Abstract
Dendritic spines receive the vast majority of excitatory synaptic contacts in the mammalian brain and are presumed to contain machinery for the integration of various signal transduction pathways. Protein phosphatase 1 (PP1) is greatly enriched in dendritic spines and has been implicated in both the regulation of ionic conductances and long-term synaptic plasticity. The molecular mechanism whereby PP1 is localized to spines is unknown. We have now characterized a novel protein that forms a complex with the catalytic subunit of PP1 and is a potent modulator of PP1 enzymatic activity in vitro. Within the brain this protein displays a remarkably distinct localization to the heads of dendritic spines and has therefore been named spinophilin. Spinophilin has the properties expected of a scaffolding protein localized to the cell membrane and contains a single consensus sequence in PSD95/DLG/zo-1, which implies cross-linking of PP1 to transmembrane protein complexes. We propose that spinophilin represents a novel targeting subunit for PP1, which directs the enzyme to those substrates in the dendritic spine compartment, e.g., neurotransmitter receptors, which mediate the regulation of synaptic function by PP1.
Most neurotransmitters affect the excitability of neuronal cells by binding to receptors that are coupled either directly to ionic conductances or to signaling cascades that indirectly regulate ionic conductances. Indirect coupling allows postreceptor integration of the diverse inputs received by neurons at points where signaling pathways converge, thus enabling more complex responses. Dendritic spines receive the vast majority of excitatory synapses in the mammalian brain (1) and are therefore likely to house the machinery for regulating this input. Among the enzymes implicated in postsynaptic signal integration is protein phosphatase 1 (PP1), a serine/threonine phosphatase that is enriched in the brain and is specifically localized to dendritic spines (2). PP1 has been shown to be involved in dopamine regulation of Ca2+ currents (3), neuropeptide regulation of K+ current (4), and the induction of hippocampal long-term depression (5). The enzyme is expressed ubiquitously and exhibits a broad substrate specificity in vitro. Recent work in nonneuronal tissues has shown that PP1 associates with a range of regulatory subunits. These proteins are thought to dictate the subcellular localization of the enzyme and, in some cases, to modulate the catalytic activity of the enzyme toward specific substrates at these sites (6). Although the free catalytic subunit of PP1 is soluble, the majority of the protein found in brain extracts is associated with the particulate fraction, where it is predominantly in an inactive state (7). This suggested that a proportion of the PP1 found in the particulate fraction might be associated with insoluble and inhibitory targeting subunits. However, none of the previously characterized PP1 targeting subunits have been shown to play a specific role in the nervous system. We therefore initiated a search for novel neuronal regulatory subunits.
MATERIALS AND METHODS
Library Construction and Yeast Two-Hybrid Screening.
cDNA for library construction was synthesized from 5 μg of oligo(dT)-purified total rat brain mRNA using the adapter 5′-pGACTAGTTCTAGATCGCGAGCGGCCGCCC(T)15 to prime first strand synthesis with SuperScript II reverse transcriptase (GIBCO/BRL). Second strand synthesis was achieved using nick-translational mRNA replacement. The SalI adapter 5′-TCGACCCACGCGTCCG/5′-pCGGACGCGTGGG was ligated to double-stranded DNA followed by digestion with NotI. cDNAs were then directionally subcloned into the NotI/SalI-digested GAL4 activation-domain expression vector pPC86 (8). The cDNA for PP1α was expressed in HF7c yeast cells (9) as a GAL4 fusion in the expression vector pPC62 (8).
cDNA Expression and Coimmunoprecipitation.
The expression vector pcDNA1 Neo (Invitrogen) was modified using a synthetic double-stranded oligonucleotide (5′-pAGCTTCCAGCAGCCATGGACTACAAGGACGACGATGACAAAGGTGGGTCGACGC; 5′-pGGCCGCGTCGACCCACCTTTGTCATCGTCGTCCTTGTAGTCCATGGCTGCTGGA), which was ligated into the HindIII/NotI-digested plasmid. This oligonucleotide provided an rRNA binding site, an initiation codon, and sequences encoding the FLAG epitope followed by a SalI site in-frame with SalI/NotI library inserts. Constructs were transiently expressed in 293T cells for 36 hr following calcium phosphate transfection. Cells were lysed in 0.5% Nonidet P-40, 1 mM EDTA, 50 mM Tris⋅HCl (pH 8.0), 120 mM sodium chloride, 1 mM phenylmethylsulfonyl fluoride (PMSF), 20 μg/ml leupeptin/antipain, and 5 μg/ml pepstatin/chymostatin on ice for 30 min. Protein concentrations in the 100,000 × g supernatant were determined using BCA assay (Pierce). One milligram total protein was used for immunoprecipitation with anti-FLAG monoclonal antibody M2 coupled to agarose (IBI). Following several washes in lysis buffer and one in 50 mM Tris⋅HCl (pH 7.0), proteins were separated by SDS/PAGE and subjected to Western blotting. Coimmunoprecipitation from rat brain followed the same procedure except that tissue was dispersed using a probe sonicator and lysates were incubated with a protein A-Sepharose/primary antibody complex (RU144 or RU145).
cDNA Cloning and Expression Pattern of Spinophilin.
The 5′ end of the cDNA was characterized using five clones isolated from a rat hippocampus λZAP cDNA library kindly provided by Markus Stoffel (The Rockefeller University). Sequencing of both cDNA strands was performed by dye terminator cycling and resolution on an Applied Biosystems 373 Stretch Sequencer. Analysis was performed using Geneworks (IntelliGenetics) and the blast algorithm (10). Northern blot analysis was performed using an AgeI/BamHI cDNA fragment (residues 1409–2139) and a CLONTECH rat tissue poly(A)+ RNA blot.
For Western blotting, tissues and brain regions were dissected from an adult male Sprague–Dawley rat. Samples were dispersed in 1% SDS, 1 mM PMSF, 20 μg/ml leupeptin/antipain, and 5 μg/ml pepstatin/chymostatin using a probe sonicator. Proteins were separated by SDS/PAGE and transferred to Immobilon P (Millipore). Membranes were blocked in TBS (5% nonfat milk/0.5% Tween 20), followed by incubation with primary antibody (0.1 μg/ml immunoglobulin). Detection was with anti-rabbit horseradish peroxidase-conjugated secondary antibody (Calbiochem) and development with enhanced chemiluminescence (Amersham). Immunohistochemistry was performed as described (2).
Antibody Production and Purification.
The synthetic peptide DNGRAPDMAPEEVDESKKEDFSEAC (residues 367–390) was provided by Janet Crawford (W. M. Keck Foundation, Yale University). For immunization of rabbits, 5 mg of peptide was conjugated to 33 mg thyroglobulin (Sigma) using glutaraldehyde. Crude sera were affinity purified against the immunization peptide coupled to SulfoLink (Pierce).
Preparation of Bacterial Glutathione S-Transferase (GST) Fusion Protein.
Residues 296–817 of spinophilin were fused to GST and expressed from pGEX-4T-2 (Pharmacia) in BL21DE3 Escherichia coli. The fusion protein was purified by affinity chromatography on glutathione–agarose beads (Pharmacia) using 5 mM glutathione/50 mM Tris⋅HCl (pH 8.0) for elution.
Protein Phosphatase Activity Assay.
Purified rabbit muscle PP1 and PP2A were kindly provided by Hsien-bin Huang (The Rockefeller University). Preparation and activity assays were performed as described (11) with PP1 or PP2A at ≈150 pM.
RESULTS
Isolation of a PP1 Binding Protein.
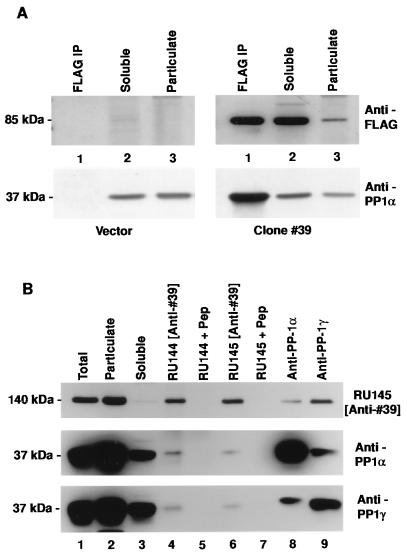
A yeast two-hybrid screen (12) was used to identify neuronal proteins capable of interacting with the α isoform of PP1. Eight distinct classes of cDNA were isolated from among 60 clones. Clone #39 was the sole representative of one class and appeared to interact specifically with PP1α, as it would not activate transcription of yeast reporter genes either independently or in combination with irrelevant baits. For an independent assessment of this protein–protein interaction, the ability of the product of clone #39 to coimmunoprecipitate with PP1α was examined in extracts from mammalian tissue culture cells. For this purpose, clone #39 was tagged at the amino terminus with a FLAG epitope for antibody recognition and expressed transiently in 293T cells. Immunoprecipitation was carried out using anti-FLAG antibody and proteins present in the pellet were solubilized, separated by SDS/PAGE, and immunoblotted sequentially using anti-FLAG and anti-PP1α antibodies (Fig. 1A). The FLAG-tagged protein, representing the product of the partial cDNA clone isolated in the library screen, migrated with an apparent Mr of 85 kDa. The overexpressed protein was largely soluble in 293T cell lysates. The strong signal seen for PP1 upon immunoprecipitation with anti-FLAG antibody indicated that the product of clone #39 existed in a complex with PP1 in 293T lysates.
Figure 1.
Clone #39 represents a PP1 binding protein. (A) Immunoblot analysis of proteins prepared from 293T cells transfected with expression vector alone (Left) or with clone #39 expression construct (Right). Lanes: 1, anti-FLAG immunoprecipitate; 2, 50 μg soluble fraction of cell lysate; 3, 50 μg particulate fraction of cell lysate. Blots were probed with anti-FLAG antibody (Upper), stripped, and reprobed with anti-PP1α antiserum (Lower). (B) Immunoblot analysis of proteins prepared from rat brain homogenate. Lanes: 1–3, 50 μg total lysate, 50 μg particulate fraction, 50 μg soluble fraction, respectively; 4–9, immunoprecipitates prepared from the soluble fraction using anti-clone #39 peptide antisera (RU144 and RU145 +/− immunogen peptide) or anti-PP1 antisera. The blot was probed sequentially with anti-clone #39 antiserum (Top), anti-PP1α antiserum (Middle), and anti-PP1γ antiserum (Bottom).
Two separate rabbit anti-peptide antisera against clone #39 were prepared (RU144/5) and found to recognize a protein present in rat brain homogenates, which migrated with an apparent Mr of ≈140 kDa (Fig. 1B). In contrast to the overexpressed recombinant protein, the endogenous full-length protein was largely insoluble in brain extracts. Coimmunoprecipitation analysis using rat brain homogenate was performed using these antibodies as well as isoform-specific antisera raised against PP1α and PP1γ (13). In each case, the immunoprecipitating antibody pulled down a complex between PP1 and the endogenous protein represented by clone #39. For the anti-clone #39 antisera, pre-absorption with peptide immunogen prior to immunoprecipitation abolished the signal seen in subsequent anti-PP1 immunoblots, demonstrating the specificity of the interaction. In addition, the blot was stripped and reprobed using antibodies against calcium/calmodulin-dependent protein kinase II α and the N-methyl-d-aspartate receptor subunit NR1. Although both of these proteins are enriched in the postsynaptic density, neither were detectable in the coimmunoprecipitates, further demonstrating the specificity of the interaction between PP1 and the ≈140 kDa protein. PP1α coimmunoprecipitated with PP1γ and vice versa. Because the two anti-PP1 antibodies are isoform-specific (13), the results suggest that the two PP1 isoforms were present in the same protein complex.
Expression Pattern of the PP1 Binding Protein.
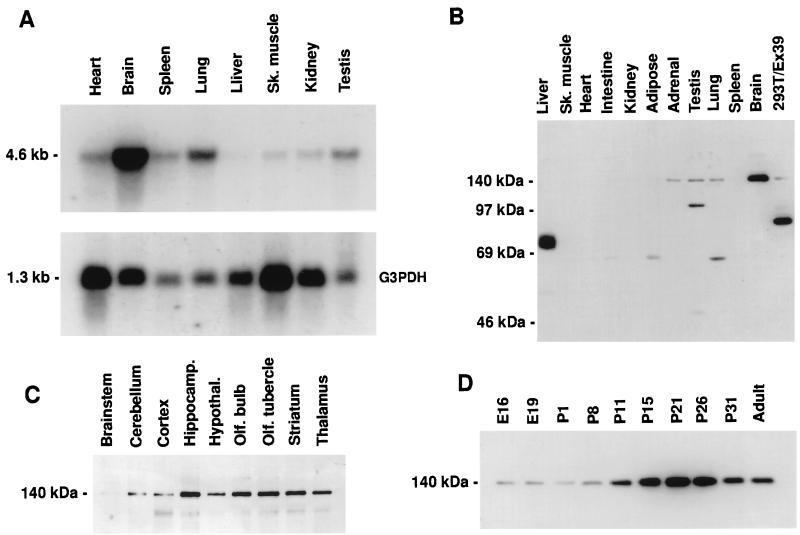
The expression pattern of mRNA for the endogenous protein represented by clone #39 was examined in various rat tissues (Fig. 2A). A 4.6 kb transcript was detected by Northern blotting in all tissues examined, but was most abundant in brain. There was no evidence for alternatively spliced transcripts. Expression of the protein was examined by immunoblotting with the anti-peptide antibodies. Both RU145 (Fig. 2B) and RU144 (not shown) recognized a single major band of ≈140 kDa in Western blots of brain extracts. With both antibodies the ≈140 kDa immunoreactivity was highest in the brain, with much lower levels in other tissues, including adrenal gland, testis, and lung. The ≈140 kDa protein showed a variation in its expression level within the brain. The highest levels were detected in hippocampus and the lowest in cortex, cerebellum, and particularly brainstem (Fig. 2C). Expression was low during embryogenesis and was followed by a distinct peak at around postnatal day 21 (Fig. 2C).
Figure 2.
Expression pattern of mRNA and distribution of the endogenous protein represented by clone #39. (A) Northern blot analysis. (Upper) Detection of a transcript of ≈4.6 kb using a clone #39 probe. (Lower) Levels of the transcript for G3PDH to indicate relative mRNA loading. (B–D) Immunoblot analysis using anti-clone #39 antiserum RU145 and 50 μg per lane rat tissue homogenate in 1% SDS. (B) Expression of a protein migrating at ≈140 kDa in various rat tissues. Far right lane, 20 μg of 1% SDS homogenate from 293T cells expressing partial cDNA clone #39. Proteins detected by RU145 migrating at less than 140 kDa were not reactive with RU144 and vice versa (data not shown). These are presumably cross-reactive species. (C) Expression levels of the ≈140 kDa protein in various adult brain structures. (D) Developmental profile of the ≈140 kDa protein present in cortical homogenates at the embryonic (E) and postnatal (P) days indicated.
Neuronal Distribution of the PP1 Binding Protein.
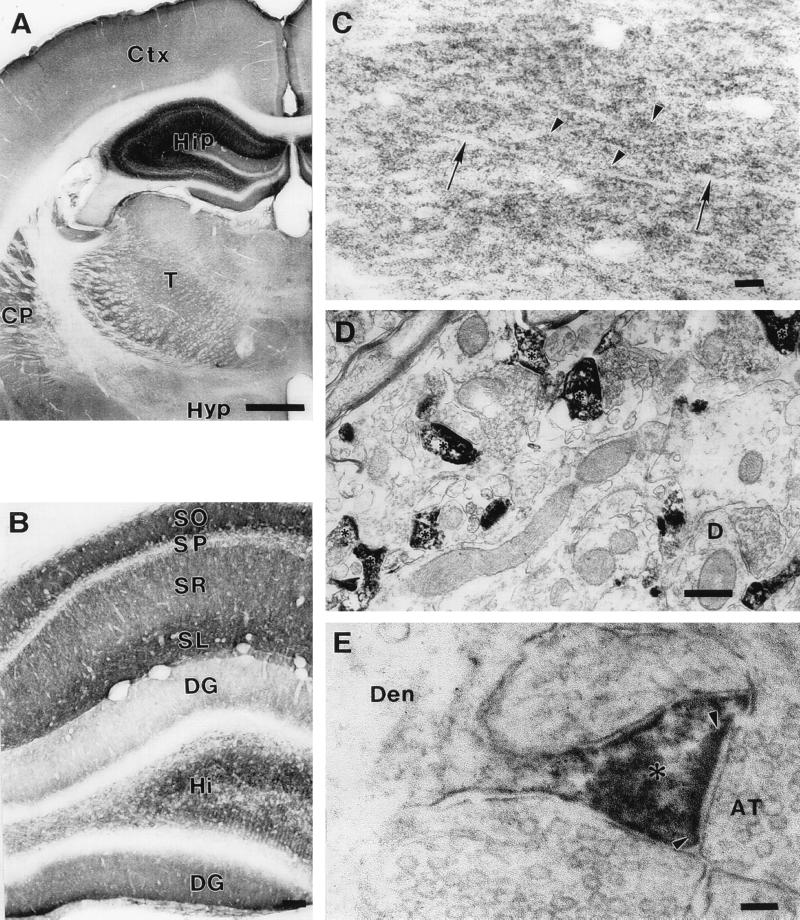
RU144 (Fig. 3) and RU145 (not shown) were used to examine the distribution of the protein in rat brain by immunohistochemistry. The two antibodies gave identical staining patterns. At the light microscopic level, immunoreactivity appeared as very numerous small puncta, 0.5–1 μm in diameter (Fig. 3C). These immunoreactive puncta were abundant in the neuropil of virtually all brain regions examined. In the areas examined by electron microscopy (hippocampus and caudatoputamen), these puncta were resolved as dendritic spines (Fig. 3 D and E). Immunoreactivity was intense in the spine neck and especially in the spine head, and disappeared abruptly at the junction between the spine neck and dendritic shaft (Fig. 3E). Within dendritic spines, immunoreaction product adhered to the cytoplasmic surface of the plasmalemma, the spine apparatus, and the postsynaptic density. Light microscopy showed that the intensity of dendritic spine immunoreactivity varied from region to region. The strongest immunoreactivity was present in the hippocampal formation and moderate levels were seen in caudatoputamen and dorsal thalamus (Fig. 3A). Within the hippocampal formation, immunoreactivity was strongest in stratum lacunosum moleculare and stratum oriens of Ammon’s horn (Fig. 3B). In stratum radiatum, immunoreactive puncta were aligned with unstained dendritic shafts, giving this lamina a striated appearance (Fig. 3C). The dentate gyrus was less strongly immunoreactive than the remainder of the hippocampal formation, creating a sharp line of demarcation between the ectal limb of the dentate gyrus and the adjacent stratum lacunosum moleculare (Fig. 3B). Preabsorption of the primary antibody with cognate peptide or omission of the primary antibody, secondary antibody, and/or avidin–biotin complex produced no immunostaining (data not shown). In view of the striking compartmentalization of this protein to the dendritic spines of neurons, we have named it spinophilin.
Figure 3.
Neuronal distribution of spinophilin. (A) Light microscopic photomicrograph of a coronal section at the level of the hippocampus and thalamus. Immunoreactivity is most intense in the hippocampus (Hip). Moderate immunoreactivity is also present in the caudatoputamen (CP), thalamus (T), and cerebral cortex (Ctx). Immunoreactivity in the hypothalamus (Hyp) is light. (Bar = 1 mm.) (B) Light microscopic photomicrograph of the laminar pattern of immunoreactivity in a coronal section of the hippocampal formation. Immunoreactivity is most intense in stratum oriens (SO), stratum lacunosum moleculare (SL), and hilus (Hi). Immunoreactivity is moderately strong in stratum radiatum (SR) and absent from stratum pyramidale (SP). The upper, or ectal, limb of the dentate gyrus (DG) shows weaker immunoreactivity than the lower, or endal, limb. (Bar = 100 μm.) (C) High power light microscopic photomicrograph of the stratum radiatum showing immunoreactive puncta (arrowheads). Dendritic shafts (arrows) are not immunoreactive. (Bar = 10 μm.) (D) Electron micrograph showing immunolabeled dendritic spines (asterisks). An example of an unlabeled dendritic shaft that receives a synaptic contact in the plane of section is shown (D). (Bar = 500 nm.) (E) Electron micrograph showing immunoreactivity in a dendritic spine neck and especially in a spine head (asterisk). Immunoreactivity (black flocculent material) lines the cytoplasmic surface of the spine plasmalemma and coats the postsynaptic density (delineated by arrowheads). Immunoreactivity is not present in axon terminals (AT) or in the dendritic shaft (Den). (Bar = 100 nm.)
Spinophilin Modulates PP1 Catalytic Activity.
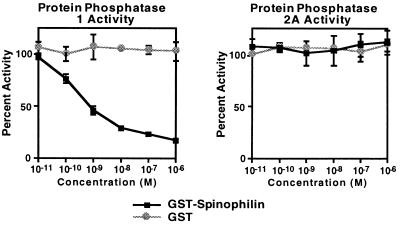
To examine the effect of spinophilin on the catalytic activity of PP1, we prepared a bacterial protein consisting of GST fused to the spinophilin residues (296–817) isolated in the two-hybrid screen. Larger bacterial fusion proteins were found to be unstable. This protein was purified by affinity chromatography on glutathione beads and the activity of purified PP1 catalytic subunit was measured in the presence of various concentrations of spinophilin using radiolabeled phosphorylase a as substrate. The GST–spinophilin fusion protein potently inhibited PP1 enzymatic activity with an IC50 of slightly less than 1 nM (Fig. 4). In contrast, GST alone had no effect on the activity of PP1, even at high concentrations. Neither GST nor GST–spinophilin affected the activity of the purified catalytic subunit of a closely related protein phosphatase, PP-2A. Together these results indicate that spinophilin specifically modulates the in vitro activity of PP1 toward phosphorylase a, either by acting as an inhibitor or by affecting substrate specificity.
Figure 4.
Effect of bacterial spinophilin fusion protein on the catalytic activity of PP1 and PP2A. Results (means ± SEM) were calculated as percent of activity in the absence of added bacterial protein.
Sequence Analysis of Spinophilin.
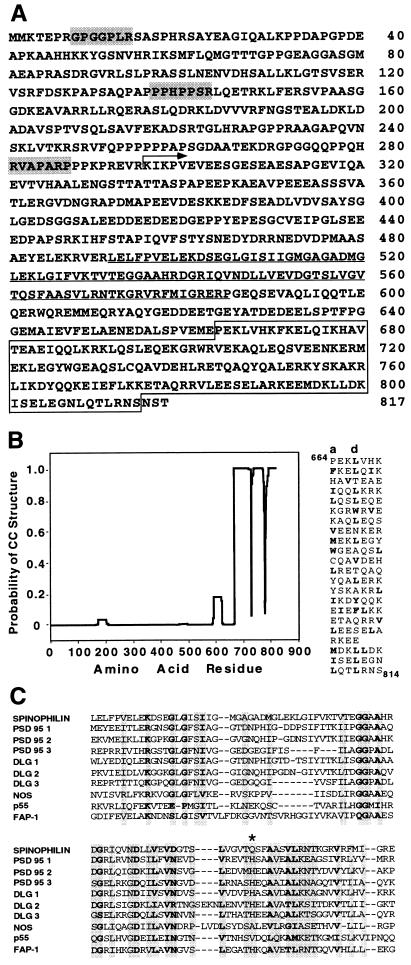
A cDNA of 4.5 kb was characterized using six clones from a rat hippocampus λZAP library. This cDNA contained a predicted ORF of 817 amino acids (Fig. 5A). The identity of the initiating methionine was confirmed by expressing 5′ end deletion mutants in 293T cells followed by Western blot analysis of the products (data not shown). The predicted molecular weight of spinophilin is 89.5 kDa, which differs significantly from the apparent Mr seen in SDS/PAGE. Both the expressed full-length cDNA (not shown) and the endogenous rat protein run at ≈140 kDa. This may be due to an extended conformation and/or low SDS binding capacity. The protein is predicted to be acidic, with a theoretical pI of 4.6. Database searches revealed a region in the carboxyl terminus of spinophilin with similarity to a broad array of filamentous myosin-like proteins. These similarities are rather diffuse: e.g., chicken nonmuscle myosin heavy chain, 20.2% similarity over 219 residues; yeast cytoskeletal protein Uso1, 20.4% similarity over 165 residues. Common amino acid signatures were not apparent upon multiple alignment. However, as many of these proteins have a coiled–coil structure, we used the algorithm of Lupas et al. (14), which predicted that the carboxyl terminus of spinophilin forms a solvent-exposed left-handed coiled–coil (Fig. 5B). This region might adopt an antiparallel organization to form an intramolecular coiled–coil or, alternatively, spinophilin might form dimers through this region, either with itself or with another protein possessing a complementary amphipathic α-helix. The filamentous coiled–coil structure may contribute to the putative “scaffolding” function within the spine cytomatrix and is likely to contribute to the relative insolubility of spinophilin. The amino terminus of spinophilin is rich in proline and contains three stretches that conform to the consensus for Src homology 3 domain binding (15), suggesting that spinophilin might target proteins other than PP1 to the spine.
Figure 5.
Sequence characteristics of spinophilin. (A) Predicted amino acid sequence of spinophilin. The region encoded by the cDNA isolated in the library screen is indicated (arrow). Shaded heptamers indicate potential Src homology 3-binding domain motifs present in the proline-rich amino terminus. Underlined sequence indicates the position of a PDZ domain. Boxed sequence indicates a region predicted to form a coiled–coil structure. (B Left) Coiled–coil prediction. The mtidk matrix algorithm was used without weighting at the a and d positions (14). (Right) Alignment of amino acids in the C terminus of spinophilin as a heptad repeat. Hydrophobic residues are in boldface type and a stagger is introduced at the point where the prediction breaks down. (C) Amino acid alignment of the PDZ domain (492–583) of spinophilin with selected examples of other PDZ domain-containing proteins: PSD95 1(61–151)/2(156–246)/3(309–393), DLG 1(36–126)/2(150–244)/3(482–566), NOS (14–100), p55 (67–153), FAP-1 (86–179). Asterisk marks the first residue in the αB helix of the PDZ domain.
Spinophilin also contains a single consensus sequence in PSD95/DLG/zo-1 (PDZ), which suggests that it might bind to a transmembrane protein(s) present in the dendritic spine (16). Fig. 5C shows the PDZ domain alignment of spinophilin with selected members of the diverse PDZ domain-containing protein family. Outside the PDZ domain, spinophilin shows no similarity to other members of this family and therefore represents a novel subtype. A key determinant for peptide-binding specificity is predicted to reside at the first residue in the αB helix of the PDZ domain (17). Spinophilin is unusual in possessing a glutamine at this position. Peptide binding analysis predicts that the ligand for spinophilin might contain an aromatic residue at the −2 position in addition to a C-terminal hydrophobic residue (18). PP1 isoforms do not conform to these requirements and are unlikely to bind to the spinophilin PDZ domain through their C-terminal residues.
DISCUSSION
PP1 activity toward physiological substrates in nonneuronal tissue is controlled through association with noncatalytic regulatory subunits. These proteins direct the subcellular distribution of PP1 and are referred to as targeting subunits (6). By analogy, spinophilin may serve as a neuronal targeting subunit for PP1 and might be responsive to inputs to the neuron. Although the key substrates for the PP1-mediated regulation of neuronal excitability and synaptic transmission are largely unidentified, the enzyme calcium/calmodulin-dependent protein kinase II, a major constituent of the postsynaptic density, is a known substrate for PP1. In vitro studies using the autophosphorylated form of this kinase as substrate showed, as for phosphorylase a, that spinophilin inhibited PP1 activity (unpublished data). The neurotransmitter receptors and ion channels thought to be regulated by PP1 are additional candidates for regulation by spinophilin. For this reason, it is of great interest that spinophilin contains a PDZ domain. PDZ domains are commonly found in cytosolic proteins that are enriched at points of cell–cell communication. In many cases, PDZ domains have been shown to bind to the carboxyl termini of transmembrane proteins, and such interactions have been described at synapses (16). Presumably, both the PDZ domain and the Src homology 3 domain-binding motifs present in spinophilin play roles in promoting protein–protein interactions at the spine membrane, and it will be important to search for proteins present in these putative complexes. Such proteins would be in close proximity to PP1 and be candidate substrates for the phosphatase.
Spinophilin might be involved in the regulation of synaptic transmission not only through short-term changes in neurotransmitter receptor and ion channel function, but also through long-term changes in spine number and morphology; spinophilin expression in the developing rat cortex peaks at a point when spines are still being formed. Spine density and morphology have been shown to be dynamic during development and in the adult brain (1) and the requirement for a dynamic regulation of spine anatomy may explain the presence of the dense actin microfilament network within the cytoskeleton of spine heads. Previous work in fibroblasts has shown that actin microfilament assemblies are regulated by the PP1-mediated dephosphorylation of myosin light chain (19). Thus, an important function of spinophilin, in addition to regulating the state of phosphorylation and activity of ion channels and neurotransmitter receptors, might be to target PP1 to the spine microfilament network to regulate spine number and/or morphology.
Acknowledgments
We thank Jacqueline Bromberg, Linda Hsieh, Young-Guen Kwon, and Angus Nairn for helpful discussions. This work was supported by U.S. Public Health Service Grants MH 40899 and DA 10044.
ABBREVIATIONS
- PP1
protein phosphatase 1
- GST
glutathione S-transferase
- PDZ
consensus sequence in PSD95/DLG/zo-1
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF016252).
References
- 1.Harris K M, Kater S B. Annu Rev Neurosci. 1994;17:341–371. doi: 10.1146/annurev.ne.17.030194.002013. [DOI] [PubMed] [Google Scholar]
- 2.Ouimet C C, da Cruz e Silva E F, Greengard P. Proc Natl Acad Sci USA. 1995;92:3396–3400. doi: 10.1073/pnas.92.8.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Surmeier D J, Bargas J, Hemmings H C, Jr, Nairn A C, Greengard P. Neuron. 1995;14:385–397. doi: 10.1016/0896-6273(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 4.Endo S, Critz S D, Byrne J H, Shenolikar S. J Neurochem. 1995;64:1833–1840. doi: 10.1046/j.1471-4159.1995.64041833.x. [DOI] [PubMed] [Google Scholar]
- 5.Mulkey R M, Endo S, Shenolikar S, Malenka R C. Nature (London) 1994;369:486–488. doi: 10.1038/369486a0. [DOI] [PubMed] [Google Scholar]
- 6.Hubbard M J, Cohen P. Trends Biochem Sci. 1993;18:172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- 7.Sim A T, Ratcliffe E, Mumby M C, Villa-Moruzzi E, Rostas J A. J Neurochem. 1994;62:1552–1559. doi: 10.1046/j.1471-4159.1994.62041552.x. [DOI] [PubMed] [Google Scholar]
- 8.Chevray P M, Nathans D. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feilotter H E, Hannon G J, Ruddell C J, Beach D. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Cohen P, Alemany S, Hemmings B A, Resink T J, Stralfors P, Tung H Y. Methods Enzymol. 1988;159:390–408. doi: 10.1016/0076-6879(88)59039-0. [DOI] [PubMed] [Google Scholar]
- 12.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 13.da Cruz e Silva E F, Fox C A, Ouimet C C, Gustafson E, Watson S J, Greengard P. J Neurosci. 1995;15:3375–3389. doi: 10.1523/JNEUROSCI.15-05-03375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 15.Mayer B J, Eck M J. Curr Biol. 1995;5:364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- 16.Gomperts S N. Cell. 1996;84:659–662. doi: 10.1016/s0092-8674(00)81043-0. [DOI] [PubMed] [Google Scholar]
- 17.Doyle D A, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 18.Songyang Z, Fanning A S, Fu C, Xu J, Marfatia S M, Chishti A H, Crompton A, Chan A C, Anderson J M, Cantley L C. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez A, Brautigan D L, Mumby M, Lamb N J. J Cell Biol. 1990;111:103–112. doi: 10.1083/jcb.111.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]