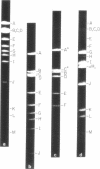
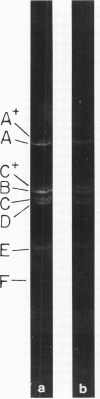
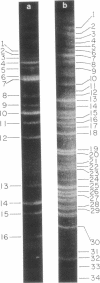
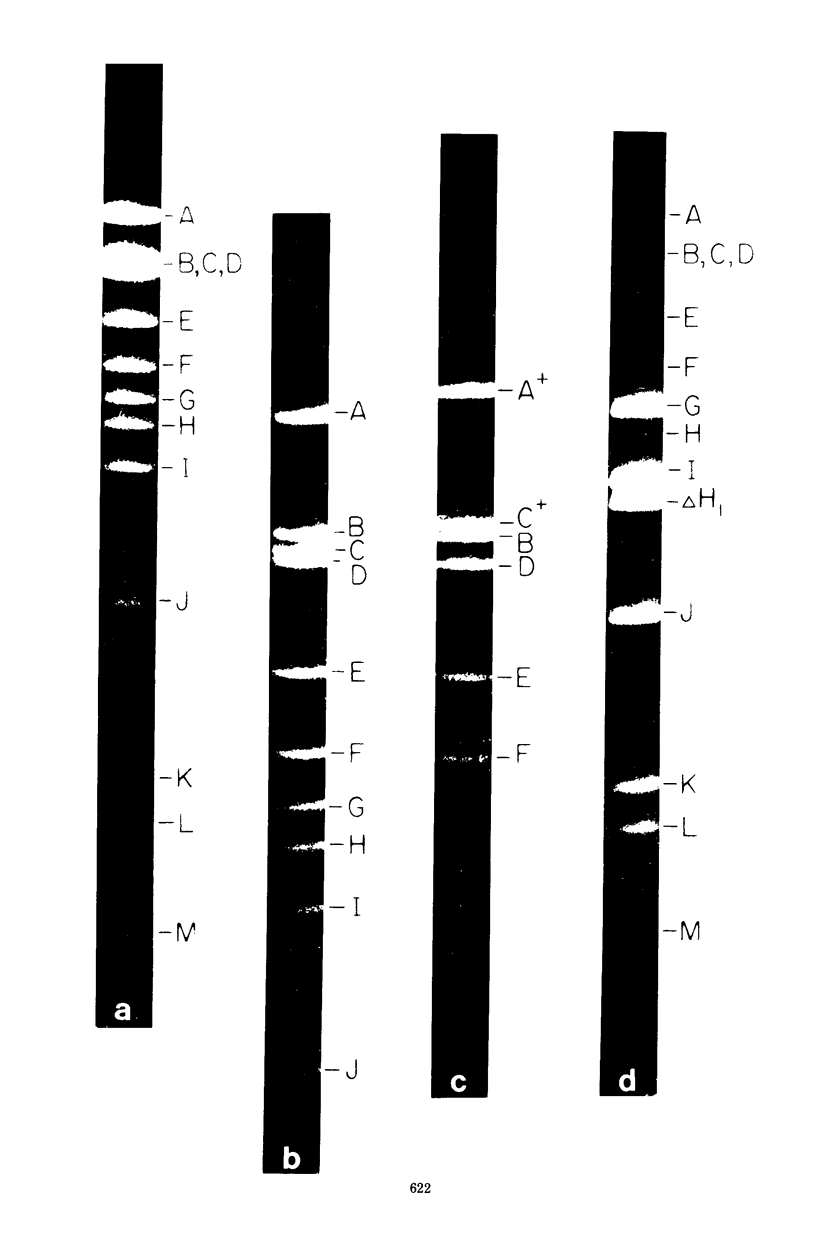
Abstract
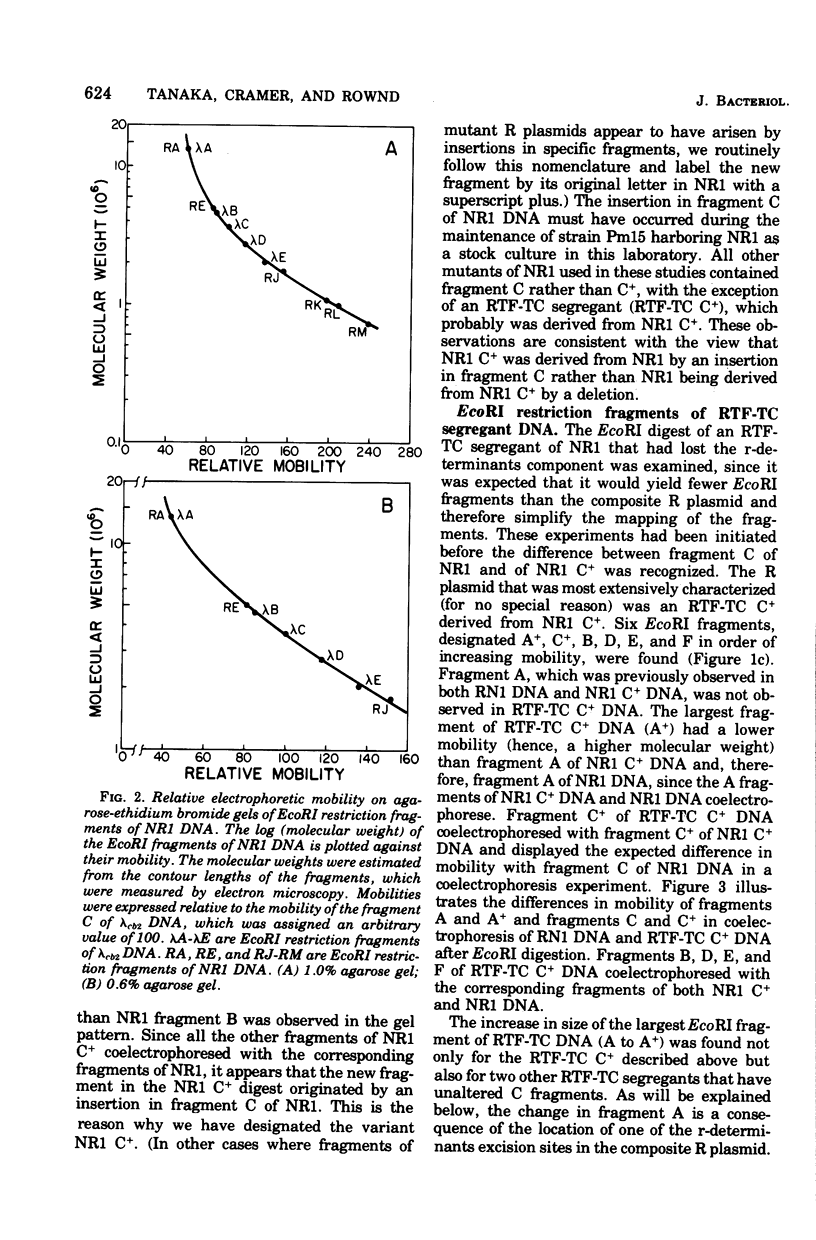
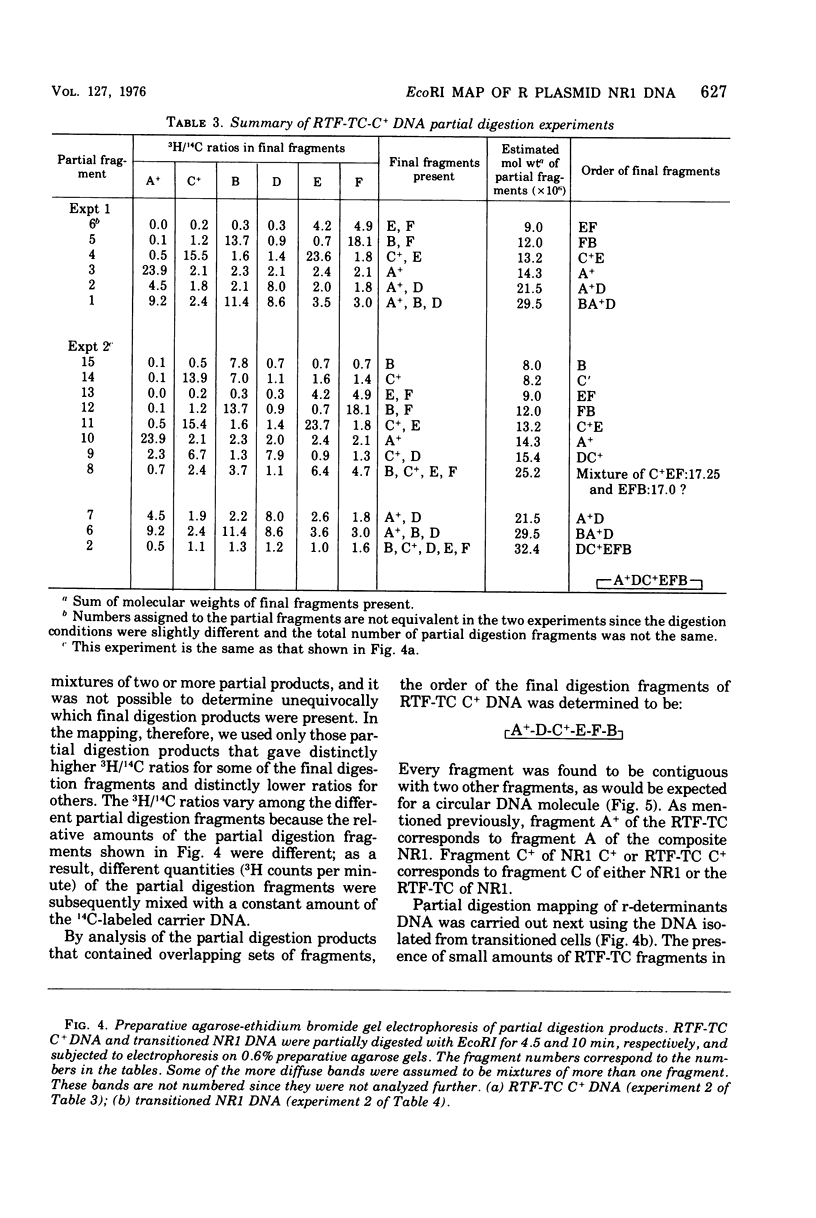
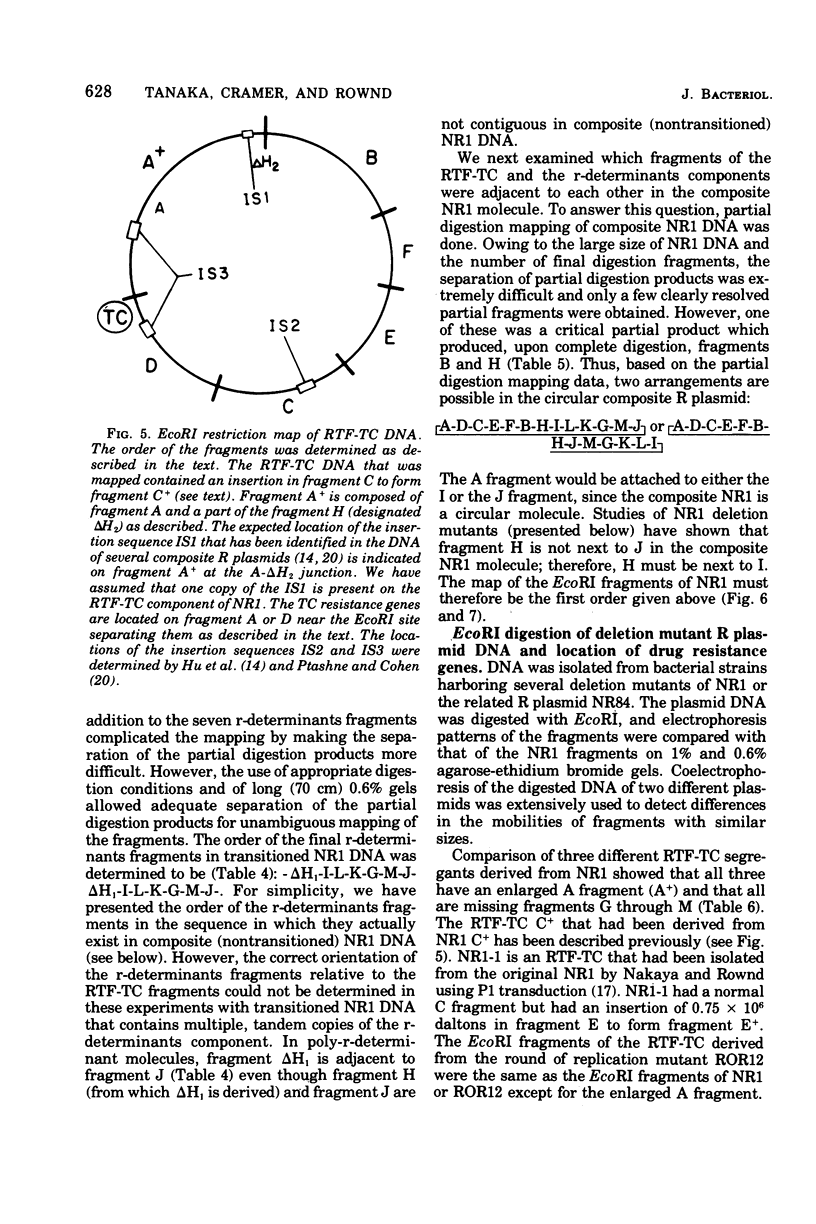
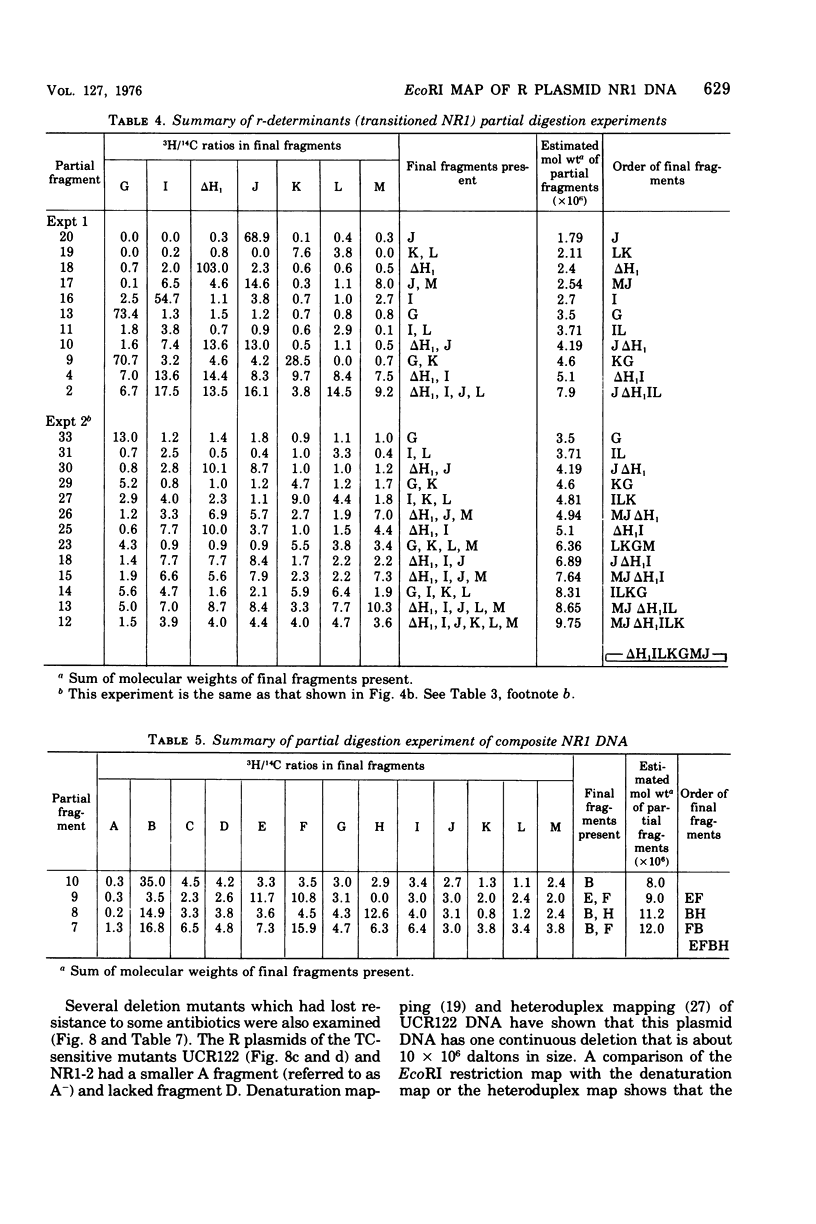
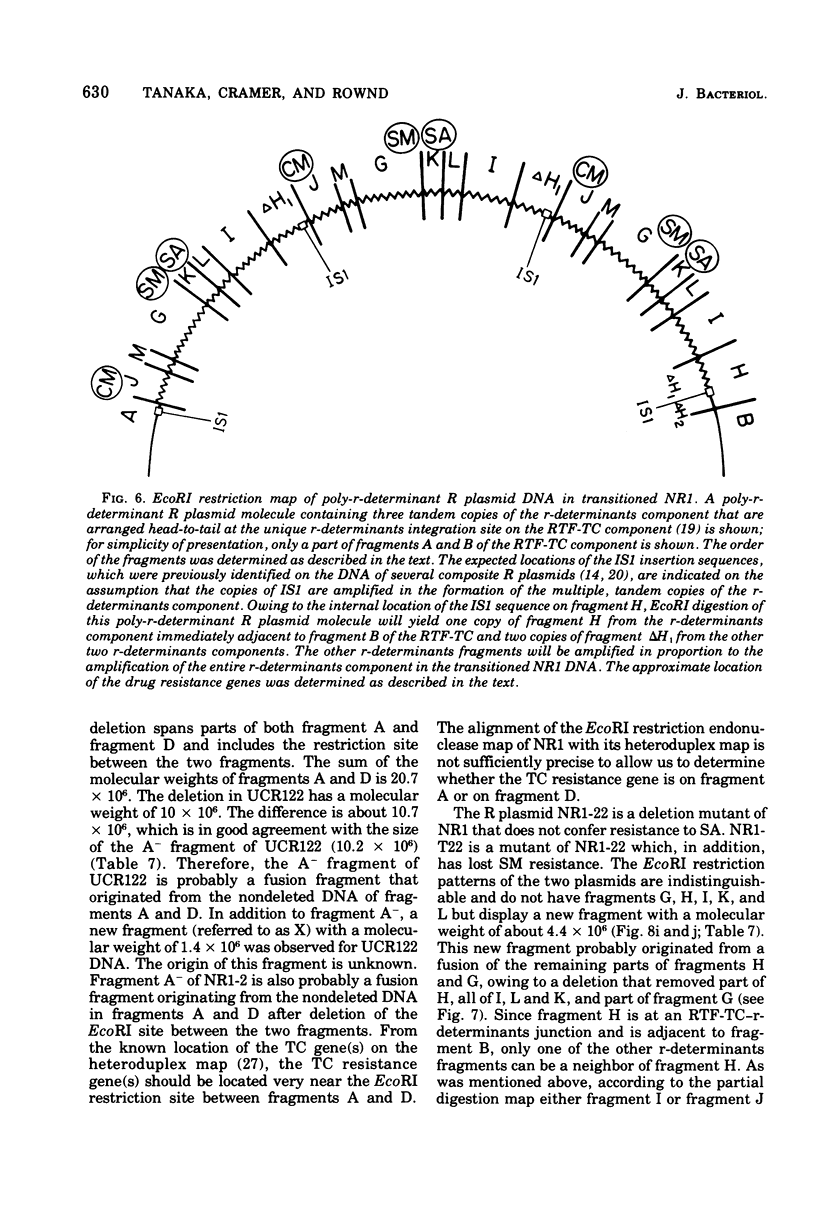
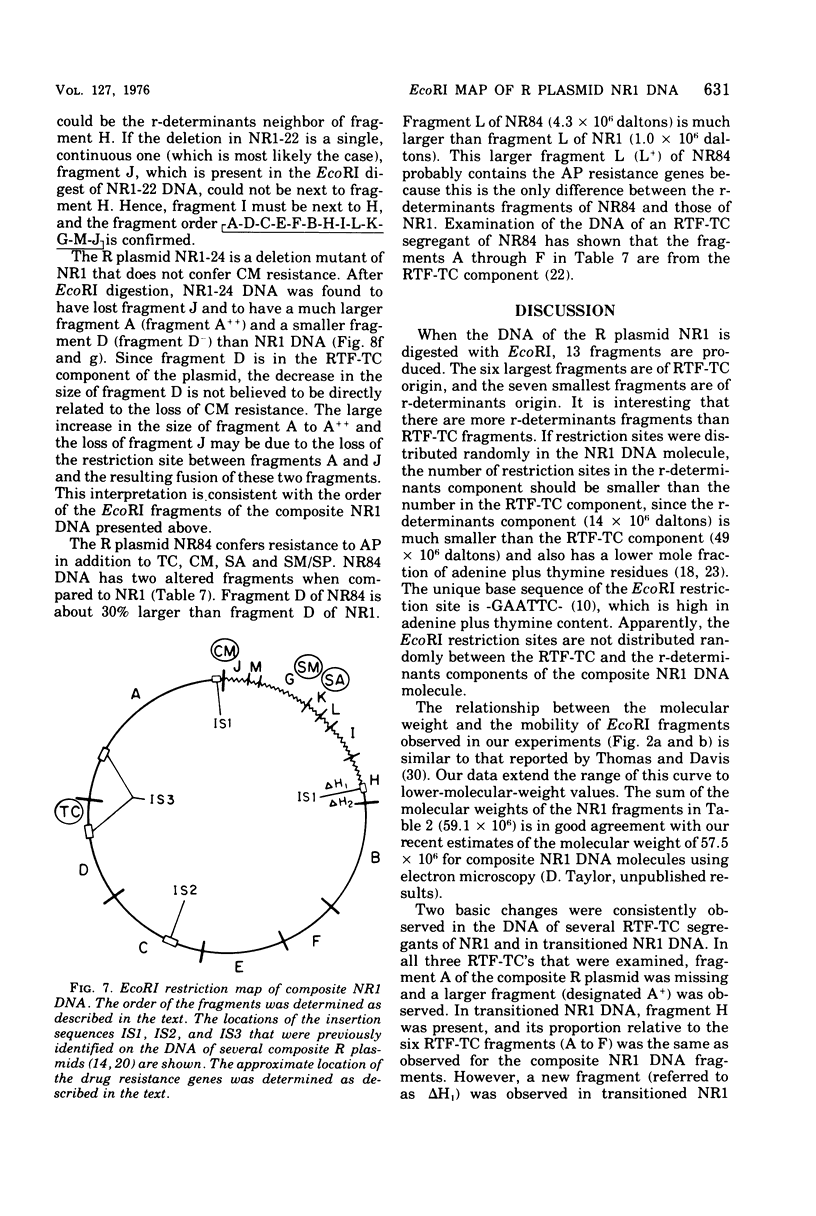
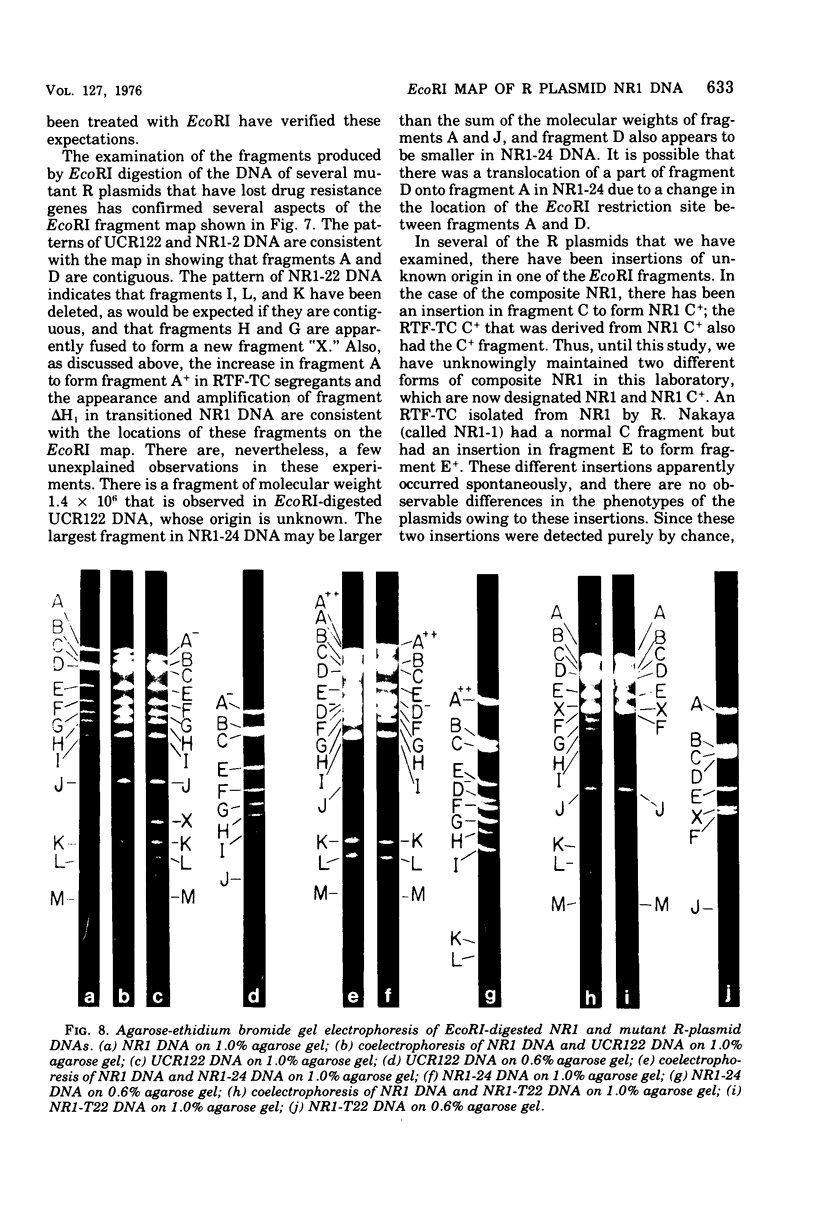
A physical map of the composite R plasmid NR1 has been constructed using specific cleavage of deoxyribonucleic acid (DNA) by the restriction endonuclease EcoR-. Digestion of composite NR1 DNA by EcoRI yields thirteen fragments. The six largest fragments (designated A to F) are from the resistance transfer factor component that harbors the tetracycline resistance genes (RTF-TC). The seven smallest fragments (designated G to M) are from the r-determinants component that harbors the chloramphenicol (CM), streptomycin-spectinomycin (SM/SP), and sulfonamide (SA) resistance genes. The largest fragment of several RTF-TC segregants of NR1 that have deleted the r-determinants component is 0.8 X 10(6) daltons larger than fragment A of composite NR1. Only a part of fragment H of the r-determinants component is amplified in transitioned NR1 DNA in Proteus mirabilis, which consists of multiple, tandem sequences of r-determinants attached to a single copy of the RTF-TC component. Both of these changes can be explained by the locations of the excision sites at the RTF-TC: r-determinants junctions that are involved in the dissociation and reassociation of the RTF-TC and r-determinants components. The thirteen fragments of composite NR1 DNA produced by EcoRI have been ordered using partial digestion techniques. The order of the fragments is: A-D-C-E-F-B-H-I-L-K-G-M-J. The approximate locations of the TC, CM, SM/SP, and SA resistance genes on the EcoRI map were determined by analyzing several deletion mutants of NR1.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clowes R. C. Molecular structure of bacterial plasmids. Bacteriol Rev. 1972 Sep;36(3):361–405. doi: 10.1128/br.36.3.361-405.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Davies J. E., Rownd R. Transmissible multiple drug resistance in Enterobacteriaceae. Science. 1972 May 19;176(4036):758–768. doi: 10.1126/science.176.4036.758. [DOI] [PubMed] [Google Scholar]
- Fiandt M., Szybalski W., Malamy M. H. Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. Mol Gen Genet. 1972;119(3):223–231. doi: 10.1007/BF00333860. [DOI] [PubMed] [Google Scholar]
- Freifelder D. Molecular weights of coliphages and coliphage DNA. IV. Molecular weights of DNA from bacteriophages T4, T5 and T7 and the general problem of determination of M. J Mol Biol. 1970 Dec 28;54(3):567–577. doi: 10.1016/0022-2836(70)90127-0. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Rownd R. H. Transition of the R factor NR1 and Proteus mirabilis: level of drug resistance of nontransitioned and transitioned cells. J Bacteriol. 1975 Jul;123(1):56–68. doi: 10.1128/jb.123.1.56-68.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132(1):31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- Hedgpeth J., Goodman H. M., Boyer H. W. DNA nucleotide sequence restricted by the RI endonuclease. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3448–3452. doi: 10.1073/pnas.69.11.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., Sublett R., Hedges R. W., Jacob A., Falkow S. Origin of the TEM-beta-lactamase gene found on plasmids. J Bacteriol. 1975 Apr;122(1):250–256. doi: 10.1128/jb.122.1.250-256.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch H. J., Starlinger P., Brachet P. Two kinds of insertions in bacterial genes. Mol Gen Genet. 1972;119(3):191–206. doi: 10.1007/BF00333858. [DOI] [PubMed] [Google Scholar]
- Hoar D. I. Fertility regulation in F-like resistance transfer factors. J Bacteriol. 1970 Mar;101(3):916–920. doi: 10.1128/jb.101.3.916-920.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman R. B., Schnös M. Partial denaturation of thymine- and 5-bromouracil-containing lambda DNA in alkali. J Mol Biol. 1970 Apr 14;49(1):93–98. doi: 10.1016/0022-2836(70)90378-5. [DOI] [PubMed] [Google Scholar]
- Morris C. F., Hashimoto H., Mickel S., Rownd R. Round of replication mutant of a drug resistance factor. J Bacteriol. 1974 Jun;118(3):855–866. doi: 10.1128/jb.118.3.855-866.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya R., Rownd R. Transduction of R factors by a Proteus mirabilis bacteriophage. J Bacteriol. 1971 Jun;106(3):773–783. doi: 10.1128/jb.106.3.773-783.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Rownd R. H. Transition of R factor NR1 in Proteus mirabilis: molecular structure and replication of NR1 deoxyribonucleic acid. J Bacteriol. 1975 Sep;123(3):1013–1034. doi: 10.1128/jb.123.3.1013-1034.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Twose T. M., Holland M. J., Rownd R. H. Denaturation mapping of R factor deoxyribonucleic acid. J Bacteriol. 1975 Sep;123(3):1035–1042. doi: 10.1128/jb.123.3.1035-1042.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne K., Cohen S. N. Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. J Bacteriol. 1975 May;122(2):776–781. doi: 10.1128/jb.122.2.776-781.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rownd R., Mickel S. Dissociation and reassociation of RTF and r-determinants of the R-factor NR1 in Proteus mirabilis. Nat New Biol. 1971 Nov 10;234(45):40–43. doi: 10.1038/newbio234040a0. [DOI] [PubMed] [Google Scholar]
- Rownd R., Perlman D., Hashimoto H., Mickel S., Applebaum E., Taylor D. Dissociation and reassociation of the transfer factor and resistance determinants of R factors as a mechanism of gene amplification in bacteria. Johns Hopkins Med J Suppl. 1973;2:115–128. [PubMed] [Google Scholar]
- Rownd R. Replication of a bacterial episome under relaxed control. J Mol Biol. 1969 Sep 28;44(3):387–402. doi: 10.1016/0022-2836(69)90368-4. [DOI] [PubMed] [Google Scholar]
- Saedler H., Heiss B. Multiple copies of the insertion-DNA sequences IS1 and IS2 in the chromosome of E. coli K-12. Mol Gen Genet. 1973 May 9;122(3):267–277. doi: 10.1007/BF00278602. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- WATANABE T., OGATA C., SATO S. EPISOME-MEDIATED TRANSFER OF DRUG RESISTANCE IN ENTEROBACTERIACEAE. 8. SIX-DRUG-RESISTANCE R FACTOR. J Bacteriol. 1964 Oct;88:922–928. doi: 10.1128/jb.88.4.922-928.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younghusband H. B., Egan J. B., Inman R. B. Characterization of the DNA from bacteriophage P2-186 hybrids and physical mapping of the 186 chromosome. Mol Gen Genet. 1975 Sep 29;140(2):101–110. doi: 10.1007/BF00329778. [DOI] [PubMed] [Google Scholar]