Abstract
The affinity between molecules depends both on the nature and presentation of the contacts. Here, we observe coupling of functional and structural elements when a protein binding domain is evolved to a smaller functional mimic. Previously, a 38-residue form of the 59-residue B-domain of protein A, termed Z38, was selected by phage display. Z38 contains 13 mutations and binds IgG only 10-fold weaker than the native B-domain. We present the solution structure of Z38 and show that it adopts a tertiary structure remarkably similar to that observed for the first two helices of B-domain in the B-domain/Fc complex [Deisenhofer, J. (1981) Biochemistry 20, 2361–2370], although it is significantly less stable. Based on this structure, we have improved on Z38 by designing a 34-residue disulfide-bonded variant (Z34C) that has dramatically enhanced stability and binds IgG with 9-fold higher affinity. The improved stability of Z34C led to NMR spectra with much greater chemical shift dispersion, resulting in a more precisely determined structure. Z34C, like Z38, has a structure virtually identical to the equivalent region from native protein A domains. The well-defined hydrophobic core of Z34C reveals key structural features that have evolved in this small, functional domain. Thus, the stabilized two-helix peptide, about half the size and having one-third of the remaining residues altered, accurately mimics both the structure and function of the native domain.
Keywords: protein A, minimization, NMR, protein design
Protein domains are independent folding units considered to be the minimal length polypeptide sequence necessary to dictate a particular three-dimensional structure. Monomeric, structured protein domains smaller than 50 residues that lack metal binding sites and disulfides are extremely rare in nature; two that have been described previously are a 35-residue subdomain within the villin headpiece (1) and the 43-residue peripheral subunit-binding domain of dihydrolipoamide acetyltransferase from pyruvate dehydrogenase (2).
We are interested in minimizing natural protein domains to small, structured peptides that maintain the native domain’s function. Such peptides provide important structural and functional information and could serve as useful synthetic templates for protein and drug design. The IgG binding domains of protein A consist of ≈60 residues that form three-helix bundle structures (3–5). Even though only the first two helices make relevant contact with Fc as seen in the crystal structure of the B-domain/Fc complex (6), the third helix is essential for domain structure and function (7). Recently we have shown that a domain from protein A could be “minimized.” A truncated fragment containing residues from only the first two α-helices was subjected to multiple rounds of amino acid randomization and selection using phage display methods (7). Ultimately, a 38-residue sequence was identified that binds IgG >10,000-fold tighter than the starting two-helix template. This variant, called Z38, contains 13 substitutions and binds with affinity only 10-fold weaker than the native 3-helix domain.
The hydrophobic residues at the interface of the B-domain/Fc complex were conserved throughout the phage selection, suggesting the minimized variant would bind Fc in essentially the same way as the native three-helix domain. Thus, the majority of selected mutations presumably influence structural stability. Previously, circular dichroism (CD) measurements demonstrated that these mutations progressively induced more helical structure in the peptide. To further explore the role of selected residues and to understand better how the minimized variant has “evolved” high affinity Fc binding ability, we determined the solution structure of Z38. We show that Z38 folds independently into a structure essentially indistinguishable from that observed for helices 1 and 2 of B-domain in the B-domain/Fc complex. In addition we have used the structure of Z38 to design a disulfide-bonded variant, Z34C, with dramatically enhanced stability and increased Fc binding affinity. The solution structures of Z38 and Z34C are presented and compared with previously determined structures for two native protein A domains: the E-domain free in solution (4) and the B-domain in complex with Fc (6).
MATERIALS AND METHODS
Peptide Synthesis.
Z38 and Z34C were prepared using an Applied Biosystems peptide synthesizer (model 431A) with standard N-fluorenylmethoxycarbonyl chemistry on Wang resin. The sulfur groups of the cysteine residues in Z34C were introduced as the p-methoxybenzyl derivative. The N and C termini of both Z38 and Z34C were not blocked. Peptides were cleaved from the resin using 5% triethylsilane in trifluoroacetic acid and purified using reverse-phase HPLC eluting with a water/acetonitrile gradient containing 0.1% trifluoroacetic acid. Z34C was initially purified as the p-methoxybenzyl derivative, then treated with hydrofluoric acid at 0°C for 1 h, oxidized using aqueous potassium ferricyanide at pH 8.5, and further purified by HPLC. Peptides were characterized by electrospray mass spectrometry and quantitative amino acid analysis. Z-domain was prepared as described (7).
Circular Dichroism.
CD spectra were recorded on an Aviv Associates (Lakewood, NJ) model 60DS spectropolarimeter in the wavelength range of 190–250 nm as the average of three scans in 0.2-nm increments with 10-s averaging times at 8 ± 0.5°C in 25 mM Tris (pH 7.5) in a thermostatted circular cuvette with a 0.050-cm path length. Thermal denaturation was measured by monitoring the change in signal at 222 nm over a temperature range of 6–90°C. Peptide concentration was 10 μM in 25 mM Tris (pH 7.5). Measurements were made in one degree increments with 20-s equilibration time and 20-s averaging time at each increment. Denaturation was fully reversible as determined by comparing spectra before and after heating.
NMR Spectroscopy.
NMR samples were prepared by dissolving lyophilized peptide in 50 mM sodium acetate-d3 (pH 5.1) 0.1 mM 3-(trimethylsilyl)-1-propane-1,1,2,2,3,3-d6 sulfonic acid (DSS) to give a peptide concentration of 1–3 mM. All spectra were acquired on a Bruker (Billerica, MA) AMX-500 spectrometer at 8°C. Standard pulse sequences and phase cycling were employed to record two-dimensional (2D) correlated spectroscopy (COSY), total correlation spectroscopy (TOCSY) (80-ms τm), and nuclear Overhauser effect spectroscopy (NOESY) (Z38: 150-ms τm; Z34C: 75-, 100-, 150-, and 200-ms τm) spectra on both Z38 and Z34C in 92% H2O/8% D2O solution essentially as described (4). In addition, COSY-35, double quantum, and NOESY (Z38: 150-ms τm; Z34C: 75-, 150-, and 200-ms τm) spectra were recorded in 99.99% D2O solution. Amide protons protected from solvent exchange were identified by running 2D TOCSY spectra (24-min acquisition time, 45-ms τm) on a sample that had been lyophilized from H2O solution and resuspended in D2O. Chemical shifts were referenced to internal DSS at 0.0 ppm.
Distance and Dihedral Restraints.
Assigned NOE crosspeaks from the 150-ms NOESY spectra acquired in H2O and D2O were characterized as strong, medium, or weak, corresponding to upper bound distance restraints of 3.3, 3.9, and 5.0 Å, respectively. Treatment of overlapped crosspeaks, application of pseudoatom corrections and use of 〈r−6〉 averaging were as described (4).
3JHN-Hα and 3JHα-Hβ coupling constants were measured as described (4). φ angle restraints of −90° < φ < −40° and −150° < φ < −90° were imposed for residues with 3JHN-Hα < 6.0 Hz and > 8.5 Hz, respectively. For residues in which 3JHN-Hα could not be measured or was in the range of 6.0–8.5 Hz, φ was restrained to be negative if the intraresidue Hα–HN NOE was less intense than the sequential Hα–HN NOE (8). χ1 restraints, stereospecific assignments for β-methylene groups, and hydrogen bond restraints were determined as described (4).
Structure Calculations.
Distance geometry calculations using the program dgii (9) within insight ii and restrained molecular dynamics using discover (Molecular Simulations, Waltham, MA) and the all-atom amber force field were carried out as described (4). The NOE distance restraints, hydrogen bond distance restraints, and dihedral angle restraints were applied using a square-well potential function (10) with force constants of 25 kcal⋅mol−1⋅Å−2, 25 kcal⋅mol−1⋅Å−2, and 100 kcal⋅mol−1⋅rad−2, respectively. Additional ω restraints (−170° < ω < 170°, force constant 200 kcal⋅mol−1⋅rad−2) were included as a supplement to the force field to maintain peptide planarity, and were not included in the violation analysis given in Table 1.
Table 1.
Z38 and Z34C structural statistics
| Z38 | Z38mean* | Z34C | Z34Cmean* | |
|---|---|---|---|---|
| rms deviation from exptl distance restraints, Å | ||||
| NOE (228, 328)† | 0.011 ± 0.003 | 0.010 | 0.006 ± 0.002 | 0.004 |
| H-bond (16, 32) | 0.002 ± 0.005 | 0.000 | 0.000 ± 0.000 | 0.000 |
| rms deviation from experimental dihedral restraints, ° | ||||
| φ (11, 18), χ1 (3, 10) | 0.01 ± 0.03 | 0.24 | 0.24 ± 0.08 | 0.33 |
| NOE distance violations | ||||
| 0.1 Å > no. > 0.01 Å | 11.8 ± 2.8 | 14 | 9.0 ± 2.8 | 6 |
| No. > 0.1 Å | 0.5 ± 0.7 | 0 | 0.0 ± 0.2 | 0 |
| Maximum, Å | 0.10 ±0.03 | 0.09 | 0.06 ± 0.02 | 0.05 |
| Dihedral angle violations | ||||
| 1° > no. > 0.1° | 0.2 ± 0.4 | 1 | 2.3 ± 0.9 | 2 |
| No. > 1° | 0.0 ± 0.0 | 0 | 0.6 ± 0.7 | 1 |
| Maximum, ° | 0.0 ± 0.1 | 0.9 | 1.0 ± 0.3 | 1.6 |
| rms deviations from idealized covalent geometry‡ | ||||
| Bonds, Å (634, 580) | 0.0050 ± 0.0001 | 0.0051 | 0.0049 ± 0.0001 | 0.0048 |
| Angles, ° (1135, 1041) | 1.43 ± 0.05 | 1.45 | 1.36 ± 0.05 | 1.27 |
| Planes, ° (131, 120) | 1.81 ± 0.19 | 1.74 | 1.44 ± 0.16 | 1.64 |
| Energies (kcal⋅mol−1) | ||||
| Erestraint§ | 0.76 ± 0.31 | 0.58 | 0.34 ± 0.16 | 0.24 |
| Ebond | 5.36 ± 0.25 | 5.50 | 4.70 ± 0.16 | 4.42 |
| Eangle | 31.4 ± 2.1 | 32.5 | 25.8 ± 2.1 | 22.6 |
| Etorsion | 34.3 ± 2.1 | 35.9 | 35.1 ± 2.0 | 34.0 |
| Eplane | 0.78 ± 0.15 | 0.74 | 0.50 ± 0.09 | 0.62 |
| EvdW¶ | −132.3 ± 5.3 | −136.6 | −132.1 ± 4.2 | −136.5 |
| Eelec‖ | −121.1 ± 5.0 | −129.4 | −111.2 ± 2.2 | −112.0 |
| Etotal | −199.4 ± 7.3 | −211.0 | −195.1 ± 6.0 | −204.8 |
| Stereochemical quality,** % | ||||
| Most favored | 75.3 ± 7.3 | 91.4 | 89.6 ± 5.0 | 87.1 |
| Acceptable | 23.2 ± 6.5 | 5.7 | 9.0 ± 4.9 | 12.9 |
| Generous | 2.6 ± 2.6 | 2.9 | 0.7 ± 1.3 | 0.0 |
| Disallowed | 1.9 ± 2.0 | 0.0 | 0.7 ± 1.3 | 0.0 |
| rms deviations to mean coordinates, Å | ||||
| Residues 10–36 | ||||
| Backbone (N, Cα, C) | 1.00 ± 0.16 | 0.87 | 0.38 ± 0.09 | 0.38 |
| Heavy | 1.73 ± 0.17 | 1.26 | 1.09 ± 0.12 | 0.94 |
| Residues 6–39 | ||||
| Backbone | 0.54 ± 0.09 | 0.58 | ||
| Heavy | 1.16 ± 0.12 | 1.04 |
The minimized mean is the structure obtained by restrained minimization of the mean coordinates of the ensemble calculated using a best fit superposition of residues 10–36.
Relevant numbers are listed in order for Z38 then Z34C throughout the table.
Idealized covalent geometry is defined by the amber force field as implemented within discover.
The final values of the square-well NOE and dihedral angle potentials are calculated with force constants of 25 kcal⋅mol−1⋅Å−2, and 100 kcal⋅mol−1⋅rad−2, respectively.
EvdW is the Lennard–Jones van der Waals energy calculated with the all-atom amber force field and a 10-Å cutoff.
Eelec is calculated using a distance-dependent dielectric constant, ɛ = 4r.
Percent of residues found in a given region of φψ space as defined in procheck (17).
Binding Measurements.
Association and dissociation rate constants were measured by surface plasmon resonance using a BIAcore 1000 (Pharmacia Biosensor) at a flow rate of 25 μl per min. A monoclonal IgG1 was immobilized on the biosensor chip nonspecifically by N-hydroxysuccinimide ester coupling. Dissociation rate constants were measured on a flowcell derivatized at a low coupling density of 4,700 RU to minimize rebinding effects, using 30-μl injections of 25-μM ligand. Residual plots of the curve fits were within ± 1 RU. Association rate constants were measured on a flowcell derivatized with 7,500 RU of IgG1 using 30-μl injections of either 0.5-μM or 1.0-μM ligand. Measurement of koff for peptides Z38 and Z34C is influenced by rebinding effects; the values reported are equivalent to the values obtained when free IgG is injected at the end of the peptide injection to prevent rebinding of peptide. The kon and koff values reported for Z38 differ slightly from those reported previously (7); this difference is attributed to the difficulties associated with measuring fast koff values, and the values contained herein are to be considered more accurate and are certainly accurate for comparison of relative values for Z38, Z34C, and full-length Z-domain.
RESULTS
Structure of Z38.
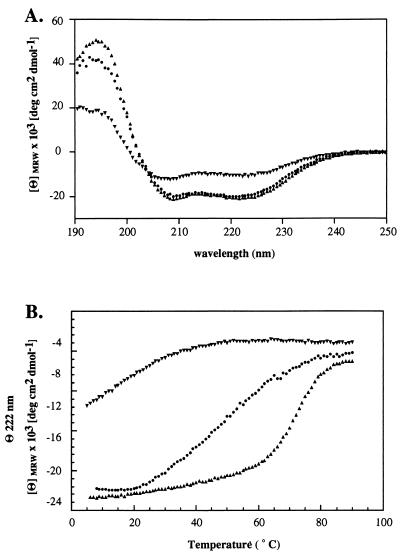
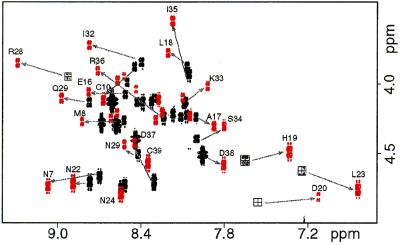
The CD spectrum of Z38 has significant α-helical character with two minima centered at 208 and 222 nm (Fig. 1A). The NMR spectrum is reasonably well dispersed (Figs. 2 and 3A), and complete 1H resonance assignments were obtained by standard 2D NMR methods (11). A total of 228 NOE-derived distance restraints nonredundant with covalent geometry [including 61 medium-range (1 < ‖i − j‖ < 5) and 25 long-range (‖i − j‖ > 5)] were obtained from analysis of 2D NOESY spectra. Backbone 3JHN-Hα coupling constants were measured for 22 resolved HN–Hα crosspeaks out of the 36 total; however, all had values between 6 and 8 Hz indicating significant conformational averaging was occurring under the experimental conditions used. Side chain 3JHα-Hβ coupling constants were measured for 8 of the 30 β-methylene containing residues from which 3 χ1 dihedral angle restraints were derived and 2 β-methylene groups were stereospecifically assigned. Eleven amide protons with slowed proton/deuterium exchange rates were identified at 8°C (pH 4.1). Thus, structures were calculated from a total of 228 NOE-derived distance restraints, 11 negative φ and 3 χ1 dihedral angle restraints, and 16 restraints for 8 α-helical H-bonds totaling 6.8 restraints per residue.
Figure 1.
(A) CD spectra at 8°C (pH 7.5) and (B) temperature dependence of the ellipticity at 222 nm (θ222) for Z-domain (▴), Z38 (▾), and Z34C (•).
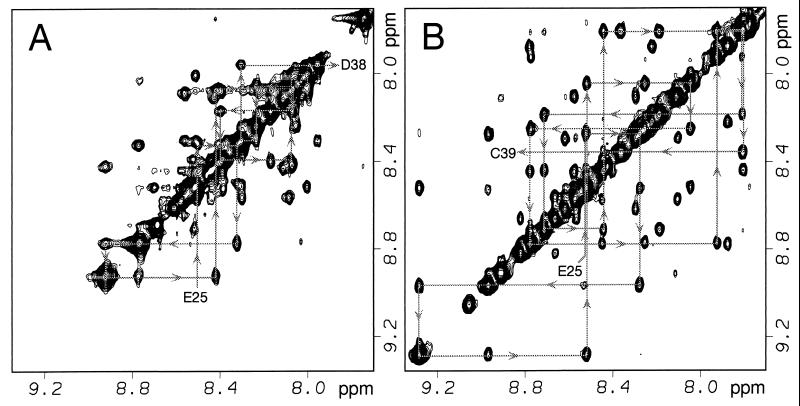
Figure 2.
Overlay of the fingerprint region of COSY spectra for Z38 (black) and Z34C (red). Spectra were acquired under identical conditions at 8°C (pH 5.1). Selected crosspeaks are labeled for Z34C; arrows show the location of the same crosspeak in Z38. Complete 1H resonance assignments are available from the authors upon request.
Figure 3.
Amide region of the 2D NOESY spectra of (A) Z38 and (B) Z34C acquired with a 150-ms mixing time. NOEs involving neighboring amide protons from residues in helix-2 are indicated.
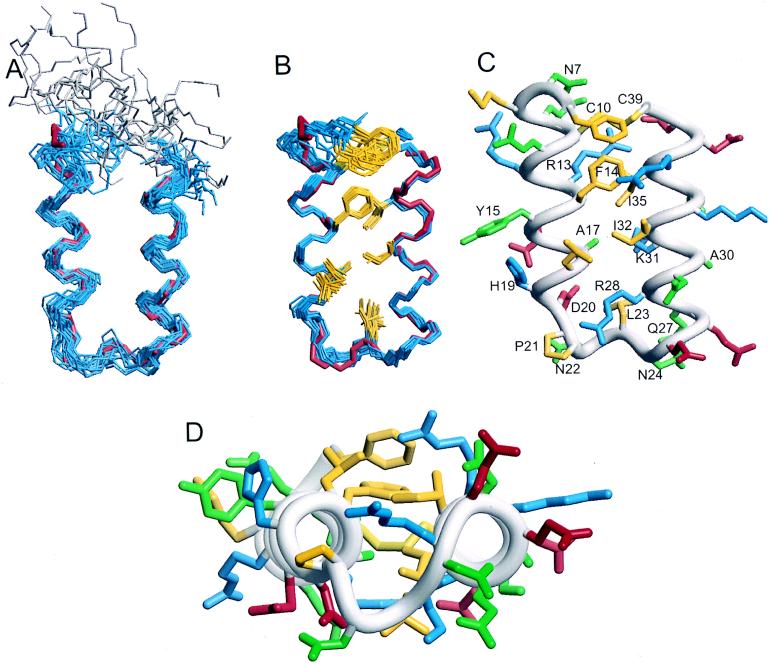
The Z38 structure ensemble is shown in Fig. 4A. The structures satisfy the input data very well as evident from the low restraint violation energy and no structure having any distance or dihedral angle violation greater than 0.14 Å or 0.3°, respectively (Table 1). The precision is limited by the low number of input restraints; nevertheless backbone atomic RMSDs for residues 10–36 are 1.00 ± 0.16 Å. Thus, there are clearly sufficient NOEs to define a unique tertiary fold. Z38 is composed of two antiparallel α-helices connected by a turn; the N-terminal 5 residues are disordered. Comparison with the crystal structure of B-domain bound to Fc indicates that although this molecule is 21 residues shorter and 13 of the remaining 38 residues are different, the backbone structure of residues 10–36 is indistinguishable within the precision of the structure determinations (Fig. 4A).
Figure 4.
NMR structure ensembles for (A) Z38 and (B) Z34C. In each case, 24 models are shown aligned using the backbone atoms of residues 10–36. Shown also are the coordinates for residues 7–38 from the crystal structure of the B-domain/Fc complex (red) (6). The disordered N-terminal residues 1–5 from Z38 are colored gray. Side chains from the hydrophobic core of Z34C are shown (Phe-6, Cys-10, Phe-14, Ala-17, Leu-18, Leu-23, Ile-32, Ile-35, and Cys-39). (C) The minimized mean structure of Z34C showing all side chains colored by amino acid type: hydrophobic (yellow), positively charged (blue), negatively charged (red), and polar (green); only those that are labeled are well defined in the NMR ensemble with χ1 angular order parameters >0.9. Note that only the χ1 = −60° orientation for Phe-14 is shown, but this side chain also exists in the χ1 = 180° conformation in solution. (D) Same as C, but rotated 90° with the Fc binding surface on top.
Z34C Design.
Although Z38 appears to have a unique fold, the structure is not very stable. The temperature dependence of the CD spectrum indicates that even at 8°C, where the NMR data were collected, Z38 is undergoing thermal denaturation (Fig. 1B). In an effort to stabilize the inherent structure of Z38, Cα–Cα and Cβ–Cβ distances were evaluated to identify locations suited to the introduction of a disulfide bond that would not significantly perturb the structure (12). The disulfide was positioned as close to the ends of the peptide as possible to provide maximum stabilization due to entropic effects on the denatured state (13). A mutant containing a disulfide between an added cysteine at the C terminus (which is proline in the native sequence) and a cysteine substituting for Gln-10 was predicted to maintain the same tertiary fold as seen for this region in the B-domain/Fc crystal structure. In addition, five residues that were shown previously to be unnecessary for binding (7) and unstructured by NMR were removed from the N terminus. Thus, Z34C contains 34 residues beginning with Phe-6 and ending with Cys-39.
The temperature dependence of the CD spectrum for Z34C is shown in Fig. 1B. The disulfide leads to a dramatic gain in thermal stability resulting in a greater than 40°C change in midpoint of the thermal transition. The temperature dependence of the CD signal for Z34C displays a more shallow transition than that for native Z-domain, suggesting that denaturation of Z34C is less cooperative than for the native domain; monitoring thermal denaturation by NMR chemical shift perturbation shows a similarly shaped curve for Z34C, although the range of temperatures that could be studied was smaller (data not shown).
A surprisingly large increase in chemical shift dispersion is observed for Z34C compared with Z38 (Figs. 2 and 3). The majority of amide and alpha proton resonances move away from random coil positions and undergo changes in chemical shift as large as 0.49 and 0.37 ppm for HN and Hα, respectively. The greater stability is not only evidenced by CD and chemical shifts, but also in backbone coupling constants and hydrogen/deuterium exchange behavior; Z34C has 14 3JHN-Hα coupling constants with values less than 6 Hz and 25 amide protons protected from solvent exchange at 8°C (pH 6.3). The greater stability and chemical shift dispersion allowed for nearly double the number of NMR structural restraints to be obtained for Z34C compared with that obtained for Z38.
Structure of Z34C.
Complete proton resonance assignments for Z34C were obtained and structures were calculated from a total of 328 (90 medium-range and 63 long-range) NOE-derived distance restraints, 18 φ and 10 χ1 dihedral angle restraints, and 32 H-bond restraints for 16 α-helical H-bonds totaling 11.4 restraints per residue. The resulting ensemble satisfies the experimental restraints very well, having no distance or dihedral angle violations greater than 0.10 Å or 1.6°, respectively. The final ensemble of 24 structures (Fig. 4B) has good geometry with 90% of the residues in the most favored region of the Ramachandran plot (Table 1). The ensemble is of high precision with a backbone atomic RMSD for residues 10–36 of 0.36 ± 0.09 Å. Unlike Z38, the ends of the molecule are ordered with a backbone atomic RMSD for all residues of 0.54 ± 0.09 Å.
Binding Properties.
The binding kinetics of Z34C were evaluated by surface plasmon resonance (Table 2). Not only is Z34C more stable than Z38, but its affinity for IgG is 9-fold greater than that for Z38. This improvement in affinity is predominately manifested in the association rate of Z34C, which is approximately 6-fold faster than that of Z38. The improved structural stability apparently results in Z34C being better ordered for binding and hence promotes faster binding.
Table 2.
Binding kinetics for Z38 and Z34C
| koff, ×10−3 (s−1) | kon, ×105 (M−1⋅s−1) | Kd (nM) | |
|---|---|---|---|
| Z38 | 40.6 ± 0.8 | 2.20 ± 0.18 | 185 ± 15 |
| Z34C | 30.2 ± 0.2 | 15.1 ± 0.6 | 20.0 ± 0.8 |
| Z-domain | 6.81 ± 0.01 | 4.56 ± 0.02 | 14.9 ± 0.1 |
Surface plasmon resonance is described in Materials and Methods. Errors reflect the standard deviations from two individual measurements using the same biosensor chip.
The 20 nM dissociation constant for Z34C binding to IgG is essentially identical to that measured for the native three-helix domain. However, Z34C and Z-domain differ in their relative association and dissociation rates, such that the faster dissociation rate for Z34C is compensated by a similarly faster association rate.
DISCUSSION
Description of the Structure.
We have determined the solution structure for two minimized protein A domain variants, Z38 and Z34C. Both peptides are highly soluble and monomeric in solution as determined by equilibrium ultracentrifugation (data not shown). The two peptides were found to share very similar structures composed of two antiparallel α-helices.
The NOEs observed for Z38 define a specific alignment and orientation for the two α-helices in this peptide generating a reasonably well-defined structure ensemble that satisfies the NOE distance restraints (Fig. 4A and Table 1). However, this ensemble of 24 structures does not fully represent the range of conformations explored by this peptide in solution. If it did, we would expect the values of the backbone coupling constants for the helical residues to be much lower (≈5 Hz) than was observed (>6 Hz). Therefore, this ensemble should be considered to represent an average conformation of Z38 in solution, with the understanding that this molecule is dynamic and undergoing denaturation under these conditions (Fig. 1B).
The disulfide in Z34C does not appear to perturb the structure, but rather stabilizes the inherent structure of Z38. Because Z38 is less well defined, we will focus detailed discussion on the Z34C structure. Z34C is composed of two α-helices, helix-1 (residues 8–18) and helix-2 (residues 25–39), connected by a type I turn (residues 20–23). Nineteen of the 34 residues have well-defined side chain conformations with χ1 angular order parameters > 0.9 (Fig. 4C) (14).
Hydrophobic side chains (Cys-10, Phe-14, Ile-32, Ile-35, Cys-39) line the interface between the two α-helices (Fig. 4 B and C). Interestingly, three residues that were chosen during the phage selection, Arg-13, Lys-31, and Arg-36, pack along the outer side of this “core.” These residues effectively perform two functions, both increasing the extent of hydrophobic interactions through the aliphatic portion of their side chains while at the same time providing solubilizing charges. Arg-28 appears to have a similar role and was conserved from the native domain. The N-cap of helix-2 was also conserved: the amide proton of Asn-24 H-bonds to the side chain Oɛ1 of Gln-27 in 7 of 24 structures and the amide proton of Gln-27 H-bonds to the side chain Oδ1 of Asn-24 in 16 of 24 structures.
Introduction of the disulfide in Z34C had a surprisingly large effect on the stability of the minidomain (Fig. 1). The disulfide closes a 30-residue loop which undoubtedly leads to a significant decrease in the entropy of the denatured state (13). Furthermore, having one of the cysteines located at the end of the molecule limits any strain that might be induced by the strict geometric constraints of the disulfide bond. Additional stabilization could result from hydrophobic interactions between the disulfide and Phe-6 at the N terminus, although the exact positioning of this side chain is not well defined (Fig. 4B).
Comparison with Other Protein A Domain Structures.
Z38 and Z34C have structures that compare very well with the solution structure of E-domain (4), a native three-helix protein A domain. Backbone atomic RMSDs for residues 11–36 from the mean coordinates for E-domain are 1.3 ± 0.3 and 0.79 ± 0.09 Å for Z38 and Z34C, respectively. Thus, within the precision of the structure determinations, Z38 and Z34C adopt backbone structures indistinguishable from E-domain in solution. Furthermore, seven of eight side chains that are well defined and conserved in both E-domain and Z34C have similar orientations.
Surprisingly, Phe-14 of Z34C appears to have conserved the property of occupying more than one conformation in solution as was seen for E-domain. The analogous phenylalanine side chain of E-domain was previously shown to exist in a 2:1 ratio of −60° and 180° χ1 rotamers (4). In the complex with Fc, this residue is fully buried in the Fc interface and only the 180° rotamer is compatible with Fc binding; the −60° rotamer would sterically clash with Ile-253 of the Fc. Thus the majority of molecules must reorient Phe-14 upon binding. Phe-14 of Z34C has 3JHα-Hβ coupling constants that are inconsistent with a single conformation in solution (6.1 and 9.3 Hz). Assuming that the coupling constants represent a population average of a distribution of the three staggered rotamers indicates that Phe-14 populates both −60° and 180° χ1 rotamers (15). The pattern of NOEs between Phe-14 and Leu-18, Ile-32, and Ile-35 are similar to those observed for the E-domain and similarly, cannot all be satisfied by a single χ1 orientation of Phe-14 (4). The ensemble shown in Fig. 4 has Phe-14 exclusively in the −60° rotamer for simplicity; two NOEs between Phe-14 and Leu-18 were removed from the calculation generating this ensemble as those distances are only satisfied in the 180° χ1 conformer. Thus, the population of molecules having Phe-14 in the −60° χ1 orientation must undergo the same kind of conformational change as was invoked for three-helix E-domain upon Fc binding.
The solution structures of uncomplexed Z38 and Z34C are also nearly indistinguishable from the Fc-bound structure of B-domain (Fig. 4 A and B) with backbone atomic RMSDs for residues 11–36 of 1.1 ± 0.2 and 0.77 ± 0.10 Å, respectively. The only significant difference is in the orientation of the side chain of Phe-14 where it is found exclusively in the 180° χ1 rotamer in the Fc-bound state as discussed above. Thus, phage selection resulted in the identification of a sequence that folds into a two-helix structure indistinguishable from that region of a three-helix domain.
The introduction of the disulfide not only increased the stability of Z34C relative to Z38, but also the IgG binding affinity. The fact that the improvement in binding comes about largely from a faster association rate is consistent with the disulfide having stabilized a structure very similar to that seen in the Fc-bound state of B-domain and thereby minimizing entropic losses upon binding. Furthermore, the association rate of Z34C is about 3-fold faster than that of native three-helix Z-domain. A slightly faster association rate is expected due to the smaller size of this peptide; possibly a more facile Phe-14 reorientation in Z34C compared with the native domain also contributes to the faster association rate.
Z38 was selected based purely on binding affinity, yet structural stability increased concomitantly. Further improvements to generate Z34C created a novel stable antiparallel helix scaffold with functional activity. Z34C represents one of the smallest functional peptides known that exhibits features typical of a folded protein; it is composed of regular secondary structural elements, has a well-defined hydrophobic core protected by polar and charged residues, and displays a flat surface for binding. Interestingly, Z34C does not show ideal interhelical packing: the interhelical angle (−175 ± 4°) is not optimal (16), and the hydrophobic residues are clustered on one edge of the helical interface instead of packing tightly between the two helices (Fig. 4D). These are characteristics that one might try to avoid when designing a stable structure, yet are essential to the function of Z38 and Z34C. These results emphasize the challenge of designing a functional, stably folded peptide and the importance of diversity techniques such as phage display for surmounting these difficulties.
In summary, we have determined the structure of a variant protein A domain that is about half the size but maintains the same structure and function of the native molecule. Furthermore, we have improved on the minidomain selected by phage display by shortening the N terminus and introducing a disulfide bond to stabilize the inherent structure. The disulfide-bonded variant Z34C has structure, stability, and binding affinity comparable to the native three-helix domain. In using phage display to select for binding activity, we have effectively selected for a stable structure in a novel context. In the process we have created a stable antiparallel α-helix scaffold that could be useful as a starting template in the design of other novel binding peptides.
Acknowledgments
We thank Nicholas Skelton and Wayne Fairbrother for many valuable discussions and critical reading of the manuscript and Jun Liu for ultracentrifugation measurements.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: 2D, two dimensional; COSY, correlated spectroscopy; NOESY, nuclear Overhauser effect spectroscopy.
Data deposition: The atomic coordinates for the ensembles and minimized mean structures have been deposited in the Protein Data Bank, Biochemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (references 1Zda and 1Zdb for Z38 and 1Zdc and 1Zdd for Z34c).
References
- 1.McKnight C J, Doering D S, Matsudaira P T, Kim P S. J Mol Biol. 1996;260:126–134. doi: 10.1006/jmbi.1996.0387. [DOI] [PubMed] [Google Scholar]
- 2.Kalia Y N, Brocklehurst S M, Hipps D S, Appella E, Sakaguchi K, Perham R N. J Mol Biol. 1993;230:323–341. doi: 10.1006/jmbi.1993.1145. [DOI] [PubMed] [Google Scholar]
- 3.Gouda H, Torigoe H, Saito A, Sato M, Arata Y, Shimada I. Biochemistry. 1992;31:9665–9672. doi: 10.1021/bi00155a020. [DOI] [PubMed] [Google Scholar]
- 4.Starovasnik M A, Skelton N J, O’Connell M P, Kelley R F, Reilly D, Fairbrother W J. Biochemistry. 1996;35:15558–15569. doi: 10.1021/bi961409x. [DOI] [PubMed] [Google Scholar]
- 5.Jendeberg L, Tashiro M, Tejero R, Lyons B A, Uhlen M, Montelione G T, Nilsson B. Biochemistry. 1996;35:22–31. doi: 10.1021/bi9512814. [DOI] [PubMed] [Google Scholar]
- 6.Deisenhofer J. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 7.Braisted A C, Wells J A. Proc Natl Acad Sci USA. 1996;93:5688–5692. doi: 10.1073/pnas.93.12.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clubb R T, Ferguson S B, Walsh C T, Wagner G. Biochemistry. 1994;33:2761–2772. doi: 10.1021/bi00176a004. [DOI] [PubMed] [Google Scholar]
- 9.Havel T F. Prog Biophys Mol Biol. 1991;56:43–78. doi: 10.1016/0079-6107(91)90007-f. [DOI] [PubMed] [Google Scholar]
- 10.Kessler H, Griesinger C, Lautz J, Müller A, van Gunsteren W F, Berendsen H J C. J Am Chem Soc. 1988;110:3393–3396. [Google Scholar]
- 11.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 12.Srinivasan N, Sowdhamini R, Ramakrishnan C, Balaram P. Int J Pept Protein Res. 1990;36:147–155. doi: 10.1111/j.1399-3011.1990.tb00958.x. [DOI] [PubMed] [Google Scholar]
- 13.Matsumura M, Becktel W J, Levitt M, Matthews B W. Proc Natl Acad Sci USA. 1989;86:6562–6566. doi: 10.1073/pnas.86.17.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyberts S G, Goldberg M S, Havel T F, Wagner G. Protein Sci. 1992;1:736–751. doi: 10.1002/pro.5560010606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kessler H, Griesinger C, Kerssebaum R, Wagner K, Ernst R R. J Am Chem Soc. 1987;109:607–608. [Google Scholar]
- 16.Chothia C, Levitt M, Richardson D. J Mol Biol. 1981;145:215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]
- 17.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]