOI] [PubMed] [Google Scholar]
- 17.Schier R, McCall A, Adams G P, Marshall K W, Merritt H, Yim M, Crawford R S, Weiner L M, Marks C, Marks J D. J Mol Biol. 1996;263:551–567. doi: 10.1006/jmbi.1996.0598. [DOI] [PubMed] [Google Scholar]
- 18.Guo J C, Huang W, Scanlan T S. J Am Chem Soc. 1994;116:6062–6069. [Google Scholar]
- 19.Zhou G W, Guo J C, Huang W, Fletterick R J, Scanlan T S. Science. 1994;265:1059–1064. doi: 10.1126/science.8066444. [DOI] [PubMed] [Google Scholar]
- 20.Ulrich H D, Patten P A, Yang P L, Romesberg F E, Schultz P G. Proc Natl Acad Sci USA. 1995;92:11907–11911. doi: 10.1073/pnas.92.25.11907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baca M, Presta L G, O’Connor S, Wells J A. J Biol Chem. 1997;278:10678–10684. doi: 10.1074/jbc.272.16.10678. [DOI] [PubMed] [Google Scholar]
- 22.Carter P, Presta L, Gorman C M, Ridgway J B, Henner D, Wong W L, Rowland A M, Kotts C, Carver M E, Shepard H M. Proc Natl Acad Sci USA. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller C. Sequences of Proteins of Immunological Interest. 5th Ed. National Institutes of Health, Bethesda, MD: Public Health Service; 1991. [Google Scholar]
- 24.Chothia C, Lesk A M. J Mol Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- 25.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 26.Kunkel T A, Bebenek K, McClary J. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham B C, Lowe D G, Li B, Bennett B D, Wells J A. EMBO J. 1994;13:2508–2515. doi: 10.1002/j.1460-2075.1994.tb06540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter P, Kelley R F, Rodrigues M L, Snedecor B, Covarrubias M, Velligan M D, Wong W L, Rowland A M, Kotts C E, Carver M E, Yang M, Bourell J H, Shepard H M, Henner D. Bio/Technology. 1992;10:163–167. doi: 10.1038/nbt0292-163. [DOI] [PubMed] [Google Scholar]
- 29.Wells J A. Biochemistry. 1990;29:8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- 30.Stewart J D, Roberts V A, Thomas N R, Getzoff E D, Benkovic S J. Biochemistry. 1994;33:1994–2003. doi: 10.1021/bi00174a004. [DOI] [PubMed] [Google Scholar]
- 31.Patten P A, Gray N S, Yang P L, Marks C B, Wedemayer G J, Boniface J J, Stevens R C, Schultz P G. Science. 1996;271:1086–1091. doi: 10.1126/science.271.5252.1086. [DOI] [PubMed] [Google Scholar]
- 32.Tawfik D S, Green B S, Chap R, Sela M, Eshhar Z. Proc Natl Acad Sci USA. 1993;90:373–377. doi: 10.1073/pnas.90.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janda K D, Lo L C, Lo C H L, Sim M M, Wang R, Wong C H, Lerner R A. Science. 1997;275:945–948. doi: 10.1126/science.275.5302.945. [DOI] [PubMed] [Google Scholar]