Abstract
The scrapie prion protein (PrPSc) is the major, and possibly the only, component of the infectious prion; it is generated from the cellular isoform (PrPC) by a conformational change. N-terminal truncation of PrPSc by limited proteolysis produces a protein of ≈142 residues designated PrP 27–30, which retains infectivity. A recombinant protein (rPrP) corresponding to Syrian hamster PrP 27–30 was expressed in Escherichia coli and purified. After refolding rPrP into an α-helical form resembling PrPC, the structure was solved by multidimensional heteronuclear NMR, revealing many structural features of rPrP that were not found in two shorter PrP fragments studied previously. Extensive side-chain interactions for residues 113–125 characterize a hydrophobic cluster, which packs against an irregular β-sheet, whereas residues 90–112 exhibit little defined structure. Although identifiable secondary structure is largely lacking in the N terminus of rPrP, paradoxically this N terminus increases the amount of secondary structure in the remainder of rPrP. The surface of a long helix (residues 200–227) and a structured loop (residues 165–171) form a discontinuous epitope for binding of a protein that facilitates PrPSc formation. Polymorphic residues within this epitope seem to modulate susceptibility of sheep and humans to prion disease. Conformational heterogeneity of rPrP at the N terminus may be key to the transformation of PrPC into PrPSc, whereas the discontinuous epitope near the C terminus controls this transition.
Keywords: NMR structure, conformational change
Prions cause neurodegenerative illnesses in humans and animals (1). Those illnesses in humans include kuru, Creutzfeldt–Jakob disease (CJD), Gerstmann–Sträussler–Scheinker disease (GSS), and fatal familial insomnia (FFI) (2–4). Familial CJD, GSS, and FFI are autosomal dominant diseases caused by mutations in the PrP gene and are transmissible to experimental animals. In animals, bovine spongiform encephalopathy, or “mad cow” disease, has caused more than 160,000 cattle deaths in Great Britain. It is thought to be caused by a meat and bone meal dietary supplement containing prion-contaminated offal from sheep and cattle (5). Recent reports suggest that bovine prions may have been transmitted to humans (6, 7).
In contrast to viruses and viroids, prions do not contain a nucleic acid genome encoding their progeny. Rather, prions are composed largely, if not entirely, of a modified host-encoded glycoprotein denoted PrPSc. Through a posttranslational process, PrPSc is formed from the normal, cellular prion protein (PrP) isoform designated PrPC. No posttranslational chemical modification responsible for conversion of PrPC into PrPSc has been identified. Both PrPC and PrPSc possess a glycosylphosphatidyl inositol (GPI) anchor at the C terminus and are glycosylated at Asp181 and Asp197, but no covalent chemical differences between the two isoforms have been found (8). The GPI anchor of PrPC apparently targets it to caveolar-like structures within or adjacent to the plasma membrane where PrPC is either degraded or converted into PrPSc (9). Differences between PrPSc and PrPC lie in their properties: PrPC is soluble in nondenaturing detergents, and PrPSc is not, and whereas PrPC is largely α-helical and readily degraded by proteases, PrPSc has substantial β-sheet structure and a proteolytically stable core, termed PrP 27–30. Prediction efforts suggested that PrPC could form a four-helix bundle (H1-H4), whereas PrPSc would lose two of the four helices in favor of a substantial β-sheet (10, 11). Preparations containing this truncated PrP retain scrapie infectivity (12), and similarly truncated recombinant PrP (rPrP) promotes formation of PrPSc in cultured cells and in transgenic mice (13, 14). Recombinant antibody fragments (rFabs) that bind the N terminus (residues 90–112) of PrP 27–30 recognize native PrPC but not PrPSc, whereas other rFabs to epitopes in the C-terminal region bind to both native PrPC and PrPSc (15, 16). Because both N- and C-terminal rFabs bind to denatured PrPSc, we conclude that as PrPSc is formed, epitopes exposed in PrPC become buried (17).
Production of rPrP in large quantities for structural studies recently has been successful with expression in Escherichia coli of a 142-residue polypeptide corresponding to the Syrian hamster (SHa) sequence of PrP 27–30 (18). While this protein was being investigated, smaller segments of PrP were studied extensively by NMR spectroscopy. A 56-residue peptide consisting of PrP residues 90–145 was found to exist as an α-helical structure or one with intermolecular β-sheets depending on the microenvironment (19). In a hydrophobic environment, chemical shift and nuclear Overhauser effect (NOE) connectivities confirmed the existence of helices in the predicted H1 region and, more weakly, in the H2 region. A 111-residue polypeptide spanning mouse (Mo) PrP residues 121–231 [MoPrP(121–231)] was expressed in E. coli and found to contain H3 and H4 as predicted, with these helices stabilized by a disulfide bond joining the two Cys residues in PrP (20). An additional α-helix and two antiparallel four-residue β-strands were observed (20). One β-strand corresponds to a portion of H2 that was predicted to participate in a β-sheet of PrPSc (10). Both PrPC and PrPSc possess a disulfide bond (21). We report here on the NMR structure of rPrP(90–231) corresponding to the sequence of PrP 27–30 in an α-helical form that appears to resemble PrPC (18, 22).
MATERIALS AND METHODS
Isotopic Labeling of rPrP.
Uniform isotopic labeling of the protein was done by minimal modifications of the protocol (described by ref. 18). The bacteria were grown overnight in a shaker flask containing 100 ml of Celtone microbial growth media either 15N- or 15N/13C-labeled. The culture then was used for inoculation of a 1-liter fermentation where the following components were substituted for the isotopically labeled ones: isoleucine, ammonium sulfate, ammonium hydroxide, glucose, and 10% reconstituted Celtone powdered media for the 20% yeast extract and NZ amines. The fermentation was allowed to proceed for either 24 hr (15N) or 12–14 hr (15N/13C) depending on the reagents that were limiting. Wet cell paste yield was between 40 and 50 g, ultimately obtaining a minimum of 100 mg of purified protein.
Expression and Purification of rPrP.
SHa rPrP was expressed using an alkaline phosphatase promoter in a protease-deficient strain of E. coli (27C7), as described (18). Insoluble particles containing rPrP were extruded by a microfluidizer and pelleted by centrifugation. The extruded material was solubilized in 8 M guanidinium hydrochloride (GdnHCl)/100 mM DTT, pH 8.0 and subjected to purification by two sequential chromatographic procedures: size-exclusion chromatography (Pharmacia Superdex 200) eluted by 6 M GdnHCl/50 mM Tris⋅acetate, pH 8.0/1 mM EDTA, followed by reversed-phase chromatography using a C-4 column (Vydac) eluted by a gradient of acetonitrile/trifluoroacetic acid/water. Purified rPrP subsequently was lyophilized before refolding. Lyophilized rPrP was solubilized at 1 mg/ml in 8 M GdnHCl and rapidly diluted into 20 mM Tris⋅acetate, pH 8.0/5 mM EDTA to a final concentration of 0.1 mg/ml. The refolded protein was dialyzed against 20 mM sodium acetate, pH 5.0/0.005% sodium azide. Insoluble material was removed by filtration through a 0.2-μm filter. Solutions were concentrated by Centriplus 10 (Amicon) to a final protein concentration of 0.7–1.3 mM for NMR studies. Deuterated buffer exchange was done simultaneously with the final concentration step. Samples were analyzed by mass spectroscopy, Fourier transform infrared spectroscopy, and circular dichroism spectroscopy.
NMR Spectral Acquisition and Analysis.
NMR spectra for resonance assignments were acquired at 30°C, with 1 mM uniformly 15N-labeled and 15N/13C-labeled rPrP at pH 5.2, in 10% 2H2O on the following spectrometers: a Bruker DMX750, a Bruker AMX500, or a Varian UnityPlus600, each equipped with a 5-mm 13C/15N/1H triple resonance probe. Backbone and Cβ assignments were made using CBCA(CO)NH, HNCACB, and HNCA experiments (23). Side-chain 13C and 1H assignments were obtained using (H)C(CO)NH-total correlated spectroscopy (24) and HCCH-total correlated spectroscopy (25) spectra. NOE spectroscopy (NOESY) experiments were run at 750 MHz on a Bruker DMX750 spectrometer. Distance restraints were obtained from the NOESY data, using a 15N-labeled sample, from a three-dimensional 15N-resolved NOESY with a 100-ms mixing time (modified from ref. 26) and, using a 13C/15N-labeled sample, from a three-dimensional 13C-resolved NOESY with a 100-ms mixing time (27).
Structure Determination.
Spectra were processed using the program nmrpipe (28). Spectra were analyzed, and assignments managed using the locally written program sparky (29). All spectra were referenced relative to 3-(trimethylsilyl)-tetradeutero-sodium propionate. The backbone 1H, 15N, and 13Cα resonance assignments of rPrP(90–231) are complete. Ninety-two percent of the side-chain resonances are assigned, with the unassigned resonances mainly concerning residues with aromatic rings. From the NOE crosspeaks, 2,401 experimental distance restraints were used to generate low-resolution structures via the program diana (30), followed by minimization with amber 4.1 (31).
RESULTS
To investigate the basis of the PrP structural transitions, we prepared rPrP with the SHaPrP sequence corresponding to residues 90–231 of PrP 27–30 (18, 22). The rPrP was uniformly labeled with 15N or with both 15N and 13C; it was refolded into a conformer that resembles PrPC based on optical spectroscopic and immunochemical measurements (15, 32). The pH of the rPrP solution was found to be critical: N-terminal epitopes (residues 90–112) in rPrP that were observed to be partially buried at pH 5.2–8.0 by ELISA using N-terminal rFabs (17) (Y. Matsunaga and S.B.P., unpublished work) became completely exposed at pH 4.8 or lower. Whether the pH-dependent conformational transition detected by rFabs extends to other parts of the protein remains to be established, but it may be pertinent to structural differences between rPrP and MoPrP(121–231), because the latter structure was determined at pH 4.5 (20) (vide infra). When rPrP (0.9 mM) was poised in the middle of this structural transition at pH 5.0 in 20 mM Na acetate and 0.005% Na azide, its α-helical state was stable for at least 15 days at temperatures from 4°C to 30°C as judged by circular dichroism. In contrast, 1 day at 35°C led to a substantial loss of α-helix and a concomitant acquisition of β-sheet that was concentration-dependent (data not shown). At 35°C, incremental increases in the concentration of rPrP from 0.03 mM to 0.75 mM steadily increased the rate of α-helix to β-sheet conversion.
Multidimensional heteronuclear NMR studies were performed with rPrP. Signal linewidths and spectral dispersion indicated that most of the protein is well structured at the concentration (ca. 1 mM) and solution conditions used: 20 mM sodium acetate, pH 5.2, 30°C. Analytical sedimentation indicated that the protein was essentially monomeric at 25° (22). However, 15N spin-lattice and spin-spin relaxation time measurements indicate that the protein is undergoing rapid interconversion between a weak dimer and monomer. No specific intermolecular interactions have been identified to date.
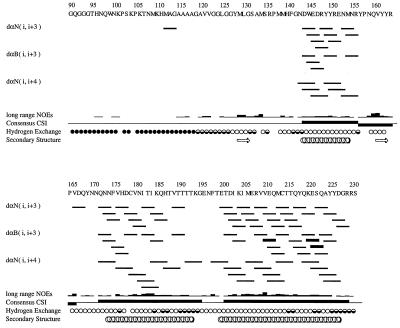
Essentially three parts to the protein are readily reflected in simple NMR spectral features: residues 90–112 are characterized by narrow (18 Hz) 15N heteronuclear single quantum coherence (HSQC) spectral signals and few long-range NOE crosspeaks; residues 113–126 have relatively narrow (ca. 18 Hz) 15N HSQC spectral (ca. 18Hz) signals and many NOE crosspeaks; and most of the remaining residues exhibit ca. 6 Hz broader HSQC signals and numerous NOE crosspeaks. The consensus chemical shift indices (33), as well as the proton NOE connectivities evident in NOESY spectra, consistently indicate that rPrP contains three α-helical regions (Fig. 1). The locations of these correspond largely, but not entirely, to those found for MoPrP(121–231) under similar solution conditions (0.8 mM protein, pH 4.5, no buffer, 20°C) (20) and to two of the four helices predicted for the entire sequence (10).
Figure 1.
Secondary structure diagram for rPrP. NOE connectivities are denoted by lines, where the thickness qualitatively represents the relative intensity (weak, medium, or strong) of the NOE crosspeaks, and i designates the residue number for rPrP. dαN(i, i+3) denotes an NOE between the α-proton of residue i and the amide proton of residue i+3. The long-range NOE line indicates by height the relative number of NOE crosspeaks between residues i → i + ≥4. In the consensus chemical shift index (59), contiguous up bars designate α-helix and down bars designate β-strand. Regions of secondary structure are depicted by helices for α-helices and broad arrows for β-strands. Hydrogen exchange was calculated from the intensity of proton NOE crosspeak between the amide and water: open circles for slow, filled for fast, and half-filled circles for medium exchange rate. No circle indicates spectral overlap or proline. The secondary structure diagram was created using the program vince (60).
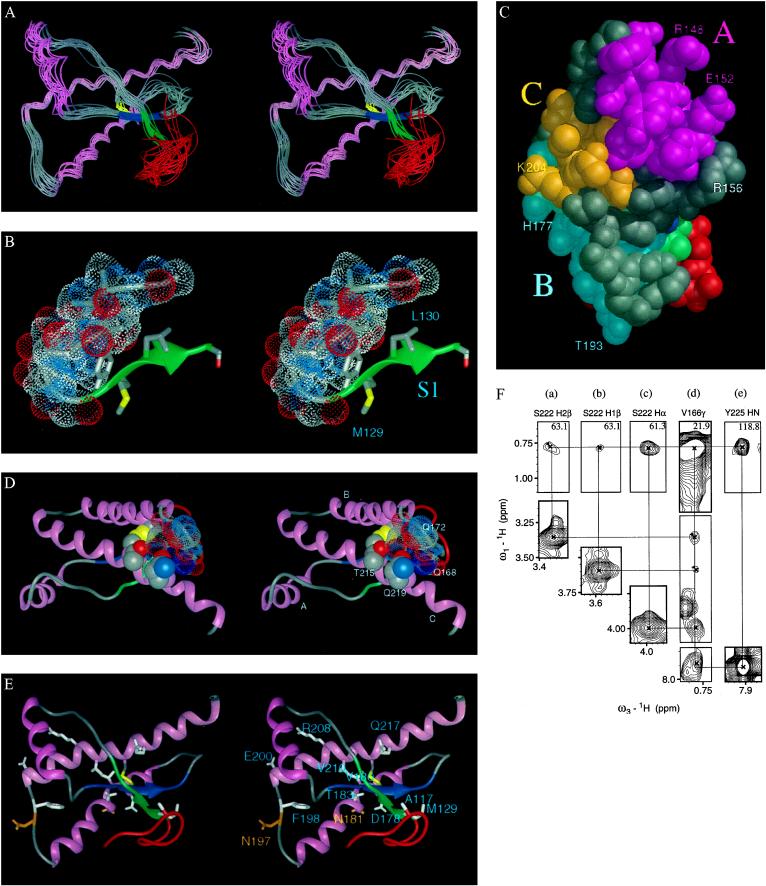
A best-fit superposition of backbone atoms for residues 113–228 of rPrP is shown in Fig. 2A. To distinguish the α-helices found in rPrP by NMR from those predicted by molecular modeling, we provisionally designate these helices A, B, and C. Helix A spans residues 144–156 with the last turn distorted, corresponding to helix 144–154 found for MoPrP(121–231). Helix B spans residues 172–193, with the first turn irregular at the present stage of structure refinement. This helix is about two turns longer than the 179–193 helix found for MoPrP(121–231), which agrees well with predicted helix H3 (179–191). Helix C extends from residues 200 to 227 with the 225–227 turn irregular. This helix is about three turns longer than the helix corresponding to residues 200–217 in MoPrP(121–231). It is notable that predicted helix H4 (residues 202–218) corresponds well with that found in MoPrP(121–231). Two four-residue β-strands (128–131 and 161–164) were identified in the MoPrP(121–231) structure. We found a similar antiparallel β-sheet, with S2 spanning residues 161–163 and S1 spanning 129–131 possessing β-sheet characteristics, but the two strands do not manifest standard β-sheet geometry. In fact, a β-bridge occurs only between Leu130 and Tyr162, although extensive cross-strand connectivities of residues are in segment 129–134 with proximate residues on the antiparallel segment 159–165.
Figure 2.
NMR structure of SHa rPrP(90–231). (A) Comparison of the 15 best-scoring structures of rPrP shown with a best-fit superposition of backbone atoms for residues 113–227 (stereoview). In all figures except C, the color scheme is: disulfide between Cys179 and Cys214, yellow; sites of glycosidation in PrPC, i.e., Asn181 and Asn197, gold; hydrophobic cluster composed of residues 113–126, red; helices, pink; loops, gray; residues 129–134, green, encompassing strand S1 and residues 159–165, blue, encompassing strand S2; the arrows span residues 129–131 and 161–163, as these show a closer resemblance to β-sheet. The structures were generated with the program diana (30), followed by energy minimization with amber 4.1 (31). Structure generation parameters are as follows: 2,401 distance restraints (intraresidue, 858; sequential (i → i +1), 753; (i → i +2), 195; (i → i +3), 233; (i → i +4), 109; and (i → i + ≥5), 253 for amino acid i); hydrogen bond restraints, 44; distance restraint violations >0.5 Å per structure, 30; amber energy, −1,443 ± 111 kcal/mol. Precision of structures: atomic rms deviation for all backbone heavy atoms of residues 128–227, <1.9 Å. The distance restraint violations and precision in some molecular moities reflect the conformational heterogeneity of rPrP. (B) Residues 113–132 illustrating (stereoview) in one representative structure the interaction of the hydrophobic cluster, with van der Waals rendering of atoms in residues 113–127, with the first β-strand. (C) Van der Waals surface of rPrP turned approximately 180° from A, illustrating the interaction of helix A with helix C. Helices A, B, and C are colored magenta, cyan, and gold, respectively. (D) Stereoview, using ribbonjr, illustrating the proximity of helix C to the 165–171 loop and the end of helix B, where residues Gln168 and Gln172 are depicted with a low-density van der Waals rendering and helix C residues Thr215 and Gln219 are depicted with a high-density van der Waals rendering. (E) Stereoview, highlighting in white the residues corresponding to point mutations that lead to human prion diseases. Illustrations were generated with midasplus. (F) Portion of the three-dimensional 13C-NOESY spectrum corresponding to 13C planes of the unresolved Val166 methyl resonances and the Ser222 resonances (a–d) and the 15N plane showing the Tyr225 amide interaction with Val166 (e). The diagonal peaks and mirrored crosspeaks for each 1H-1H connectivity are shown. The solid lines connecting peaks designate NOE connectivities.
The loop between S2 and helix B (i.e., residues 165–171) yields resonances clearly exhibiting long-range as well as medium-range restraints, which were not seen for the backbone atoms of residues 167–176 in the shorter MoPrP(121–231). Our results indicate that the loop is reasonably ordered, whereas this region is disordered in MoPrP(121–231) (20). Fig. 2F shows only one example: 1H-1H crosspeaks between the unresolved methyl protons of Val166 with Ser222 and Tyr225. The methyl protons of Val166 also exhibit 15 long-range crosspeaks with protons in the same loop, e.g., Tyr169, and in the extension of helix C, e.g., Tyr218 and Tyr225. Connectivities of Val166 to residues two turns apart on helix C suggest that the loop may exist in at least two conformations. Apparently, the interaction of the 165–171 loop with the helix C extension is important in stabilizing the structure (see Fig. 2A).
The mature human, Mo, and SHa prions manifest >90% sequence homology (34). Nevertheless, we must consider whether variations in sequence might cause the differences in structures observed for MoPrP(121–231) and SHa rPrP. Only four sequence variations are in the region 121–231. Whereas three appear to be conservative with no apparent structural effect, we note that Thr215 in the SHa sequence is Val in the mouse-A and Ile in the human PrP. Although it is conceivable that this may account for the differences in the length of helix C, it seems unlikely.
The α-proton and α-carbon chemical shifts for residues 90–127 are consistent with the region having α-helical content, but the extent of the chemical shifts relative to that of random coil values was generally not enough to indicate α-helix formation via tripartite chemical shift indices. Insufficient NOE connectivities exist to conclude that an α-helix is formed. The few medium-range connectivities in the segment 90–112 demonstrate sparse elements of structure. For example, for residues 95–100, we have so far identified nine nonsequential NOE connectivities. This implies that some structure exists at least transiently. The small number of long-range connectivities for the N-terminal segment 90–112 implies that it is largely disordered.
We have so far identified 36 long-range NOE crosspeaks involving side-chain resonances for the hydrophobic residues in the segment 113–125. An uncommon combination of glycines and hydrophobic residues leads to an unusual and dynamic structural feature. Most of the NOE connectivities indicate that these residues form a hydrophobic cluster with substantial backbone reversals permitted by the many glycines; indeed Val121, Val122, and Leu125 each exhibit 10 ± 3 long-range connectivities. As seen in Fig. 2A, the backbone for this cluster is not well defined in spite of the many connectivities. This may reflect the true dynamic nature of such a hydrophobic cluster, as the 15N HSQC spectral linewidths for these residues were also about 6 Hz smaller than for the core of the protein (ca. 18 vs. 24–25 Hz). Apparently, the combination of glycines with hydrophobic residues permits many alternative conformations with comparable free energies.
Some long-range connectivities place the hydrophobic cluster adjacent to the β-sheet in contact with the S1 strand (Fig. 2B). The weak, broadened 15N HSQC spectral signals for S1 residues Met129 (32 Hz), Leu130 (32 Hz), and Gly131 (44 Hz), as well as for S2 residues Tyr162 (33 Hz) and Arg164 (33 Hz), may well reflect conformational exchange effects in the interacting hydrophobic cluster pervading the adjacent irregular β-sheet. Taken together, these results suggest that the hydrophobic cluster and adjacent β-strands constitute a domain with marginally stable polymorphic structure.
DISCUSSION
The apparent conformational heterogeneity of the N-terminal region of rPrP may reflect the process by which PrPC is converted into PrPSc. Transgenetic studies suggest that PrPSc formation requires the substrate PrPC to bind to the product PrPSc at an intermediate stage of the conversion process (35). PrPC is thought to be in equilibrium with a metastable intermediate, designated PrP*, which binds to PrPSc in the conversion process (36). In fact, destabilization of PrPC has been shown to be necessary for it to bind to PrPSc in vitro (37–39). Further evidence for the conformational plasticity of PrP comes from unfolding studies of rPrP using guanidinium chloride (22). The free energy difference ΔG2 of 6.5 ± 1.2 kcal/mol between an intermediate state and the unfolded state was found to be comparable to literature values (5–15 kcal/mol) for protein unfolding (40, 41). However, the completely refolded rPrP, as used for the present NMR studies, is only marginally more stable (ΔG1 = 1.9 ± 0.4 kcal/mol) than the folding intermediate. This is consistent with the extensive conformational flexibility evident in the current NMR studies for part of the protein.
The NMR results for rPrP, compared with the structure reported for MoPrP(121–231) (20), support the notion that the core of the PrPC structure is formed by parts of helices B and C, corresponding largely to the predicted H3 and H4 regions (10), and is stabilized by the disulfide, which is essential for α-helical folding (18, 22). As seen in Fig. 2, helices B and C essentially form one side of the protein structure. This core is further stabilized by helix A, which lies across helix C with side chains interacting between the two helices (Fig. 2C). Strand S2 also lies on this side of the protein and interacts predominantly with helices B and C as well as S1. With or without S2 and S1, we presume this relatively stable folding core is associated with the second unfolding transition. Attempts to prepare MoPrP(108–231) resulted in proteolytic cleavage producing MoPrP(121–231) (20), indicative of a stable core beginning with residue 121 (41). Conclusions about the “stable core” of MoPrP(121–231), however, must be considered within the context of the unstructured loop (165–171) and the shortened helices B and C. It is likely that the structure of MoPrP(121–231) corresponds to that of PrPC-II, which is formed in caveolae during the initial degradation of PrPC (9, 42). Whether the ineligibility of PrPC-II for conversion into PrPSc is determined by the disordered structure assumed by the loop (165–171) as well as by the unraveling of helices B and C remains to be determined (43).
The presence of the additional 31 N-terminal residues of rPrP, relative to MoPrP(121–231), induces substantial changes in the structure of PrP, which include alterations in the C terminus. Helix C is extended by at least nine residues, helix B is up to seven residues longer, and the loop comprising residues 165–171 is sufficiently ordered that many long-range restraints can be observed. The hydrophobic cluster (residues 113–125) predominantly interacts with S1 in the β-sheet (Fig. 2) and may serve to stabilize the observed extension of helix B from 179 in MoPrP(121–231) to 172 in rPrP. Stability also may be conferred by hydrophobic interactions of Tyr128 with Tyr163 in the β-sheet, which, in turn, interact with Val176. The relative stability of the 165–171 loop and the three additional helical turns in helix C presumably are connected to stabilization of the other structural elements.
Strains of prions exhibit different incubation times before symptoms of disease appear and different patterns of PrPSc accumulation. Recent work indicates that the properties of prion strains might be manifestations of different conformers of PrPSc (44, 45). Studies on the transmission of human prions to transgenic mice suggest that a species-specific factor, provisionally designated protein X, might function like a molecular chaperone in PrPSc formation (46). Our current working hypothesis is that protein X forms a transient complex with the metastable intermediate PrP*, diminishing the activation energy barrier between PrPC and PrPSc and facilitating formation of PrPSc (36, 46). Our analysis comparing the full-length helix C and ordering of the 165–171 loop in rPrP with the truncation of helix C at Gln217 in MoPrP(121–231) is consistent with this concept. Recent work suggests SHa residues Gln168, Gln172, Thr215, and Gln219 are at the site of protein X binding (47); the glycosylation sites, Asn181 and Asn197, are apparently quite distant from this putative binding site (Fig. 2E). As seen in Fig. 2D, Thr215 and Gln219 lie in register one turn apart on helix C and interact with the residues in the 165–171 loop. SHa residue 168 is a Gln in most species and corresponds to sheep PrP codon 171, which is polymorphic, encoding either Gln or Arg. Almost all Suffolk sheep with scrapie were found to be Gln/Gln, indicating that heterozygosity for Arg conferred resistance (48–56). Equally important is the observation that ≈12% of PrP alleles in Japanese encode Lys instead of Glu at position 219 (57). No cases of Creutzfeldt–Jakob disease have been found in people with Lys219, which, like Arg, is basic. These findings and data on the conversion of mutagenized PrP into PrPSc in ScN2a cells suggest that protein X binds to a discontinuous epitope, incorporating residues 168, 172, 215, and 219 in rPrP that is disordered in MoPrP(121–231).
Residues where point mutations lead to human diseases are highlighted in Fig. 2E. The D178N point mutation in the PrP gene causes fatal familial insomnia if residue 129 is Met (4). A potentially important difference between the structures of rPrP and MoPrP(121–231) lies in the proximity of residues 178 and 129. The side chains of these residues determine the phenotypes of two inherited human prion diseases (58). In rPrP, residue 178 lies within helix B and is located opposite residue 129 with strand S2 partially intervening. Such geometry suggests that the D178N mutation destabilizes PrP through partially unraveling helix B and that the conformation of mutant PrPSc is modulated by the side chain of residue 129. The particular conformation adopted by mutant PrPSc might determine in which regions of the central nervous system PrPSc is deposited, and thus, be responsible for whether patients present with insomnia or dementia (58). When residue 129 is Val, then patients present with a dementing illness called familial Creutzfeldt–Jakob disease. In MoPrP(121–231), residues 178 and 129 are apparently distant from each other; in fact, residue 178 does not even form part of helix B.
Our structural studies of rPrP underscore the conformational plasticity evident in the N-terminal region (19) and define important structural features not evident in a smaller C-terminal fragment (20). Previous studies, including those most recently performed with rFabs, indicate that the region corresponding to the N-terminal 30–40 residues of rPrP undergoes a profound conformational change during formation of PrPSc (17). This conformational change seems to be mediated by protein X, whose binding site on PrPC was delineated by the structure of rPrP reported here and by site-directed mutagenesis (47).
Acknowledgments
We are especially grateful to P. E. Wright and H. J. Dyson for support and for use of the Skaggs NMR Facility. For help with some of the initial spectra, we thank V. J. Basus, M. A. Kennedy, D. L. Mattiello, S. Mayo, S. Ross, and G. M. Wolfe. We thank C. Kojima for helpful discussions and the University of California, San Francisco Computer Graphics Laboratory for use of its facilities. This work was supported by National Institutes of Health Grants AG10770 and GM39247 as well as by gifts from the Leila and Harold G. Mathers, Sherman Fairchild, and the Bernard Osher foundations.
ABBREVIATIONS
- PrP
prion protein
- PrPSc
scrapie isoform of PrP
- PrPC
cellular isoform of PrP
- PrP 27–30
N-terminally truncated fragment of PrPSc
- rPrP
recombinant PrP consisting of residues 90–231
- SHa
Syrian hamster
- Mo
mouse
- rFabs
recombinant antibody fragments
- NOE(SY)
nuclear Overhauser effect (spectroscopy)
- HSQC
heteronuclear single quantum coherence
Footnotes
Data deposition: The coordinates from the NMR studies have been deposited in the Protein Data Bank, Brookhaven National Laboratory, Upton, NY 11973.
References
- 1.Prusiner S B. Trends Biochem Sci. 1996;252:482–487. doi: 10.1016/s0968-0004(96)10063-3. [DOI] [PubMed] [Google Scholar]
- 2.Gajdusek D C. Science. 1977;197:943–960. doi: 10.1126/science.142303. [DOI] [PubMed] [Google Scholar]
- 3.Masters C L, Gajdusek D C, Gibbs C J., Jr Brain. 1981;104:559–588. doi: 10.1093/brain/104.3.559. [DOI] [PubMed] [Google Scholar]
- 4.Medori R, Tritschler H-J, LeBlanc A, Villare F, Manetto V, Chen H Y, Xue R, Leal S, Montagna P, Cortelli P, Tinuper P, Avoni P, Mochi M, Baruzzi A, Hauw J J, Ott J, Lugaresi E, Autilio-Gambetti L, Gambetti P. N Engl J Med. 1992;326:444–449. doi: 10.1056/NEJM199202133260704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson R M, Donnelly C A, Ferguson N M, Woolhouse M E J, Watt C J, Udy H J, MaWhinney S, Dunstan S P, Southwood T R E, Wilesmith J W, Ryan J B M, Hoinville L J, Hillerton J E, Austin A R, Wells G A H. Nature (London) 1996;382:779–788. doi: 10.1038/382779a0. [DOI] [PubMed] [Google Scholar]
- 6.Chazot G, Broussolle E, Lapras C I, Blättler T, Aguzzi A, Kopp N. Lancet. 1996;347:1181. doi: 10.1016/s0140-6736(96)90638-8. [DOI] [PubMed] [Google Scholar]
- 7.Will R G, Ironside J W, Zeidler M, Cousens S N, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofman A, Smith P G. Lancet. 1996;347:921–925. doi: 10.1016/s0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 8.Stahl N, Baldwin M A, Teplow D B, Hood L, Gibson B W, Burlingame A L, Prusiner S B. Biochemistry. 1993;32:1991–2002. doi: 10.1021/bi00059a016. [DOI] [PubMed] [Google Scholar]
- 9.Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner S B. J Cell Biol. 1995;129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Z, Gabriel J-M, Baldwin M A, Fletterick R J, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 1994;91:7139–7143. doi: 10.1073/pnas.91.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Z, Prusiner S B, Cohen F E. Folding Design. 1996;1:13–19. doi: 10.1016/S1359-0278(96)00007-7. [DOI] [PubMed] [Google Scholar]
- 12.Prusiner S B, Bolton D C, Groth D F, Bowman K A, Cochran S P, McKinley M P. Biochemistry. 1982;21:6942–6950. doi: 10.1021/bi00269a050. [DOI] [PubMed] [Google Scholar]
- 13.Rogers M, Yehiely F, Scott M, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:3182–3186. doi: 10.1073/pnas.90.8.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 15.Williamson R A, Peretz D, Smorodinsky N, Bastidas R, Serban H, Mehlhorn I, DeArmond S J, Prusiner S B, Burton D R. Proc Natl Acad Sci USA. 1996;93:7279–7282. doi: 10.1073/pnas.93.14.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peretz D, Williamson A, Matsunaga Y, Burton D R, Prusiner S B. J Allergy Clin Immunol. 1997;99:S222. [Google Scholar]
- 17.Peretz, D., Williamson, R. A., Matsunaga, Y., Serban, H., Pinilla, C., Bastidas, R., Rozenshteyn, R., James, T. L., Houghten, R. A., Cohen, F. E., Prusiner, S. B. & Burton, D. R. (1997) J. Mol. Biol., in press. [DOI] [PubMed]
- 18.Mehlhorn I, Groth D, Stöckel J, Moffat B, Reilly D, Yansura D, Willett W S, Baldwin M, Fletterick R, Cohen F E, Vandlen R, Henner D, Prusiner S B. Biochemistry. 1996;35:5528–5537. doi: 10.1021/bi952965e. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Kaneko K, Nguyen J T, Livshits T L, Baldwin M A, Cohen F E, James T L, Prusiner S B. J Mol Biol. 1995;250:514–526. doi: 10.1006/jmbi.1995.0395. [DOI] [PubMed] [Google Scholar]
- 20.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 21.Turk E, Teplow D B, Hood L E, Prusiner S B. Eur J Biochem. 1988;176:21–30. doi: 10.1111/j.1432-1033.1988.tb14246.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Stöckel J, Mehlhorn I, Groth D, Baldwin M A, Prusiner S B, James T L, Cohen F E. Biochemistry. 1997;36:3543–3553. doi: 10.1021/bi961965r. [DOI] [PubMed] [Google Scholar]
- 23.Muhandiram D R, Kay L E. J Magn Reson B. 1994;103:203–216. [Google Scholar]
- 24.Logan T M, Olejniczak E T, Xu R X, Fesik S W. J Biomol NMR. 1993;3:225–231. doi: 10.1007/BF00178264. [DOI] [PubMed] [Google Scholar]
- 25.Kay L E, Xu G-Y, Singer A U, Muhandiram D R, Forman-Kay J D. J Magn Reson B. 1993;101:333–337. [Google Scholar]
- 26.Marion D, Driscoll P C, Kay L E, Wingfield P T, Bax A, Gronenborn A M, Clore G M. Biochemistry. 1989;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 27.Muhandiram D R, Farrow N A, Xu G Y, Smallcombe S H, Kay L E. J Magn Reson B. 1993;102:317–321. [Google Scholar]
- 28.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 29.Kneller D, Kuntz I D, Goddard T. sparky. San Francisco: University of California; 1996. [Google Scholar]
- 30.Güntert P, Braun W, Wüthrich K. J Mol Biol. 1991;217:517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- 31.Pearlman D A, Case D A, Caldwell J, Ross W S, Cheatham TE, III, Ferguson D N, Seibel G L, Singh U C, Weiner P K, Kollman P A. amber 4.1. San Francisco: University of California; 1995. [Google Scholar]
- 32.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wishart D S, Sykes B D, Richards F M. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 34.Schätzl H M, Da Costa M, Taylor L, Cohen F E, Prusiner S B. J Mol Biol. 1995;245:362–374. doi: 10.1006/jmbi.1994.0030. [DOI] [PubMed] [Google Scholar]
- 35.Prusiner S B, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson G A, Hoppe P C, Westaway D, DeArmond S J. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 36.Cohen F E, Pan K-M, Huang Z, Baldwin M, Fletterick R J, Prusiner S B. Science. 1994;264:530–531. doi: 10.1126/science.7909169. [DOI] [PubMed] [Google Scholar]
- 37.Kocisko D A, Come J H, Priola S A, Chesebro B, Raymond G J, Lansbury P T, Jr, Caughey B. Nature (London) 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko K, Peretz D, Pan K-M, Blochberger T, Wille H, Gabizon R, Griffith O H, Cohen F E, Baldwin M A, Prusiner S B. Proc Natl Acad Sci USA. 1995;32:11160–11164. doi: 10.1073/pnas.92.24.11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaneko K, Wille H, Mehlhorn I, Zhang H, Ball H, Cohen F E, Baldwin M A, Prusiner S B. J Mol Biol. 1997;270:574–586. doi: 10.1006/jmbi.1997.1135. [DOI] [PubMed] [Google Scholar]
- 40.Pace C N. Methods Enzymol. 1995;259:538–554. doi: 10.1016/0076-6879(95)59060-9. [DOI] [PubMed] [Google Scholar]
- 41.Hornemann S, Glockshuber R. J Mol Biol. 1996;262:614–619. doi: 10.1006/jmbi.1996.0487. [DOI] [PubMed] [Google Scholar]
- 42.Pan K-M, Stahl N, Prusiner S B. Protein Sci. 1992;1:1343–1352. doi: 10.1002/pro.5560011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muramoto T, Scott M, Cohen F, Prusiner S B. Proc Natl Acad Sci USA. 1996;93:15457–15462. doi: 10.1073/pnas.93.26.15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bessen R A, Marsh R F. J Virol. 1992;66:2096–2101. doi: 10.1128/jvi.66.4.2096-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Telling G C, Parchi P, DeArmond S J, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner S B. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 46.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 47.Kaneko, K., Zulianello, L., Scott, M., Cooper, C. M., Wallace, A. C., James, T. L., Cohen, F. E. & Prusiner, S. B. (1997) Proc. Natl. Acad. Sci. USA 10069–10074. [DOI] [PMC free article] [PubMed]
- 48.Hunter N, Goldmann W, Benson G, Foster J D, Hope J. J Gen Virol. 1993;74:1025–1031. doi: 10.1099/0022-1317-74-6-1025. [DOI] [PubMed] [Google Scholar]
- 49.Goldmann W, Hunter N, Smith G, Foster J, Hope J. J Gen Virol. 1994;75:989–995. doi: 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- 50.Westaway D, Zuliani V, Cooper C M, Da Costa M, Neuman S, Jenny A L, Detwiler L, Prusiner S B. Genes Dev. 1994;8:959–969. doi: 10.1101/gad.8.8.959. [DOI] [PubMed] [Google Scholar]
- 51.Belt P B G M, Muileman I H, Schreuder B E C, Ruijter J B, Gielkens A L J, Smits M A. J Gen Virol. 1995;76:509–517. doi: 10.1099/0022-1317-76-3-509. [DOI] [PubMed] [Google Scholar]
- 52.Clousard C, Beaudry P, Elsen J M, Milan D, Dussaucy M, Bounneau C, Schelcher F, Chatelain J, Launay J M, Laplanche J L. J Gen Virol. 1995;76:2097–2101. doi: 10.1099/0022-1317-76-8-2097. [DOI] [PubMed] [Google Scholar]
- 53.Ikeda T, Horiuchi M, Ishiguro N, Muramatsu Y, Kai-Uwe G D, Shinagawa M. J Gen Virol. 1995;76:2577–2581. doi: 10.1099/0022-1317-76-10-2577. [DOI] [PubMed] [Google Scholar]
- 54.Hunter N, Moore L, Hosie B D, Dingwall W S, Greig A. Vet Rec. 1997;140:59–63. doi: 10.1136/vr.140.3.59. [DOI] [PubMed] [Google Scholar]
- 55.Hunter N, Cairns D, Foster J D, Smith G, Goldmann W, Donnelly K. Nature (London) 1997;386:137. doi: 10.1038/386137a0. [DOI] [PubMed] [Google Scholar]
- 56.O’Rourke K I, Holyoak G R, Clark W W, Mickelson J R, Wang S, Melco R P, Besser T E, Foote W C. J Gen Virol. 1997;78:975–978. doi: 10.1099/0022-1317-78-4-975. [DOI] [PubMed] [Google Scholar]
- 57.Kitamoto T, Tateishi J. Philos Trans R Soc London B. 1994;343:391–398. doi: 10.1098/rstb.1994.0034. [DOI] [PubMed] [Google Scholar]
- 58.Goldfarb L G, Petersen R B, Tabaton M, Brown P, LeBlanc A C, et al. Science. 1992;258:806–808. doi: 10.1126/science.1439789. [DOI] [PubMed] [Google Scholar]
- 59.Wishart D S, Bigam C G, Yao J, Abildgaard F, Dyson H J, Oldfield E, Markley J L, Sykes B D. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 60.La Rosa E, Stern A, Hoch J. vince. Cambridge, MA: Rowland Institute for Science; 1996. [Google Scholar]