Abstract
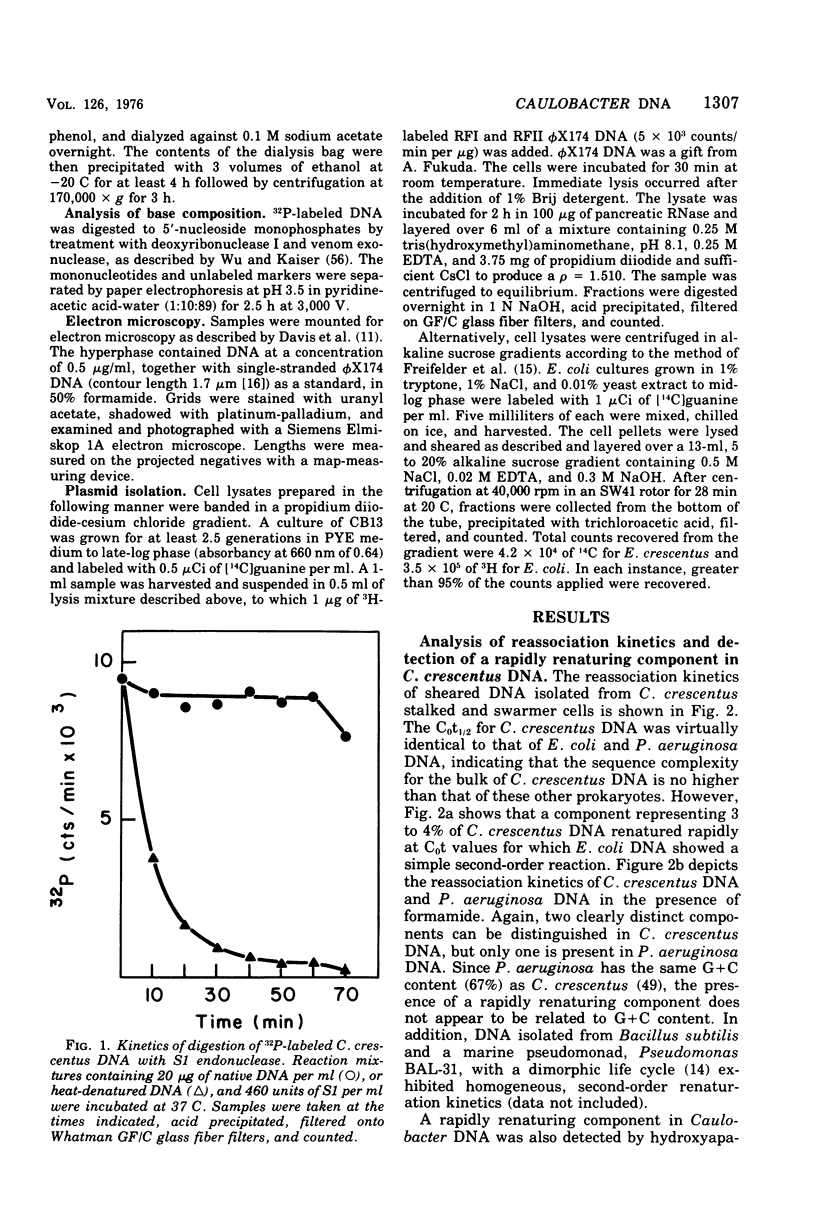
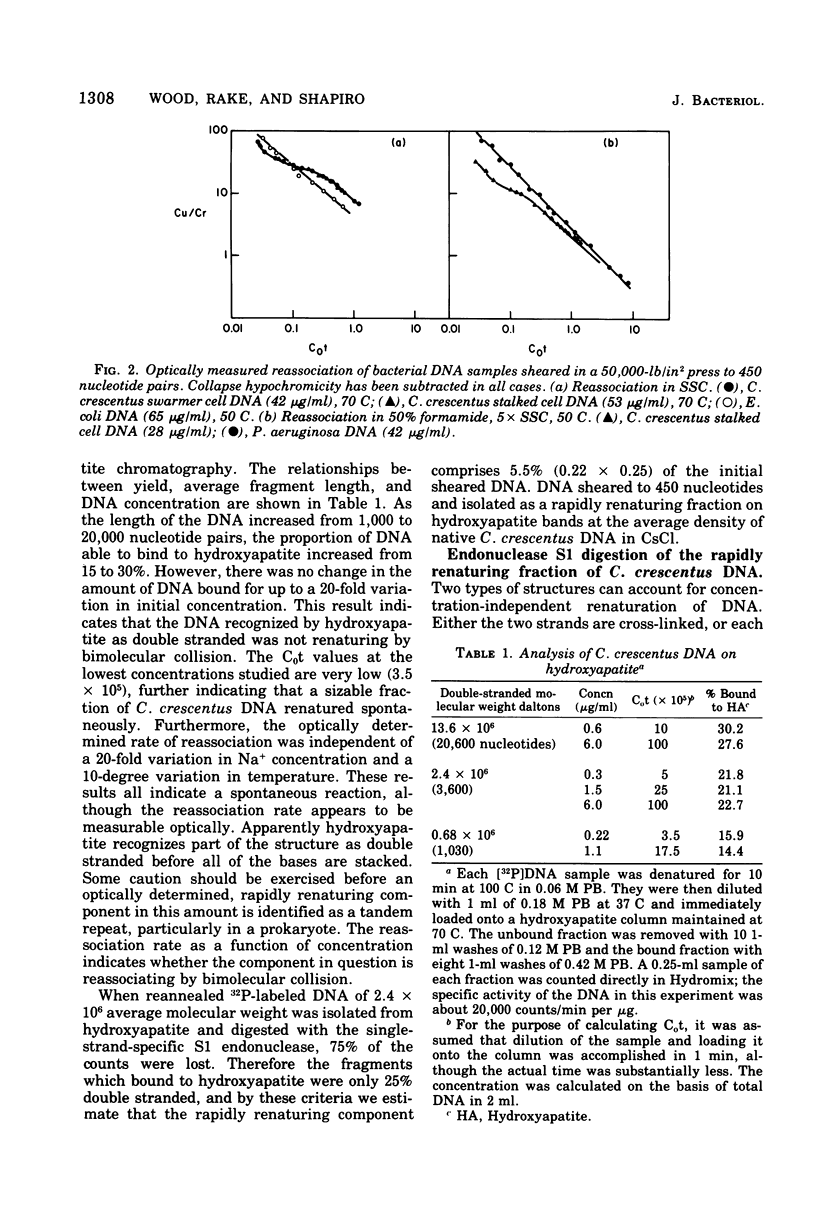
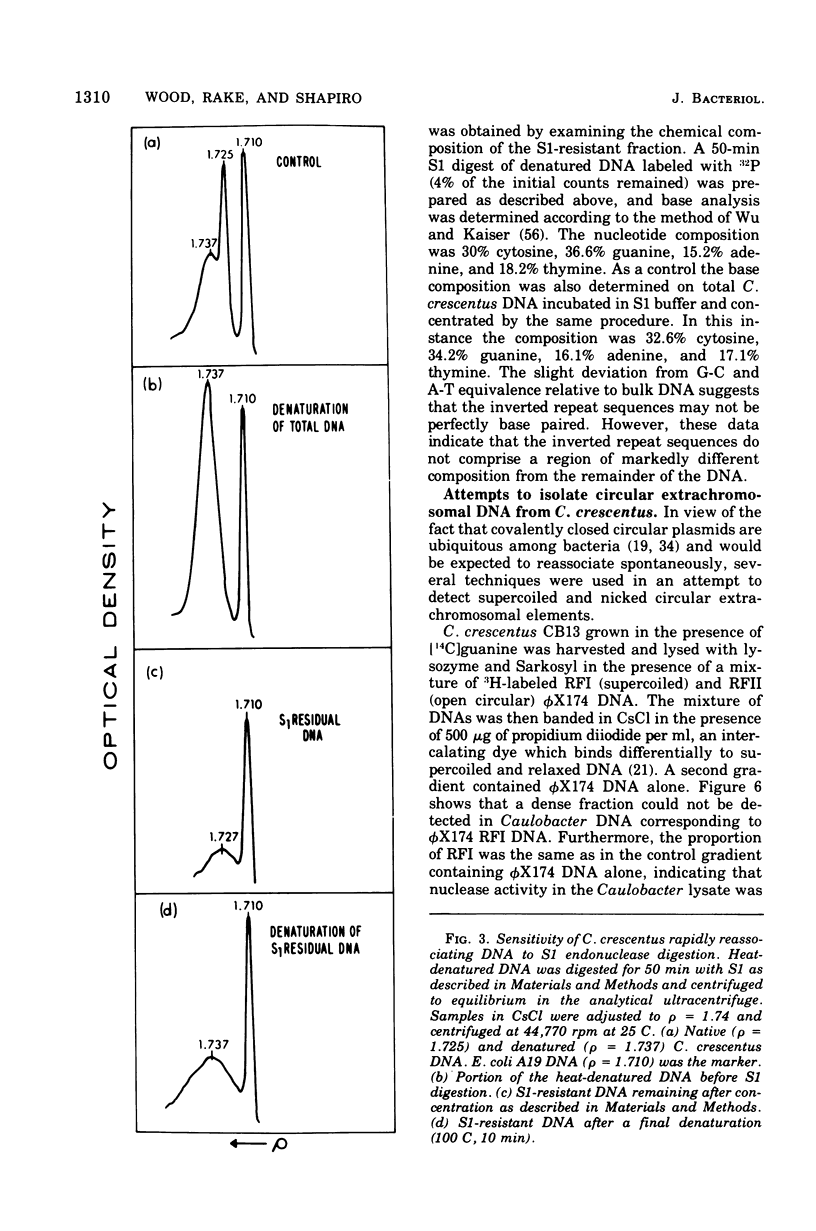
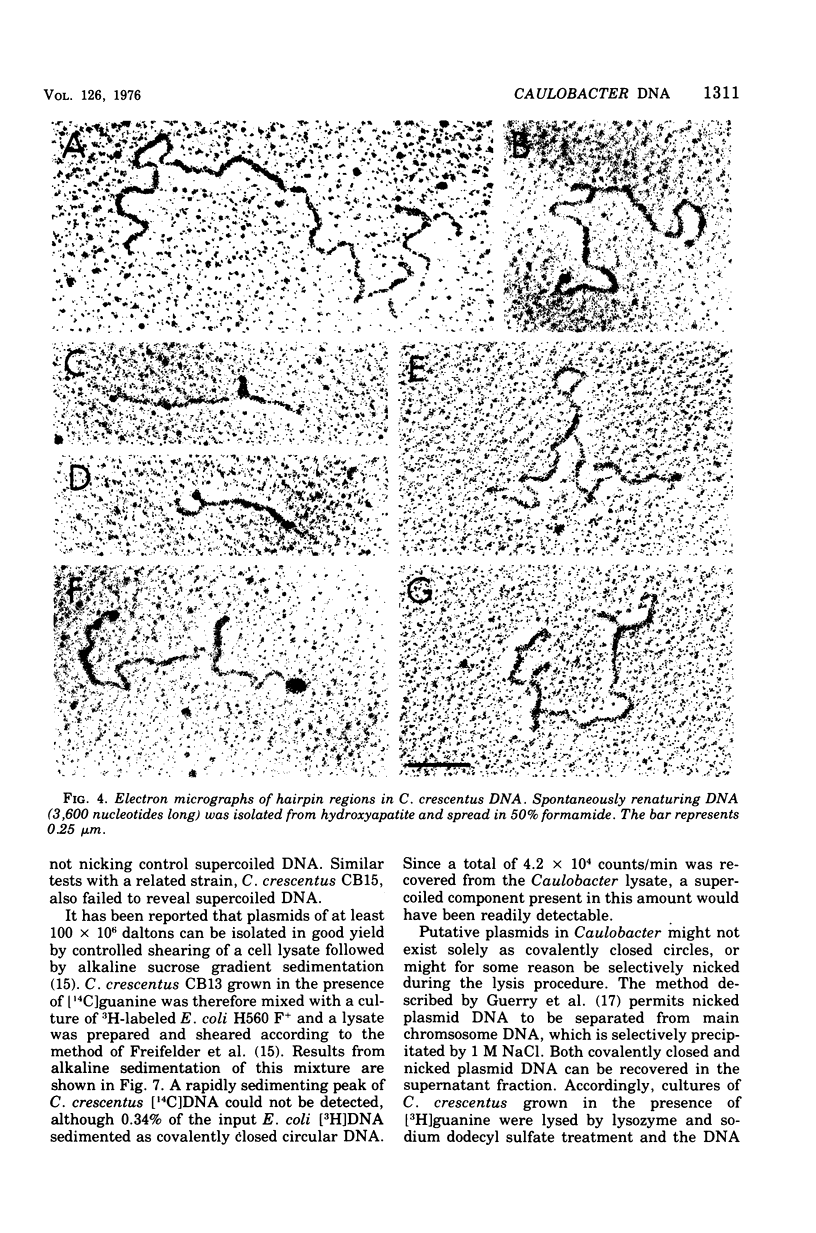
The deoxyribonucleic acid of the dimorphic bacterium Caulobacter crescentus contains a component that renatures with rapid, unimolecular kinetics. This component was present in both swarmer and stalked cells and exhibited the sensitivity to endonuclease S1 expected for hairpin loops. Double-stranded side branches between 100 and 600 nucleotide pairs in length were visible in electron micrographs of rapidly reassociating deoxyribonucleic acid isolated by hydroxyapatite chromatography. No extrachromosomal elements were found in spite of systematic attempts to detect their presence. These results indicate that the rapidly reassociating fraction derives from inverted repeat sequences within the chromosome and not from cross-links or plasmids. We estimate that there are approximately 350 inverted repeat regions per Caulobacter genome. The kinetic complexity of Caulobacter deoxyribonucleic acid, however, is no greater than that of other bacteria.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M. Characterization of a naturally occurring, cross-linked fraction of DNA. II. Origin of the cross-linkage. J Mol Biol. 1968 Mar 14;32(2):405–421. doi: 10.1016/0022-2836(68)90018-1. [DOI] [PubMed] [Google Scholar]
- Alberts B. M., Doty P. Characterization of a naturally occurring, cross-linked fraction of DNA. 1. Nature of the cross-linkage. J Mol Biol. 1968 Mar 14;32(2):379–403. doi: 10.1016/0022-2836(68)90017-x. [DOI] [PubMed] [Google Scholar]
- Barzilai R., Thomas C. A., Jr Spontaneous renaturation of newly-synthesized bacteriophage T7 deoxyribonucleic acid. J Mol Biol. 1970 Jul 14;51(1):145–155. doi: 10.1016/0022-2836(70)90276-7. [DOI] [PubMed] [Google Scholar]
- Bendis I., Shapiro L. Properties of Caulobacter ribonucleic acid bacteriophage phi Cb5. J Virol. 1970 Dec;6(6):847–854. doi: 10.1128/jvi.6.6.847-854.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. O. Molecular hybridization of ribonucleic acid with a large excess of deoxyribonucleic acid. Biochem J. 1972 Jan;126(1):171–185. doi: 10.1042/bj1260171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Rosenfeld A., Hearst J. E. Characterization of the most rapidly renaturing sequences in mouse main-band DNA. J Mol Biol. 1973 Dec 15;81(3):299–325. doi: 10.1016/0022-2836(73)90143-5. [DOI] [PubMed] [Google Scholar]
- Chan H. W., Wells R. D. Structural uniqueness of lactose operator. Nature. 1974 Nov 15;252(5480):205–209. doi: 10.1038/252205a0. [DOI] [PubMed] [Google Scholar]
- Chevallier M. R., Bernardi G. Residual transforming activity of denatured Haemophilus influenzae DNA. J Mol Biol. 1968 Mar 14;32(2):437–451. doi: 10.1016/0022-2836(68)90020-x. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Degnen S. T., Newton A. Chromosome replication during development in Caulobacter crescentus. J Mol Biol. 1972 Mar 14;64(3):671–680. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Abelson J., Barnes W. M., Reznikoff W. S. Genetic regulation: the Lac control region. Science. 1975 Jan 10;187(4171):27–35. doi: 10.1126/science.1088926. [DOI] [PubMed] [Google Scholar]
- Espejo R. T., Canelo E. S. Properties of bacteriophage PM2: a lipid-containing bacterial virus. Virology. 1968 Apr;34(4):738–747. doi: 10.1016/0042-6822(68)90094-9. [DOI] [PubMed] [Google Scholar]
- FREIFELDER D., KLEINSCHMIDT A. K., SINSHEIMER R. L. ELECTRON MICROSCOPY OF SINGLE-STRANDED DNA: CIRCULARITY OF DNA OF BACTERIOPHAGE PHI-X174. Science. 1964 Oct 9;146(3641):254–255. doi: 10.1126/science.146.3641.254. [DOI] [PubMed] [Google Scholar]
- Freifelder D., Folkmanis A., Kirschner I. Studies on Escherichia coli sex factors: evidence that covalent circles exist within cells and the general problem of isolation of covalent circles. J Bacteriol. 1971 Mar;105(3):722–727. doi: 10.1128/jb.105.3.722-727.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helinski D. R., Clewell D. B. Circular DNA. Annu Rev Biochem. 1971;40:899–942. doi: 10.1146/annurev.bi.40.070171.004343. [DOI] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M. T., Davidson N. Electron microscope heteroduplex study of the heterogeneity of Mu phage and prophage DNA. Virology. 1974 Mar;58(1):229–239. doi: 10.1016/0042-6822(74)90157-3. [DOI] [PubMed] [Google Scholar]
- Hudson B., Upholt W. B., Devinny J., Vinograd J. The use of an ethidium analogue in the dye-buoyant density procedure for the isolation of closed circular DNA: the variation of the superhelix density of mitochondrial DNA. Proc Natl Acad Sci U S A. 1969 Mar;62(3):813–820. doi: 10.1073/pnas.62.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecko D. J., Cohen S. N. Site specific recA--independent recombination between bacterial plasmids: involvement of palindromes at the recombinational loci. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1373–1377. doi: 10.1073/pnas.72.4.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A. Active center of DNA polymerase. Science. 1969 Mar 28;163(3874):1410–1418. doi: 10.1126/science.163.3874.1410. [DOI] [PubMed] [Google Scholar]
- Kurn N., Ammer S., Shapiro L. A pleiotropic mutation affecting expression of polar development events in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3157–3161. doi: 10.1073/pnas.71.8.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker J., Rabinowitz M., Getz G. S. Tandem inverted repeats in mitochondrial DNA of petite mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1366–1370. doi: 10.1073/pnas.71.4.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Ptashne M., Barrell B. G., Donelson J. Sequence of a repressor-binding site in the DNA of bacteriophage lamda. Nature. 1974 Aug 2;250(465):394–397. doi: 10.1038/250394a0. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Mulder C., Doty P. Residual activity of denatured transforming DNA of Haemophilus influenzae: a natrually occurring cross-linked DNA. J Mol Biol. 1968 Mar 14;32(2):423–435. doi: 10.1016/0022-2836(68)90019-3. [DOI] [PubMed] [Google Scholar]
- Newton A., Allebach E. Gene transfer in Caulobacter crescentus: polarized inheritance of genetic markers. Genetics. 1975 May;80(1):1–11. doi: 10.1093/genetics/80.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Sequence of the OR operator of phage lambda. Nature. 1975 Mar 13;254(5496):114–117. doi: 10.1038/254114a0. [DOI] [PubMed] [Google Scholar]
- Pouwels P. H., Knijnenburg C. M., van Rotterdam J., Cohen J. A. Structure of the replicative form of bacteriphage phi X174. VI. Studies on alkali-denatured double-stranded phi X DNA. J Mol Biol. 1968 Mar 14;32(2):169–182. doi: 10.1016/0022-2836(68)90002-8. [DOI] [PubMed] [Google Scholar]
- Ptashne K., Cohen S. N. Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. J Bacteriol. 1975 May;122(2):776–781. doi: 10.1128/jb.122.2.776-781.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rownd R., Green D. M., Sternglanz R., Doty P. Origin of the residual transforming activity of denatured Bacillus subtilis DNA. J Mol Biol. 1968 Mar 14;32(2):369–377. doi: 10.1016/0022-2836(68)90016-8. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Manning J. E., Davidson N. Inverted repeat sequences in the Drosophila genome. Cell. 1975 Jun;5(2):159–172. doi: 10.1016/0092-8674(75)90024-0. [DOI] [PubMed] [Google Scholar]
- Schmidt J. M. Observations on the adsorption of Caulobacter bacteriophages containing ribonucleic acid. J Gen Microbiol. 1966 Nov;45(2):347–353. doi: 10.1099/00221287-45-2-347. [DOI] [PubMed] [Google Scholar]
- Schmidt J. M., Stanier R. Y. The development of cellular stalks in bacteria. J Cell Biol. 1966 Mar;28(3):423–436. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T., Khorana H. G. Nucleotide sequence in the promoter region of the Escherichia coli tyrosine tRNA gene. Proc Natl Acad Sci U S A. 1974 Aug;71(8):2978–2982. doi: 10.1073/pnas.71.8.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L., Agabian-Keshishian N., Bendis I. Bacterial differentiation. Science. 1971 Sep 3;173(4000):884–892. doi: 10.1126/science.173.4000.884. [DOI] [PubMed] [Google Scholar]
- Shapiro L., Agabian-Keshishian N., Hirsch A., Rosen O. M. Effect of dibutyryladenosine 3':5'-cyclic monophosphate on growth and differentiation in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1972 May;69(5):1225–1229. doi: 10.1073/pnas.69.5.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L., Agabian-Keshishian N. Specific Assay for Differentiation in the Stalked Bacterium Caulobacter crescentus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):200–203. doi: 10.1073/pnas.67.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L., Maizel J. V., Jr Synthesis and structure of Caulobacter crescentus flagella. J Bacteriol. 1973 Jan;113(1):478–485. doi: 10.1128/jb.113.1.478-485.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Okamoto T., Sugisaki H., Takanami M. The nucleotide sequence of an RNA polymerase binding site on bacteriophage fd DNA. Nature. 1975 Feb 6;253(5491):410–414. doi: 10.1038/253410a0. [DOI] [PubMed] [Google Scholar]
- Sutton W. D. A crude nuclease preparation suitable for use in DNA reassociation experiments. Biochim Biophys Acta. 1971 Jul 29;240(4):522–531. doi: 10.1016/0005-2787(71)90709-x. [DOI] [PubMed] [Google Scholar]
- Wesley R. D. Inverted repetitious sequences in the macronuclear DNA of hypotrichous ciliates. Proc Natl Acad Sci U S A. 1975 Feb;72(2):678–682. doi: 10.1073/pnas.72.2.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Hydroxyapatite chromatography of short double-helical DNA. Biochim Biophys Acta. 1973 Dec 21;331(3):333–340. doi: 10.1016/0005-2787(73)90019-1. [DOI] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Palindromes in chromosomes. J Mol Biol. 1974 Mar 25;84(1):115–138. doi: 10.1016/0022-2836(74)90216-2. [DOI] [PubMed] [Google Scholar]
- Wu R., Kaiser A. D. Mapping the 5'-terminal nucleotides of the DNA of bacteriophage lambda and related phages. Proc Natl Acad Sci U S A. 1967 Jan;57(1):170–177. doi: 10.1073/pnas.57.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]




