Abstract
Intestinal colonic crypts are derived from a stem cell population located at the base of each crypt. A new analysis of mitochondrial function and of the rates of mitochondrial DNA (mtDNA) mutation in individual crypts shows that mtDNA mutations arise in stem cells — and at a surprisingly high frequency. Because crypts turn over extremely rapidly (about once per week), somatic mtDNA mutations can “take over the system” and even become homoplasmic, in a manner similar to what has been shown to occur in tumors.
Stem cells are the progenitors of specific cell lineages that become the body’s organs and tissues during embryonic development. After birth, however, stem cells continue to play an equally important role in tissue maintenance, as they are called upon to repopulate cells that turn over constantly. Hematopoietic stem cells were among the earliest identified exemplars of this role, but stem cells exist even in long-lived tissues — for example, muscle “satellite” cells — and, with the discovery in the last few years of stem cell lineages in brain and heart, our whole view of the idea of a “terminally differentiated” tissue has undergone a complete overhaul.
Mitochondrial dysfunction in stem cells
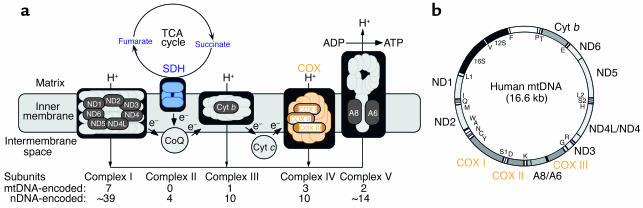
Mitochondria are semiautonomous organelles that are present in essentially all cells of the body. They contain their own DNA (mtDNA) and are the seat of a number of important housekeeping functions. Foremost among these is the production of energy via the respiratory chain and oxidative phosphorylation, an intricate system composed of five complexes and two electron carriers (Figure 1a). The mtDNA (Figure 1b), a tiny 16.6 kb maternally inherited circular genome present in multiple copies in each organelle (there are about 10,000 mtDNAs in a typical cell), encodes 2 rRNAs, 22 tRNAs, and only 13 polypeptides, all of which are subunits of the respiratory complexes. In the last 15 years, mutations in mtDNA, all of which impair oxidative energy metabolism, have been found to cause a wide spectrum of disorders (1). In these patients, the mutations are typically heteroplasmic; that is, mutated mtDNAs coexist with wild-type mtDNAs in varying proportions, resulting in a mosaic pattern of respiratorily competent and incompetent cells. Respiratorily deficient cells must typically contain at least 80% mutated mtDNA to initiate dysfunction.
Figure 1.
(a) The respiratory chain. Nuclear DNA-encoded subunits are light gray; mtDNA-encoded subunits (see panel b) are dark gray. (b) Map of the human mitochondrial genome. Polypeptide-coding gene products (outside the circle) specify 7 subunits of NADH dehydrogenase-CoQ oxidoreductase (ND), 1 subunit of CoQ-cytochrome b oxidoreducase (Cyt b), 3 subunits of COX, and 2 subunits of ATP synthase (A). Protein synthesis gene products (inside the circle) specify 12S and 16S rRNAs, and 22 tRNAs (one-letter code). Figure modified from Schon and Manfredi (10).
Heteroplasmic populations of mtDNA mutations can also arise randomly in somatic cells and can accumulate at low levels in individual cells during the course of normal aging (2). Even more intriguingly, somatic mtDNA mutations arise and are amplified in solid tumors, such as colon cancers (3), although a causative relationship between mtDNA mutations and tumorigenesis has not yet been established.
Mitochondria in every cell, even those that do not divide, turn over, because they replicate their DNA and divide independently of the cell cycle. In a sense, then, as befitting an organelle that is derived evolutionarily from bacteria, mitochondria are each cell’s own “stem” population, continually dividing and replacing themselves within their “hosts.”
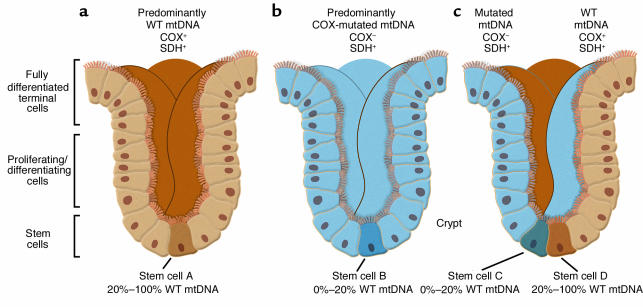
If mtDNA is always replicating within cells, it stands to reason that somatic mtDNA mutations could arise in stem cells as well, but the confirmation of this supposition, both qualitatively (does it happen?) and quantitatively (at what rate?), has been lacking. This question is not an academic one, as it goes to the heart of issues relating to the accumulation of mutated mtDNAs in disease (especially in the brain, a particularly susceptible tissue), in aging, and in tumorigenesis. It is also a hard question to answer for the simple reason that it has been extremely difficult to identify a stem cell population amenable to be studied easily and in sufficient quantity — until now. In this issue of the JCI, Taylor et al. (4) provide convincing evidence that at least one population of stem cells — those giving rise to intestinal colonic crypts — do indeed harbor somatic mtDNA mutations, which, even more surprisingly, arise at a relatively high frequency. The decision to examine colonic crypts may have been inspired by earlier work on the segregation of mtDNA haplotypes in “transmitochondrial mice” by Shoubridge’s group (5). The advantage of studying a colonic crypt is that it is a macroscopically observable clonal population of cells derived from one or two single cells — the stem cells — located at the base of each crypt (Figure 2). Thus, any mtDNA mutation found in the crypt in toto must perforce have been amplified from that very mutation in the stem cell itself, as the crypt is the surrogate of the stem cell.
Figure 2.
Two-color histochemistry for SDH and COX reveals three types of crypts, with normal (a), deficient (b), and “mosaic” (c) patterns of mitochondrial function, reflecting the phenotype in the stem cells that gave rise to each type of crypt.
The assumptions underlying the approach of Taylor et al. (4) were based on a simple syllogism: if (i) a stem cell harbors an mtDNA mutation that disrupts respiratory chain function and if (ii) that mutation expands (in the stem cell) to a level exceeding the threshold for dysfunction (typically >80% mutated mtDNA), then (iii) one ought to be able to observe both the dysfunction (by histochemical and/or biochemical means) and the mtDNA mutation (by genetic means) in the daughter population comprising the entire crypt. In order to assess function, Taylor et al. (4) applied a powerful method of mitochondrial analysis on serial transverse sections from individual human crypts: two-color histochemistry (6) to detect simultaneously the enzymatic activities of cytochrome c oxidase (COX; complex IV of the respiratory chain), which contains both mtDNA- and nuclear DNA (nDNA)-encoded subunits; and succinate dehydrogenase (SDH; complex II of the respiratory chain), which contains only nDNA-encoded subunits. Importantly, when stained for both COX (brown) and SDH (blue) simultaneously, crypts with normal COX activity stained brown (the blue was hidden by the brown stain), whereas those with impaired COX activity stained blue (the brown was absent, revealing the blue). The authors then performed PCR coupled with mtDNA sequencing on the same transverse sections to search for somatic mutations that might correlate with the histochemistry. The two-color approach also allowed them to make a three-dimensional reconstruction of the COX activity in the entire crypt (Figure 2).
What Taylor et al. (4) found was, literally most illuminating. They observed three types of crypts: a majority of all-brown crypts, plus a minority consisting of both all-blue crypts and “mosaic” crypts containing ribbons of brown and blue cells. The first two patterns are consistent with the idea that the crypt has been repopulated from a single stem cell, either COX-normal (i.e., brown) or COX-deficient (blue). The most likely explanation for the third pattern (brown/blue mosaicism) is that at least two stem cells — one COX-normal and the other COX-deficient — were involved in crypt formation. Thus, all three patterns are consistent with the notion that a colonic crypt is indeed a clonal population derived from one or more stem cells.
Upon sequencing the sectioned crypts, they found many crypts with a normal mtDNA genotype (no surprise here), but in many crypts they found numerous mtDNA mutations, both in COX-negative crypts (many, but not all, mutations were in COX or protein synthesis genes) and in some of the ostensibly normal COX-positive crypts (mainly neutral mutations or mutations in non-COX genes). Interestingly, there were some COX-negative crypts in which no mtDNA mutations were found at all — presumably there were nuclear mutations in these stem cells that affected COX function. Overall, the amount of mutated mtDNA was extremely high (on average, one mtDNA point mutation per crypt), and, as has been known for a dozen years now (2), the mutational “load” increased with age.
Using mitochondrial function as a stem cell marker
So what is the significance of this work for “mitochondriacs” and for stem cell biologists? First, we now know that mtDNA mutations can indeed arise in stem cells. Second, by mathematical modeling, Taylor et al. (4) were able to estimate the rate of somatic mtDNA mutation: it is approximately 5 × 10–5 per genome per day, a rate far greater than that of nuclear DNA. Third, the accumulation of mtDNA mutations, often to homoplasmy, in crypt stem cells is highly reminiscent of the dramatic shifts in mtDNA genotypes in solid tumors (3), which, of course, are clonal expansions of a tumor “stem cell.” However, unlike stem cells, tumors are aneuploid. Moreover, they also amplify segments of their chromosomes as “double minutes,” up to 5 Mb in size, that likely contain nuclear-embedded mtDNA pseudogenes. If these are amplified by PCR, pseudogene-derived polymorphisms may be attributed erroneously to mutations in authentic mtDNA.
Finally, the use of mitochondrial markers may allow researchers to track the progeny of multiple stem cells simultaneously. In fact, one can envision the use of COX-negative cells, especially in tissues from aged individuals, as a way to scan or “map” regions that are populated by stem cells. For example, the unusual nonrandom distribution of COX-negative neurons in the hippocampus of brains of the elderly is likely due to random mtDNA somatic mutations arising in ependyma-derived single neuronal stem cells that repopulate groups of COX-negative neurons in the hippocampus (7). Similarly, the accumulation of COX-negative neurons in aging parvocellular, but not magnocellular, neurons of the lateral geniculate nucleus (8) may reflect the fact that a stem cell population exists for the former but not the latter.
Clearly, the use of colonic crypts as a stem cell “model system” allows investigators to address new types of questions in stem cell biology (9). From a mitochondrial standpoint, crypts may be of particular value, especially with regard to studying the mitochondrial population “bottleneck” that occurs during early oogenesis, and in understanding the dynamics of shifts from heteroplasmy to homoplasmy in mitochondrial disease.
Footnotes
See the related article beginning on page 1351.
Conflict of interest: The author has declared that no conflict of interest exists.
Nonstandard abbreviations used: mitochondrial DNA (mtDNA); cytochrome c oxidase (COX); nuclear DNA (nDNA); succinate dehydrogenase (SDH).
References
- 1.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 2.Khrapko K, Nekhaeva E, Kraytsberg Y, Kunz W. Clonal expansions of mitochondrial genomes: implications for in vivo mutational spectra. Mutat. Res. 2003;522:13–19. doi: 10.1016/s0027-5107(02)00306-8. [DOI] [PubMed] [Google Scholar]
- 3.Polyak K, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat. Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 4.Taylor RW, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 2003;112:1351–1360. doi:10.1172/JCI200319435. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat. Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- 6.Bonilla E, et al. New morphological approaches to the study of mitochondrial encephalomyopathies. Brain Pathol. 1992;2:113–119. doi: 10.1111/j.1750-3639.1992.tb00679.x. [DOI] [PubMed] [Google Scholar]
- 7.Bonilla E, et al. Mitochondrial involvement in Alzheimer’s disease. . Biochim. Biophys. Acta. 1999; 1410:171–182. doi: 10.1016/s0005-2728(98)00165-0. [DOI] [PubMed] [Google Scholar]
- 8.DiMauro S, Tanji K, Bonilla E, Pallotti F, Schon EA. Mitochondrial abnormalities in muscle and other aging cells: classification, causes, and effects. Muscle Nerve. 2002;26:597–607. doi: 10.1002/mus.10194. [DOI] [PubMed] [Google Scholar]
- 9.Kim KM, Shibata D. Methylation reveals a niche: stem cell succession in human colon crypts. Oncogene. 2002;21:5441–5449. doi: 10.1038/sj.onc.1205604. [DOI] [PubMed] [Google Scholar]
- 10.Schon EA, Manfredi G. Neuronal degeneration and mitochondrial dysfunction. J. Clin. Invest. 2003;111:303–312. doi:10.1172/JCI200317741. doi: 10.1172/JCI17741. [DOI] [PMC free article] [PubMed] [Google Scholar]