Abstract
Actin depolymerizing factors (ADF) are stimulus responsive actin cytoskeleton modulating proteins. They bind both monomeric actin (G-actin) and filamentous actin (F-actin) and, under certain conditions, F-actin binding is followed by filament severing. In this paper, using mutant maize ADF3 proteins, we demonstrate that the maize ADF3 binding of F-actin can be spatially distinguished from that of G-actin. One mutant, zmadf3–1, in which Tyr-103 and Ala-104 (equivalent to destrin Tyr-117 and Ala-118) have been replaced by phenylalanine and glycine, respectively, binds more weakly to both G-actin and F-actin compared with maize ADF3. A second mutant, zmadf3–2, in which both Tyr-67 and Tyr-70 are replaced by phenylalanine, shows an affinity for G-actin similar to maize ADF3, but F-actin binding is abolished. The two tyrosines, Tyr-67 and Tyr-70, are in the equivalent position to Tyr-82 and Tyr-85 of destrin, respectively. Using the tertiary structure of destrin, yeast cofilin, and Acanthamoeba actophorin, we discuss the implications of removing the aromatic hydroxyls of Tyr-82 and Tyr-85 (i.e., the effect of substituting phenylalanine for tyrosine) and conclude that Tyr-82 plays a critical role in stabilizing the tertiary structure that is essential for F-actin binding. We propose that this tertiary structure is maintained as a result of a hydrogen bond between the hydroxyl of Tyr-82 and the carbonyl of Tyr-117, which is located in the long α-helix; amino acid components of this helix (Leu-111 to Phe-128) have been implicated in G-actin and F-actin binding. The structures of human destrin and yeast cofilin indicate a hydrogen distance of 2.61 and 2.77 Å, respectively, with corresponding bond angles of 99.5° and 113°, close to the optimum for a strong hydrogen bond.
Keywords: cytoskeleton, destrin or human actin depolymerizing factor, cofilin, tertiary structure
The actin cytoskeleton forms a dynamic network in eukaryotic cells, which can reorganize itself in response to a variety of stimuli. These reorganizations are coordinated by a plethora of actin binding proteins the functions of which may be controlled by numerous signals including Ca2+, phosphoinositides, pH, or reversible phosphorylation (1–3). Few actin binding proteins have been identified in higher plants, and little is known about the regulation of actin dynamics in plant cells. One of the most dramatic examples of actin reorganization in plants occurs in pollen development. As pollen grains enter dormancy, F-actin is condensed into spiculate aggregates; during pollen germination and tube growth these are replaced by a microfilament array that is required for tip growth (4–6). The identification in pollen grains of specifically expressed isogenes of plant profilin and actin depolymerizing factor has led to the suggestion that these proteins may be specifically involved in the dynamics of this actin reorganization (7–9). Both the plant profilins and the plant actin depolymerizing factors have been shown to function in vitro in a manner similar to their animal and fungal counterparts (8–10).
The actin depolymerizing factor (ADF) group of proteins, which includes cofilin (11–14), vertebrate ADF (15) or destrin (16, 17), depactin (18), actophorin (19) and, more recently, maize ADF, ZmADF (previously named maize actin binding protein, ZmABP, in refs. 9 and 20) interact with both monomeric and filamentous actin, in vitro, and sever filaments (9, 17, 21–23). The members of the ADF group show a high conservation of primary structure and function. In yeast, the single cofilin gene is essential for cell viability, but can be replaced in gene disruption experiments by mammalian cofilin (12). Binding to actin is regulated by phosphoinositides (24) and phosphorylation (25–27). Overexpression of cofilin in mammalian cells caused disruption of pre-existing actin structures and induced cytoplasmic actin bundles (27), whereas overexpression of the endogenous cofilin in Dictyostelium discoideum stimulated cell movement and membrane ruffling (28). Thus, the ADF group of proteins performs a critical role in modulating the dynamics of the actin cytoskeleton.
Two regions in pig cofilin have been shown to be essential for actin binding. The region of cofilin Ala-105 to Met-115 can be chemically crosslinked to the N terminus of actin and a synthetic dodecapeptide containing this sequence (Trp-104 to Met-115) inhibits actin polymerization (29). Mutation analysis showed that both Lys-114 and Lys-112 play an essential role in actin binding and actin depolymerizing activity. Lys-112 has also been implicated as one of the amino acids involved in cofilin’s interaction with PIP2 (22, 30). Competition studies using a synthetic heptapeptide patterned on the sequence Asp-122 to Leu-128 suggest that this region may also constitute part of the actin binding domain (31). These actin and PIP2 binding regions are highly conserved in the equivalent plant ADF sequences (9). The tertiary structures of destrin, yeast cofilin, and Acanthamoeba actophorin have recently been reported from nuclear magnetic resonance studies (32) and x-ray analysis (33, 34). All three structures have the same basic fold and show that the two sequences, Trp-104 to Met-115 and Asp-122 to Leu-128 in destrin are located at the N and C termini, respectively, of the long α-helix starting at Leu-111 and terminating at Phe-128.
The activity of destrin and cofilin is regulated by phosphorylation of Ser-3 (26, 27), equivalent to Ser-6 in the plant ADF sequences (9). Phosphorylation inhibits interaction with actin, whereas dephosphorylation promotes interaction. Under certain conditions, cofilin and actin form intranuclear cofilin:actin rods and a nuclear localization signal has been identified between Lys-19 and Lys-34 (32, 35, 36). Cofilin is dephosphorylated prior to translocation into the nucleus. Ser-3 is located close to the actin binding long α-helix in the tertiary structure of destrin, and it has been proposed that phosphorylation of this residue may perturb the function of Lys-112 (32). The nuclear localization signal is located at the opposite side of the actin contact in the tertiary structure, and phosphorylation of Ser-3 appears to inhibit its action (32).
In this paper we describe the properties of two mutant maize ADF3 proteins, zmadf3–1 and zmadf3–2. zmadf3–1 has two amino acid substitutions; tyrosine at position 103 and alanine at 104 are replaced by phenylalanine and glycine, respectively. This mutant protein binds weakly to both G- and F-actin. zmadf3–2 has the tyrosines at positions 67 and 70, each replaced by phenylalanine. zmadf3–2 binds G-actin in a similar manner to maize ADF3, but does not bind F-actin. Both mutant proteins inhibit the assembly and disassembly of actin as compared with maize ADF3. These data are discussed in relation to the tertiary structures of destrin (32) and yeast cofilin (33).
MATERIALS AND METHODS
Amino Acid Sequence Comparisons.
All the actin depolymerizing factor-like sequences in the GenBank and EMBL databases were compared using the pileup program, and ZmADF3 was compared directly to either destrin or yeast cofilin using the bestfit program in the GCG (Madison, WI) package (37).
Site-Directed Mutagenesis.
Each of the mutant cDNAs was generated using four primers in two PCR reactions using ZmADF3 (previously named ZmABP3 in ref. 9) as template DNA, subsequent splicing together of the two resulting fragments, and cloning into a bacterial expression vector. The primers used to generate zmadf3–1 are: primer 1 (NdeI/R mutation) GCACGAGCTCGCCCCATATGGCGAACGCGCGTTCGGGT; primer 2 (BamHI/FG mutation) CTGGTTTGAGGATCCGAAAAGCATCTTGCTCTTCACCT; primer 3 (BamHI) AGAGCGAGCTCCTTTACGGATCCTCAAACCAGAAATTCAAGA; primer 4 (HindIII) ACTCGCGAAAGCTTTGCGCTAGCGTGCCCGAT.
The primers used to generate zmadf3–2 are: primer 1 (NdeI/R mutation) GCACGAGCTCGCCCCATATGGCGAACGCGCGTTCGGGT; primer 2 (EcoRI/FF mutation) AGTCGAATTCGAAGATCGCGAATCGGCAGTCATTCTCAGGGA; primer 3 (EcoRI) GCCGAGAGCTCATCTATGAATTCGACTTTGTTACTGCAGA; primer 4 (HindIII) ACTCGCGAAAGCTTTGCGCTAGCGTGCCCGAT.
The first reactions to amplify the 5′ regions used primers 1 and 2 for each mutant cDNA prepared. In each case, primer 1 includes a codon change for Arg-5 from AGA to CGT, which is more compatible for Escherichia coli expression, and primer 2 harbors the codon changes to introduce the mutations: for zmadf3–1, Tyr-103/Ala-104 to Phe-103/Gly-104; for zmadf3–2, Tyr-67/Tyr-70 to Phe-67/Phe-70. The second reaction to amplify the 3′ regions used primers 3 and 4 for each mutant cDNA. All primers included appropriate restriction enzyme sites for splicing the 5′ and 3′ regions together before cloning the final product in the correct reading frame in the E. coli expression vector pMW172. Both mutated clones were sequenced to ensure that no extraneous mutations had occurred.
PCR.
The ZmADF3 template DNA (10 ng) was amplified in the presence of 300 ng of primer, 0.5 mM dNTPs, 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris⋅HCl (pH 8.3), 0.01% (wt/vol) gelatine, and 2.5 units of AmpliTaq (Perkin–Elmer/Cetus) for 30 cycles at 60°C using an Hybaid Omnigene thermocycling apparatus. PCR products for ligations were purified on 1.5% (wt/vol) agarose gels and the DNA was electroeluted, phenol/chloroform extracted, and ethanol precipitated.
Protein Purification.
ZmADF3, zmadf3–1, and zmadf3–2 proteins were expressed in E. coli and the protein purified as described in ref. 9. Actin from rabbit skeletal muscle was prepared as described in ref. 38 and recombinant human actin depolymerizing factor prepared as in ref. 17.
Falling Ball Viscometry.
Falling ball viscometry was performed as described in ref. 38. Actin (10 μM) was mixed with different concentrations of ADF protein and volumes made to 180 μl in buffer containing 10 mM Tris⋅HCl (pH 8.0), 0.1 mM ATP, 0.2 mM CaCl2, 0.2 mM DTT, and 1 mM NaN3. Twenty microliters of 10 × KME (0.5 M KCl/10 mM MgSO4/10 mM EGTA) was added, the solution briefly vortexed, and taken up in 100 μl capillaries, which were then sealed with Plasticene. After approximately 1 hr, the viscosity was measured by timing the velocity of steel ball bearings (Atlas Ball-Bearing, Birmingham, U.K.) falling through the solutions.
G-Actin Binding Assay.
Binding to G-actin was monitored by native gel electrophoresis and using a 1:1 molar ratio of actin to ADF as described (39). Ten percent polyacrylamide gels were run at 200 V in the presence of 2 mM dithiothreitol and 0.2 mM ATP in both the gels and the running buffer. ADF and actin were each loaded at a concentration of 12 μM.
Cosedimentation Assay.
F-actin and ADF (both 10 μM) were incubated for 20 min in 12 mM Tris⋅HCl (pH 7.0), 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, 0.5 mM DTT, 0.2 mM CaCl2, and 1 mM NaN3. After sedimentation at 100,000 × g for 15 min in a Beckman TLA100 centrifuge (40), aliquots from the supernatant and pellet, resuspended in a volume of buffer equal to the supernatant, were analyzed by SDS/PAGE as described in ref. 41.
Polymerization Assay.
The effects of ADF on the rate of nucleated actin polymerization were measured as follows: ADF was mixed with 10 μM F-actin for 20–30 sec and the mixture added to 5 μM G-actin (including 0.5 μM pyrenyl actin) at 1.25 μM F-actin in buffer containing 10 mM Tris⋅HCl (pH 8.0), 0.2 mM ATP, 0.2 mM CaCl2, 1 mM DTT, and 1 mM NaN3/1 mM MgCl2/100 mM NaCl. The value for kobs (the rate constant for the exponential rise in fluorescence) was measured as described previously (17). The kobs values reflect both the number concentration of filaments (i.e., extent of any severing) and the rate constants for assembly at the two filament ends.
Depolymerization Assay.
The effects of ADF on the rate of actin depolymerization were measured as described previously (17). F-actin (8 μM) (nucleated with 80 nM gelsolin and containing 1 μM pyrenyl actin) was diluted to 200 nM actin under depolymerizing conditions in buffer containing 10 mM Tris⋅HCl (pH 8.0), 0.2 mM ATP, 0.2 mM CaCl2, 1 mM DTT, and 1 mM NaN3. ADF was added after about 60 sec and the rate constant for the exponential fluorescence decrease measured as a function of ADF concentration. The kobs values reflect both the number concentration of filaments and the depolymerization rate constants.
RESULTS
Consensus Sequence for Actin Depolymerizing Factor.
The deduced amino acid sequences of all the known actin depolymerizing factors were compared, and this revealed that only nine amino acids are fully conserved across phylogeny (Fig. 1A). This implies that these amino acids are important for ADF structure and function. Thus far, specific roles have been attributed to the residues equivalent to maize ADF3 Ser-6 and Lys-100 (22, 26, 27). We mutated the triplets encoding the two maize ADF3 tyrosines, Tyr-67 and Tyr-104, to analyze their contribution to ADF function. In the sequence alignments shown in Fig. 1B, the equivalent destrin (16) and yeast cofilin (12) tyrosines are at positions 82 and 117 and at positions 64 and 101, respectively. In addition, maize ADF3 showed 66% and 83% similarity with destrin and yeast cofilin, respectively.
Figure 1.
(A) Amino acid sequence of ZmADF3 (9), zmadf3–1, and zmadf3–2. The reversed type amino acids are those that are 100% conserved at the same or equivalent position in all currently available actin depolymerizing factor-like sequences. (B) Comparison of ZmADF3 with the destrin (16) and the yeast cofilin (12) primary structures (|, :, ., represent degrees of identity or similarity in the order | >: >.).
Generation of Two Mutant Maize ADF3 Proteins, zmadf3–1 and zmadf3–2.
In the mutant zmadf3–1, the Tyr-103 and Ala-104 were replaced by phenylalanine and glycine, respectively (Fig. 1A). These are conservative substitutions and the reason for changing the alanine to glycine was to make a convenient restriction enzyme site for cloning. Ala-104 is not conserved in all 12 actin depolymerizing factors compared; the equivalent positions in actophorin (42) and Drosophila melanogaster ADF (43) are threonine and serine, respectively. In zmadf3–2, Tyr-67 and Tyr-70 were substituted by phenylalanine (Fig. 1A). These are conservative substitutions with the principal difference being a loss of the hydroxyl group. The tyrosine at position 70 is not conserved in all actin depolymerizing factor sequences; indeed, phenylalanine is present at this position in actophorin, D. melanogaster ADF, Caenorhabditis elegans ADF1 (44), and in lily and Brassica napus ADF sequences (45). The recombinant proteins were prepared as described previously (9). The stabilities of the proteins in comparison to ZmADF3 were monitored by the effects of urea denaturation in the intrinsic fluorescence and by circular dichroism spectra to ensure that these mutations did not introduce any significant structural changes (data not shown).
Effect of Maize ADF3 and the Mutant Proteins, zmadf3–1 and zmadf3–2, on the Viscosity of Actin.
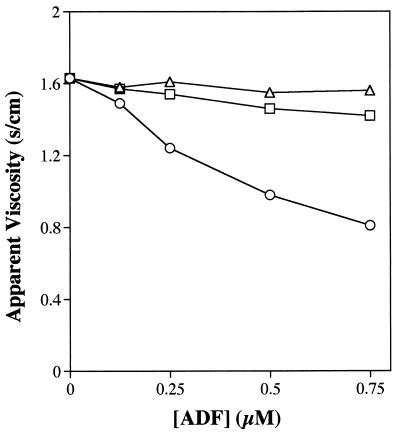
Maize ADF3 causes a dramatic reduction in the viscosity of F-actin (9) as shown in Fig. 2. In comparison, zmadf3–1 has no effect and zmadf3–2 only a small effect on the viscosity of F-actin under similar conditions. These data suggest that the mutant proteins have little or no effect in disrupting actin filaments.
Figure 2.
Effect of ZmADF3, zmadf3–1, and zmadf3–2 on the viscosity of polymerizing actin. Actin (10 μM) was polymerized for 1 hr in the presence of increasing concentrations of each of the three proteins and the viscosity measured. ZmADF3 (○) reduces the low shear viscosity of F-actin, whereas zmadf3–1 (▵) has no effect and zmadf3–2 (□) a minor effect.
Binding of Maize ADF3 and the Mutant Proteins, zmadf3–1 and zmadf3–2, to G-Actin.
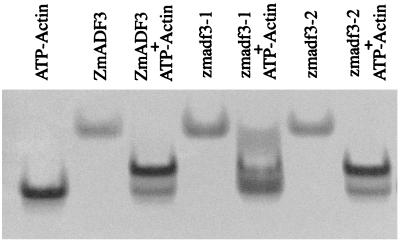
The ability of each of the three recombinant proteins to complex with G-actin was assessed using native gel electrophoresis. G-actin and each of the maize ADF proteins migrate as single bands on nondenaturing gels in the presence of ATP (Fig. 3). A mixture of G-actin with either maize ADF3 or zmadf3–2 gives a major band of intermediate mobility with little unbound actin migrating at the position of the control. In comparison, a mixture of G-actin and zmadf3–1 gives only a minor band of the complex, with most of the actin and the ADF migrating as in controls. There was some smearing of both the zmadf3–1 and actin bands toward the position of the complex, suggesting partial dissociation during electrophoresis. These data suggest that zmadf3–1 binds weakly to G-actin, whereas zmadf3–2 has an affinity for G-actin similar to maize ADF3.
Figure 3.
Native gel electrophoresis showing either ZmADF3, zmadf3–1, or zmadf3–2, and G-actin run separately or as a mixture. ZmADF3, zmadf3–2, and G-actin migrate as single bands, whereas a mixture of either ZmADF3 or zmadf3–2 with G-actin gives a major new band of intermediate mobility. The zmadf3–1 protein migrates as a single band, but a mixture of this protein and G-actin results in a minor band of intermediate mobility.
Binding of Maize ADF3 and the Mutant Proteins, zmadf3–1 and zmadf3–2, to F-Actin.
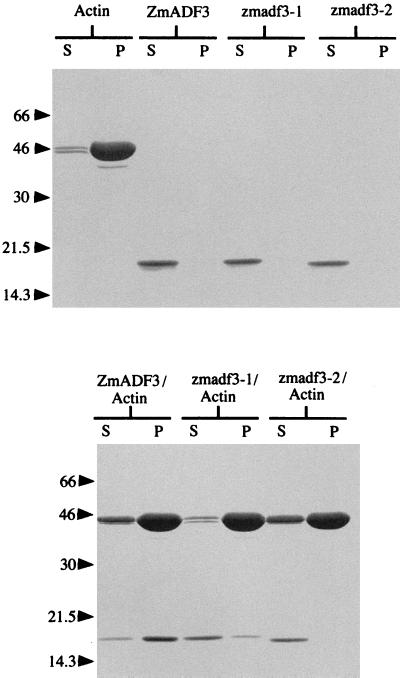
Binding of the three recombinant proteins to F-actin was compared by cosedimentation at pH 7.0. As described previously (9) and shown in Fig. 4, maize ADF3 cosediments with F-actin at 100,000 × g, although a small proportion of the ADF3 is present in the supernatant, probably complexed with G-actin. In contrast, the majority of zmadf3–1 does not cosediment with F-actin, nor is there any significant release of actin into the supernatant as compared with control actin. zmadf3–2 does not cosediment with F-actin, but there is increased actin in the supernatant together with the zmadf3–2. These data suggest that zmadf3–1 binds weakly to F-actin and zmadf3–2 does not bind at all. However, because zmadf3–2 sequesters G-actin, this would explain the release of actin into the supernatant in the sedimentation profile. Similar results were obtained when the mutant proteins were used in cosedimentation assays at pH 7.5 and pH 8.0 (data not shown).
Figure 4.
Sedimentation of either ZmADF3, zmadf3–1, or zmadf3–2, and F-actin separately or as a mixture. At 100,000 × g, ZmADF3, zmadf3–1, and zmadf3–2 alone remain in the supernatant, whereas the F-actin is pelleted (a small proportion of unpolymerized actin is present in the supernatant). When mixed together most of the ZmADF3 protein cosediments with the actin, with a small proportion released into the supernatant. Most of the zmadf-1 protein remains in the supernatant with only a little cosedimenting with actin. The zmadf3–2 protein remains solely in the supernatant with some actin being released into the supernatant.
Effects of the Different ADFs on the Kinetics of Actin Assembly and Disassembly.
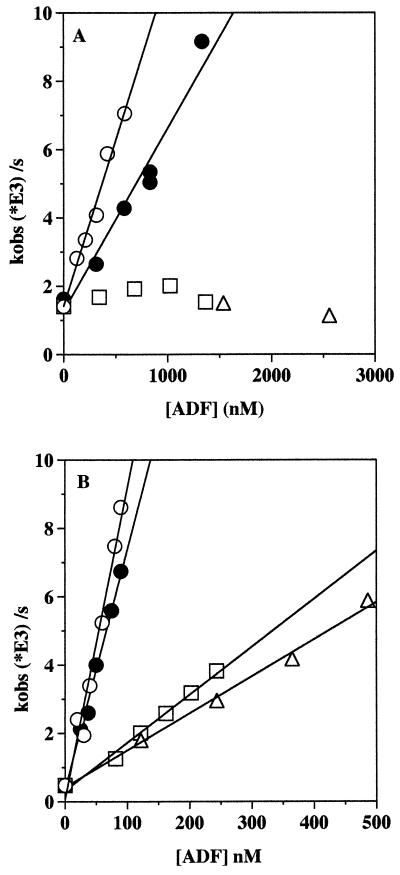
Spectrofluorimetry using pyrenyl labeled actin (17) has been used to compare the effects of maize ADF3, human ADF, and the two mutant ADFs, zmadf3–1 and zmadf3–2, on actin polymerization and depolymerization. Maize ADF3 increases the elongation rate using F-actin nuclei and shows a higher activity in this assay than human ADF (Fig. 5A). In contrast, the two maize mutant proteins had no significant effect on the elongation rate constant.
Figure 5.
(A) Effects of ADF on the rate constants for actin elongation. The second order rate constants calculated from the slopes are: maize ADF3, 0.0097 μM−1⋅s−1 (○); human ADF, 0.0053 μM−1⋅s−1 (•); neither zmadf3–1 (▵) nor zmadf3–2 (□) increase the rate of actin polymerization. (B) Effects of ADF on the rate constants for depolymerization of actin. The second order rate constants calculated from the slopes are: maize ADF3, 0.091 μM−1⋅s−1 (○); human ADF, 0.070 μM−1⋅s−1 (•); zmadf3–1, 0.014 μM−1⋅s−1 (▵); zmadf3–2, 0.011 μM−1⋅s−1 (□).
Maize ADF3 also accelerates the depolymerization rate constant of gelsolin nucleated filaments, again in a manner similar to human ADF (Fig. 5B). The two mutant maize ADFs were much less effective in this assay; zmadf3–1 gave 12% and zmadf3–2 15% the activity of maize ADF3. Taken together, these results are consistent with an ineffective interaction of either of the two mutant proteins with F-actin.
DISCUSSION
The experiments reported here show that maize ADF3 binds quantitatively to both monomeric and polymeric forms of rabbit skeletal muscle actin in a manner similar to human ADF. Furthermore, maize ADF behaves like human ADF in assays that have been developed previously to monitor both the weak severing activity of ADF and its effects on the depolymerization rate at the pointed ends of filaments (46). Indeed, in these two assays shown, the maize ADF3 showed somewhat higher activity than human ADF, but we have noted considerable variation in these assays between different ADF preparations and obtained even higher values for human ADF at pH 6.5 than those of maize ADF3 at pH 8.0 (A.G.W., unpublished data).
In contrast to the actin binding properties of maize ADF3, the two mutants, zmadf3–1 and zmadf3–2, show distinctive properties. zmadf3–1 binds weakly to both G-actin and F-actin in all assays used, whereas zmadf3–2 binds to G-actin in a manner similar to maize ADF3 but shows no binding to actin filaments in the sedimentation assay and has little effect on the low shear viscosity under polymerizing conditions. The effects of these mutants on the kinetics of actin filament assembly and disassembly are fully consistent with these conclusions. The fact that the two mutants have no effect on actin nucleating potential (Fig. 5A), which is considered a measure of severing activity (17), is consistent with the absence of stable filament binding. The small effect in increasing the depolymerization rate of capped filaments (10–15%) of ZmADF3) suggests that there may be some residual effect on filament disassembly at the pointed ends. Carlier et al. (47) have recently demonstrated that the principal effect of ADF/cofilin on F-actin is to increase the rate of treadmilling by accelerating pointed end disassembly.
The recent structural determination of destrin (32) and yeast cofilin (33) has enabled us to analyze the effects of these amino acid substitutions in relation to the three-dimensional structures of ADF. The longest region with the highest similarity between maize ADF3 and destrin (Fig. 1B) is from maize ADF3 Lys-39 to Arg-137 (equivalent to Lys-53 to Lys-151). The similarity is 83%. The major differences between the two proteins can be accounted for by three small deletions in the N-terminal 39 amino acids of maize ADF3. The N-terminal region of destrin includes a phosphorylation site (26, 27) and a nuclear localization signal (35). The putative phosphorylatable serine is conserved in maize ADF3 and we have recently demonstrated that maize ADF3 can be phosphorylated in vitro by plant cell extracts (P.J.H., unpublished data). However, the nuclear localization signal is part of one of the three deletions. In addition, the maize protein is truncated at the C terminus compared with destrin, but this truncation is beyond the end of the final helix of yeast cofilin (33) and actophorin (34).
The results of these experiments cannot be extrapolated quantitatively to the function of these proteins in plant cells, because assays have not been carried out with plant actin. Subtle differences have been reported in the binding of actophorin to Acanthamoeba actin as compared with rabbit actin muscle (48). However, these experiments demonstrate almost identical behavior of plant and human ADF controls in their effects on the dynamics of actin disassembly, and the central theme of this paper concerns specific mutations, the relative effects of which are unlikely to differ with actins from different species.
Characterization of zmadf3–1 shows that the loss of the aromatic hydroxyl on Tyr-103 (equivalent to destrin Tyr-117 and yeast cofilin Tyr-101) markedly diminishes binding of maize ADF3 to both monomeric and polymeric actins. The tyrosine residues in destrin and yeast cofilin corresponding to Tyr-103 have little surface accessibility (3 Å2 and 18 Å2, respectively). However, the adjacent alanine is surface exposed (≈70 Å2 for each protein), but has not been implicated in actin binding and is not conserved in sequence comparisons (frequently being replaced by serine or threonine). Although Tyr-117 is at the center of the long helix (Fig. 6), which Hatanaka et al. (32) have suggested may form part of the actin binding face of destrin, the hydroxyl group points away from the putative actin binding surface, inwards and towards the core of the molecule. This can readily be seen in the stereo image in figure 1 of ref. 34). An Ala to Gly mutation at position 118 should have little impact on the overall structure of the helix, although it might enhance the flexibility of the type I β-turn, which has been reported to distort the long α-helix in the actophorin structure (34). Distortions of this helix have also been reported for destrin and yeast cofilin (32, 33) and models for the cofilin binding sight on F-actin emphasize the possible significance of this distortion (49). We are unable therefore to unequivocally account for the loss of actin binding by this mutant.
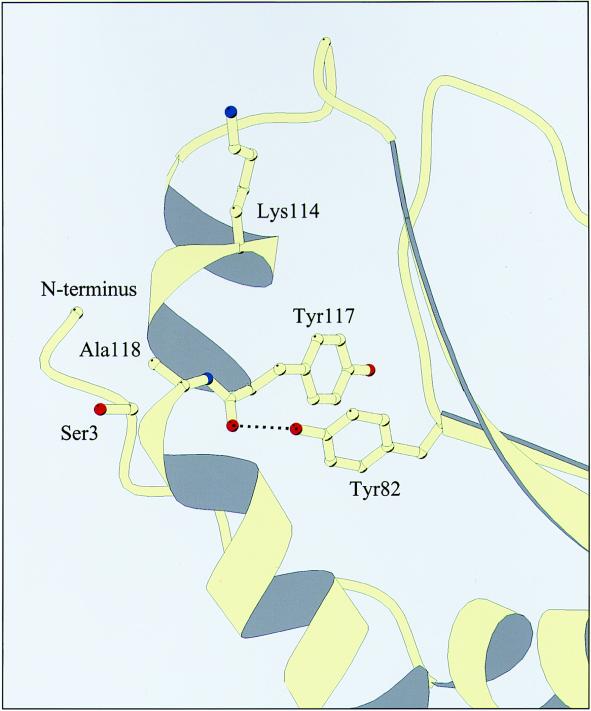
Figure 6.
Part of the tertiary structure of destrin (32) showing the position of the hydrogen bond between the hydroxyl of Tyr-82 and the carbonyl of Tyr-117. Also shown are the positions of Lys-114, essential for actin binding, and the phosphorylatable serine residue (Ser-3).
Analysis of the zmadf3–2 mutant has demonstrated that loss of the aromatic hydroxyl groups from Tyr-67 and Tyr-70 (equivalent to destrin Tyr-82 and Tyr-85 and yeast cofilin Tyr-64 and Tyr-67, respectively) results in an inability of maize ADF3 to bind F-actin, but no loss in G-actin binding. The tyrosine residue corresponding to maize Tyr-67 is totally inaccessible to the surface in both destrin and cofilin, and there is also minimal exposure of the residues corresponding to maize Tyr-70 (≈6 Å in both cases). Examination of the structure of destrin shows that the hydroxyl of Tyr-82 points toward the carbonyl oxygen of Tyr-117 (2.61 Å, 99.5° angle, Fig. 6), i.e., within hydrogen bonding distance. This is also true for the corresponding tyrosine residues in yeast cofilin (2.77 Å, 113° angle). A hydrogen bond within this position would help stabilize the location of the long α-helix in relation to the underlying β-sheet. The aromatic hydroxyl of Tyr-85 has no nearest neighbor and the R-group is directed away from the putative functional domain. This side chain is shown to form part of the hydrophobic core in the structure of actophorin, which stabilizes the central β-sheet and the overall molecular fold. In many ADF variants it is replaced by phenylalanine, which would be equally effective in this role.
The data presented here show that the F-actin and G-actin binding properties of ADF can be uncoupled by mutation of conserved tyrosine residues in zmadf3–2. Taking into account the chemical crosslinking studies, mutation analysis, and peptide competition studies that have identified the region from Trp-104 to Met-115 and Asp-122 to Leu-128 as actin binding regions (22, 29–31), we propose that both G-actin and F-actin binding require the α-helical domain determined by the primary structure from Trp-104 to Leu-128 in destrin, but only F-actin binding requires the tertiary structure afforded by the hydroxyl of Tyr-82 “holding” the α-helix in an essential conformation.
Acknowledgments
We wish to thank Drs. H. Hatanaka and F. Inagaki (Tokyo Metropolitan Institute of Medical Science) for the coordinates of destrin and Dr. S. Almo (Albert Einstein College of Medicine, New York) for providing us with the unpublished coordinates of yeast cofilin. We thank Drs. A. McCoy and T. Bullock for assistance with computer graphics and Dr. McCoy for producing Fig. 6. We also thank Sutherland Maciver (University of Edinburgh) for useful discussions through the course of this work, Brian Pope (Medical Research Council Laboratories, Cambridge) for the CD spectral analysis, Kevin Pyke (Royal Holloway University of London) for help in the preparation of figures, and Timothy Redding and James Armstrong for technical assistance. This work was supported by the Biotechnology and Biological Sciences Research Council (C.-J.J. and P.J.H.) and by the Medical Research Council (A.J.W.).
ABBREVIATION
- ADF
actin depolymerizing factor
References
- 1.Hartwig J H, Kwiatkowski D J. Curr Opin Cell Biol. 1991;3:87–97. doi: 10.1016/0955-0674(91)90170-4. [DOI] [PubMed] [Google Scholar]
- 2.Sun H-Q, Kwiatkowski K, Yin H L. Curr Opin Cell Biol. 1995;7:102–110. doi: 10.1016/0955-0674(95)80051-4. [DOI] [PubMed] [Google Scholar]
- 3.Moon A, Drubin D. Mol Biol Cell. 1995;6:1423–1431. doi: 10.1091/mbc.6.11.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heslop-Harrison J, Heslop-Harrison Y. J Cell Sci. 1989;93:299–308. [Google Scholar]
- 5.Heslop-Harrison Y, Heslop-Harrison J. Ann Bot. 1992;69:385–394. [Google Scholar]
- 6.Heslop-Harrison J, Heslop-Harrison Y. Sex Plant Reprod. 1992;5:247–255. [Google Scholar]
- 7.Staiger C J, Goodbody K C, Hussey P J, Valenta R, Drobak B K, Lloyd C W. Plant J. 1993;4:631–641. doi: 10.1046/j.1365-313x.1993.04040631.x. [DOI] [PubMed] [Google Scholar]
- 8.Valenta R, Duchenne M, Pettenburger K, Sillaber C, Valent P, Bettelheim P, Breitenbach M, Rumpold H, Kraft D, Scheiner O. Science. 1991;253:557–560. doi: 10.1126/science.1857985. [DOI] [PubMed] [Google Scholar]
- 9.Lopez I, Anthony R G, Maciver S K, Jiang C-J, Khan S, Weeds A G, Hussey P J. Proc Natl Acad Sci USA. 1996;93:7415–7420. doi: 10.1073/pnas.93.14.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staiger C J, Yuan M, Valenta R, Shaw P J, Warn R M, Lloyd C W. Curr Biol. 1994;4:215–219. doi: 10.1016/s0960-9822(00)00050-6. [DOI] [PubMed] [Google Scholar]
- 11.Nishida E, Maekawa S, Sakai H. Biochemistry. 1984;23:5307–5313. doi: 10.1021/bi00317a032. [DOI] [PubMed] [Google Scholar]
- 12.Iida K, Moriyama K, Matsumoto S, Kawasaki H, Nishida E, Yahara I. Gene. 1993;124:115–120. doi: 10.1016/0378-1119(93)90770-4. [DOI] [PubMed] [Google Scholar]
- 13.Aizawa H, Sutoh K, Tsubuki S, Kawashima S, Ishii A, Yahara I. J Biol Chem. 1995;270:10923–10932. doi: 10.1074/jbc.270.18.10923. [DOI] [PubMed] [Google Scholar]
- 14.Matsuzaki F, Matsumoto S, Yahara I, Yonezawa N, Nishida E, Sakai H. J Biol Chem. 1988;23:11564–11568. [PubMed] [Google Scholar]
- 15.Adams M E, Minamide L S, Duester G, Bamburg J R. Biochemistry. 1990;29:7414–7420. doi: 10.1021/bi00484a009. [DOI] [PubMed] [Google Scholar]
- 16.Moriyama K, Nishida E, Yonezawa N, Sakai H, Matsumoto S, Iida K, Yahara I. J Biol Chem. 1990;265:5768–5773. [PubMed] [Google Scholar]
- 17.Hawkins M, Pope B, Maciver S K, Weeds A G. Biochemistry. 1993;32:9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- 18.Mabuchi I. J Biochem. 1981;89:1341–1344. [PubMed] [Google Scholar]
- 19.Cooper J A, Blum J D, Williams R C, Jr, Pollard T D. J Biol Chem. 1986;261:477–485. [PubMed] [Google Scholar]
- 20.Rozycka M, Khan S, Lopez I, Greenland A J, Hussey P J. Plant Physiol. 1995;107:1011–1012. doi: 10.1104/pp.107.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maciver S K, Zot H G, Pollard T D. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriyama K, Yonezawa N, Sakai H, Yahara I, Nishida E. J Biol Chem. 1992;267:7240–7244. [PubMed] [Google Scholar]
- 23.Hayden S M, Miller P S, Brauweiler A, Bamburg J R. Biochemistry. 1993;32:9994–10004. doi: 10.1021/bi00089a015. [DOI] [PubMed] [Google Scholar]
- 24.Yonezawa N, Nishida E, Iida K, Yahara I, Sakai H. J Biol Chem. 1990;265:8382–8386. [PubMed] [Google Scholar]
- 25.Morgan T E, Lockerbie R O, Minamide L S, Browning M D, Bamburg J R. J Cell Biol. 1993;115:1611–1620. doi: 10.1083/jcb.122.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agnew B J, Minamide L S, Bamburg J R. J Biol Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- 27.Moriyama K, Iida K, Yahara I. Genes Cells. 1996;1:73–86. doi: 10.1046/j.1365-2443.1996.05005.x. [DOI] [PubMed] [Google Scholar]
- 28.Aizawa H, Sutoh K, Yahara I. J Cell Biol. 1996;132:335–344. doi: 10.1083/jcb.132.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yonezawa N, Homma Y, Yahara I, Sakai H, Nishida E. J Biol Chem. 1991;266:17218–17221. [PubMed] [Google Scholar]
- 30.Yonezawa N, Nishida E, Iida K, Kumagai H, Yahara I, Sakai H. J Biol Chem. 1991b;266:10485–10489. [PubMed] [Google Scholar]
- 31.Yonezawa N, Nishida E, Ohba M, Seki M, Kumagai H, Sakai H. Eur J Biochem. 1989;183:235–238. doi: 10.1111/j.1432-1033.1989.tb14918.x. [DOI] [PubMed] [Google Scholar]
- 32.Hatanaka H, Ogura K, Moriyama K, Ichikawa S, Yahara I, Inagaki F. Cell. 1996;85:1047–1055. doi: 10.1016/s0092-8674(00)81305-7. [DOI] [PubMed] [Google Scholar]
- 33.Fedorov A A, Lappalainen P, Fedorov E V, Drubin D G, Almo S C. Nat Struct Biol. 1997;4:366–369. doi: 10.1038/nsb0597-366. [DOI] [PubMed] [Google Scholar]
- 34.Leonard S A, Gittis A G, Petrulla E C, Pollard T D, Lattman E E. Nat Struct Biol. 1997;4:369–373. doi: 10.1038/nsb0597-369. [DOI] [PubMed] [Google Scholar]
- 35.Iida K, Matsumoto S, Yahara I. Cell Struct Funct. 1992;17:39–46. doi: 10.1247/csf.17.39. [DOI] [PubMed] [Google Scholar]
- 36.Abe H, Nagaoka R, Obinata T. Exp Cell Res. 1993;206:1–10. doi: 10.1006/excr.1993.1113. [DOI] [PubMed] [Google Scholar]
- 37.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maclean-Fletcher S D, Pollard T D. J Cell Biol. 1980;85:329–341. doi: 10.1083/jcb.85.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maciver S K, Weeds A G. FEBS Lett. 1994;347:251–256. doi: 10.1016/0014-5793(94)00552-4. [DOI] [PubMed] [Google Scholar]
- 40.Pope B, Weeds A G. Eur J Biochem. 1986;161:85–93. doi: 10.1111/j.1432-1033.1986.tb10127.x. [DOI] [PubMed] [Google Scholar]
- 41.Way M, Pope B, Weeds A G. J Cell Biol. 1992;119:835–842. doi: 10.1083/jcb.119.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quirk S, Maciver S K, Ampe C, Doberstein S K, Kaiser D A, VanDamme J, Vanderkerckhove J S, Pollard T D. Biochemistry. 1993;32:8525–8533. doi: 10.1021/bi00084a019. [DOI] [PubMed] [Google Scholar]
- 43.Edwards K A, Montague R A, Shepard S, Edgar B A, Erikson R L, Kiehart D P. Proc Natl Acad Sci USA. 1994;91:4589–4593. doi: 10.1073/pnas.91.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKim K S, Matheson C, Marra M A, Wakarchuk M F, Baillie D L. Mol Gen Genet. 1994;242:346–357. doi: 10.1007/BF00280425. [DOI] [PubMed] [Google Scholar]
- 45.Kim S-R, Kim Y, An G. Plant Mol Biol. 1993;21:39–45. doi: 10.1007/BF00039616. [DOI] [PubMed] [Google Scholar]
- 46.Weeds A G, Pope B, Whytock S, Maciver S K. Mol Biol Cell. 1996;7:202a. [Google Scholar]
- 47.Carlier M-F, Laurent V, Santolini J, Malki R, Didry D, Xia G-X, Hong Y, Chua N-H, Pantaloni D. J Cell Biol. 1997;136:1307–1323. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mossakowska M, Korn E D. J Muscle Res Cell Motil. 1996;17:383–389. doi: 10.1007/BF00123355. [DOI] [PubMed] [Google Scholar]
- 49.McGough, A., Pope, B., Chie, W. & Weeds, A. (1997) J. Cell Biol., in press. [DOI] [PMC free article] [PubMed]