Abstract
Internodes of deepwater rice are induced to grow rapidly when plants become submerged. This adaptation enables deepwater rice to keep part of its foliage above the rising flood waters during the monsoon season and to avoid drowning. This growth response is, ultimately, elicited by the plant hormone gibberellin (GA). The primary target tissue for GA action is the intercalary meristem of the internode. Using differential display of mRNA, we have isolated a number of genes whose expression in the intercalary meristem is regulated by GA. The product of one of these genes was identified as an ortholog of replication protein A1 (RPA1). RPA is a heterotrimeric protein involved in DNA replication, recombination, and repair and also in regulation of transcription. A chimeric construct, in which the single-stranded DNA-binding domain of rice RPA1 was spliced into the corresponding region of yeast RPA1, was able to complement a yeast rpa1 mutant. The transcript level of rice RPA1 is high in tissues containing dividing cells. RPA1 mRNA levels increase rapidly in the intercalary meristem during submergence and treatment with GA before the increase in the level of histone H3 mRNA, a marker for DNA replication.
Keywords: cell division, intercalary meristem, internodal growth, Oryza sativa
Deepwater rice (Oryza sativa L.) is a subsistence crop in regions of Southeast Asia that are flooded during the monsoon season (1). To avoid drowning, deepwater rice has evolved the capacity to elongate very rapidly when it becomes submerged. This adaptation permits deepwater rice to keep part of its foliage above the rising flood waters. In the flood plains of Bangladesh, elongation rates of up to 25 cm day−1 have been reported (2); in our laboratory, we have measured growth rates of up to 5 mm h−1 (3). Understanding the physiological and molecular basis of the growth response in deepwater rice is important for two reasons. It may help in identifying the gene(s) that could confer elongation capacity onto modern, high-yielding rice cultivars. Deepwater rice is also an excellent object in which to study basic aspects of plant growth because of its unusually high growth rate, which is under environmental and hormonal control.
In earlier work, we examined the environmental and hormonal regulation of the growth response in deepwater rice and the cellular basis of rapid internodal elongation. The plant hormone ethylene accumulates in submerged internodes because of enhanced synthesis under reduced partial pressures of O2 and because of its low rate of diffusion from the plant into the surrounding water (4). The interaction of ethylene and two other plant hormones—abscisic acid and gibberellin (GA)—determines the growth rate of the plant. Ethylene renders the internode more responsive to GA (5), at least in part by lowering the level of endogenous abscisic acid, a potent antagonist of GA action in rice (6). Growth of the internode is, ultimately, promoted by GA (5).
The primary target tissue of GA is the intercalary meristem of the internode, where GA enhances cell division activity and cell elongation (5, 7, 8). The intercalary meristem is a zone of about 3 mm in length and is located at the base of the internode (9). Cells are displaced from the intercalary meristem into the elongation zone, where they reach their final size. In GA-treated internodes, the final cell length is about three to four times longer than in control internodes (5, 7). Correspondingly, the length of the elongation zone expands from about 10 to 35 mm upon treatment with GA (8). Growth stops above the elongation zone in the differentiation zone, where lignification of developing metaxylem and cortical sclerenchyma takes place (9, 10).
We have investigated the effect of GA on cell division activity in the intercalary meristem and have correlated the progression of cells through the cell cycle to molecular events that regulate it. The fraction of meristematic cells in the G2 phase declined within 4 hr of GA treatment, indicating that these cells had entered mitosis (7). The expression of two cyclin genes in the intercalary meristem was enhanced by GA, and the time course of induction was compatible with a role for both cyclins in regulating the transition from the G2 to M phase (11). Between 4 and 7 hr of incubation in GA, the rate of [3H]thymidine incorporation into DNA doubled, showing an increase in DNA synthesis (7). In a screen for GA-regulated gene expression using differential display of mRNA (12), we identified a histone H3 gene whose transcript level increased in parallel with the GA-induced rise in DNA synthesis (13). In this report, we are describing the results of further screening, which led to the identification of a gene that encodes an ortholog of replication protein A1 (RPA1). In the internode, this gene is expressed in the intercalary meristem, and its transcript level increases as a result of GA treatment and submergence.
MATERIALS AND METHODS
Plant Material.
Deepwater rice (Oryza sativa L., cv. Pin Gaew 56) was obtained from the International Rice Research Institute, Los Baños, Philippines, and grown as described (3). For submergence experiments, 12-week-old plants were partially immersed in deionized water (14) and kept under continuous light. Stem sections containing the growing internode were excised and treated with 50 μM gibberellin A3 (gibberellic acid, GA3) for the periods indicated (4).
Differential Display of mRNA.
RNA was isolated according to Puissant and Houdeline (15) from the intercalary meristem of internodes treated for 2 hr with GA3 or distilled water as control. Before reverse transcription, the RNA was treated with DNase I (Boehringer Mannheim) to remove residual DNA. Differential display (12) was performed with RNAimage kits (GenHunter, Nashville, TN) with slight modifications of the manufacturer’s specifications. Briefly, 0.2 μg of RNA was reverse-transcribed in a total volume of 20 μl in the presence of 20 μM dNTP and 0.2 μM H-T11M for 1 hr. Two microliters of cDNA was amplified with the same H-T11M (0.2 μM) primer and 0.2 μM H-AP primer in the presence of 4 μM dNTP and 0.25 μl [α-33P]dATP (2,000 Ci/mmol, New England Nuclear) in a total volume of 20 μl. PCR conditions were: 95°C for 30 sec, 40°C for 2 min, and 72°C for 1 min over 40 cycles. PCR products were separated on a 6% DNA sequencing gel in a Genomyx programmable DNA sequencer (Foster City, CA) and visualized by autoradiography. Using the primers 5′-AAGCTTTTTTTTTTTA-3′ and 5′-AAGCTTTTGAGGT-3′, a differentially displayed cDNA band, dd12, was identified. After reamplification using the above PCR conditions, but at a dNTP concentration of 20 μM in a total volume of 40 μl, the dd12 cDNA was ligated into the pGEM-T vector (Promega). Differential expression of the dd12 transcript was confirmed by Northern blotting.
Northern Blot Analysis.
Twenty micrograms of total RNA was loaded on 1.2% agarose-formaldehyde gels (16) and transferred to Hybond N membrane (Amersham). Blots were prehybridized in 5 × standard saline citrate (SSC)/10 × Denhardt’s solution (17)/0.1% SDS/0.1 M K-phosphate, pH 6.8/100 μg/ml denatured salmon sperm DNA for 4 hr at 42°C, and hybridized in 5 × SSC/10 × Denhardt’s solution/0.1 M K-phosphate, pH 6.8/10% dextran sulfate/30% formamide/100 μg/ml denatured salmon sperm DNA overnight at 42°C with a probe prepared in the presence of [α-32P]dCTP (New England Nuclear) using a random prime labeling kit (Boehringer Mannheim). High-stringency washes were performed with 0.1 × SSC and 0.1% SDS at 65°C twice for 30 min. The radioactivity on blots was quantified by PhosphorImager analysis (Molecular Dynamics). All values were normalized for equal loading with E37, a cDNA corresponding to a transcript whose expression does not change during treatment with GA3 (13).
Sequence Analysis.
A full-length cDNA clone corresponding to the PCR product dd12 and henceforth called DD12 was isolated from a rice intercalary meristem cDNA library and cloned into the EcoRI site of pBluescript SK(−) phagemid (Stratagene). Sequence analysis was performed at the W.M. Keck facility at Yale University (New Haven, CT).
In Vitro Mutagenesis.
DD12 was subcloned into the EcoRI-SphI site of pALTERR-1 (Promega). The SphI site of DD12 was located 150 bp from the 3′ end of DD12 within the 3′ untranslated region. To create an NdeI cloning site at the 5′ end of the nucleotide sequence encoding the single-stranded DNA-binding domain SBD-A and a BamHI site at the 3′ end of the nucleotide sequence encoding SBD-B, 5′-GGCGCGGTTGCATATGACGAGAAGAGTT-3′ and 5′-GCTTGGTATGACGGATCCGGCAAGGGTACT-3′, respectively, were used as primers for mutagenesis (Fig. 1A). The internal NdeI site was eliminated by mutagenesis using 5′-AGCTAGGGCCTTATGTTGGTG-3′ as primer. The construct thus formed was called pALTER-S1. In vitro mutagenesis was performed according to manufacturer’s (Promega) specifications. Mutagenesis over the primer regions was verified by sequence analysis.
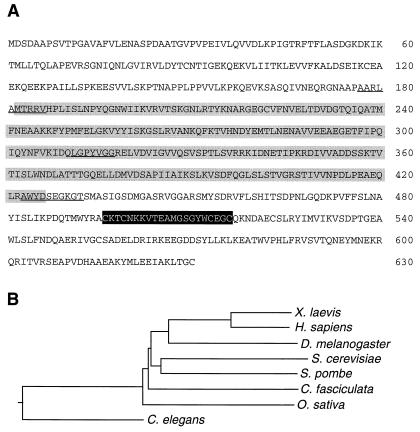
Figure 1.
(A) Amino acid sequence of DD12 (Os-RPA1). The lightly shaded amino acid sequences comprise SBD-A and SBD-B, the darker shaded sequence the zinc finger domain. The locations of the primers used for in vitro mutagenesis are underlined. (B) Phylogenetic analysis of RPA1 genes from the species indicated. The dendogram was constructed using the clustal method with the PAM250 residue weight table. The accession numbers are: Q01588 (Xenopus laevis), P27694 (H. sapiens), Z70277 (Drosophila melanogaster), P22336 (Saccharomyces cerevisiae), U59385 (Schizosaccharomyces pombe), S38458 (Crithidia fasciculata), AF009179 (O. sativa), and U41535 (C. elegans).
Yeast Complementation.
pDS1, a yeast shuttle vector containing wild-type yeast RPA1 on pRS415 (LEU2) (18), was digested with NdeI and BamHI to release the region of the yeast SBD-A and SBD-B domains. pALTER-S1 was digested with NdeI and BamHI, and the rice SBD-A and SBD-B region was cloned into the NdeI-BamHI site of pDS1. This resulted in the replacement of the yeast SBD-A and SBD-B with the rice SBD-A and SBD-B, and this construct was called pDS9. The yeast strain SBY102 (MATα, ade2–1, can1–100, leu2–3, 112, his3–11, ura3–1, Δrpa1::TRP1; ref. 18) containing wild-type RPA1 on the shuttle vector YCp50 (URA3) was transformed with the appropriate construct by a modified LiAc method (19) and selected on synthetic complete medium without leucine (20). To remove the wild-type yeast RPA1 on YCp50, the colonies were selected on the same medium containing 5-fluoroorotic acid (21).
RESULTS
Identification of a GA-Regulated Gene.
We used differential display of mRNA (12, 13) to identify genes whose transcript level in the intercalary meristem was altered within 2 hr of GA treatment. A 239-bp differentially displayed PCR product, dd12, appeared in GA-treated tissue and was further investigated. The cDNA was reamplified, cloned, and used as probe to verify by Northern blot analysis that the corresponding transcript indeed accumulated as a result of GA treatment (results not shown, see also Fig. 4).
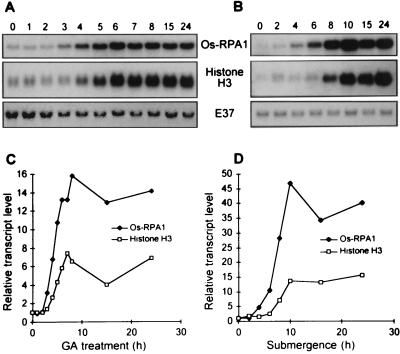
Figure 4.
Change in Os-RPA1 transcript levels in the intercalary meristem during treatment of stem sections with 50 μM GA3 and during submergence of whole plants. (A) Northern blot analysis of RNA from the intercalary meristem of stem sections treated with GA3 for the periods in hours indicated above the lanes. The same blot was hybridized to Os-RPA1, the histone H3 cDNA probe, and E37. (B) Northern blot analysis of RNA from the intercalary meristem of plants submerged for the periods in hours indicated above the lanes. The same blot was hybridized to Os-RPA1, the histone H3 cDNA probe, and E37. (C) Quantitative analysis of the Northern blot shown in A using a PhosphorImager. All values were normalized to the E37 loading control. (D) Quantitative analysis of the Northern blot shown in B.
Sequence Analysis of DD12.
A rice internode-specific cDNA library was screened with the differentially displayed and subcloned dd12 PCR product. A full-length clone of 2.3 kb was isolated whose sequence showed an ORF from position 55 to 1,944, encoding a protein of 69.6-kDa predicted molecular mass (Fig. 1A). Database searches indicated amino acid similarity to RPA1 from other organisms. RPA complexes are heterotrimers with subunits of approximately 70 (RPA1), 30 (RPA2), and 14 (RPA3) kDa (22). RPA was first identified as a factor necessary to support simian virus 40 replication (23–25). Later, it also was found to be necessary for recombination (26, 27) and for DNA repair (28, 29). DD12 encodes a protein containing two contiguous single-stranded DNA-binding domains, SBD-A and SBD-B (Fig. 1A, lightly shaded amino acid sequences), which share similarity with Escherichia coli single-stranded DNA-binding domains (18). DD12 also encodes a zinc finger motif (Fig. 1A, dark-shaded amino acid sequences), which is conserved in all RPA1 proteins but whose function is unknown. The phylogenetic relationships between all known RPA1 genes in the database are given in Fig. 1B. The percentage amino acid identity between rice and other RPA1 proteins ranges from 33.3% (Homo sapiens) to 24.5% (Crithidia fasciculata) based on pairwise comparisons using align. The percentage amino acid identity of the SBD-A and SBD-B region varies from 44.9% (H. sapiens) to 33.1% (Caenorhabditis elegans).
Yeast Complementation.
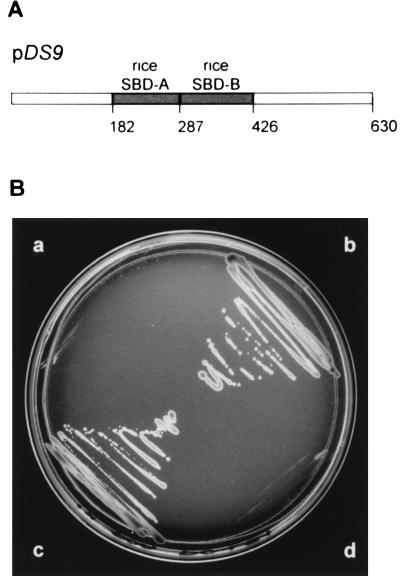
It has been suggested that species-specific interactions between RPA and other cellular components account for the inability of yeast RPA to function in simian virus 40 DNA replication (30) and for the failure of human RPA2 to complement a yeast rpa2 mutant (31). The C-terminal and N-terminal regions of RPA1 interact with other cellular factors (see below) and are less conserved among each other than are SBD-A and SBD-B. Replacement of yeast SBD-A and SBD-B with human SBD-A and SBD-B was shown before to rescue a yeast rpa1 mutant (18). We constructed a chimeric clone, pDS9, encoding a protein with the rice SBD-A and SBD-B domains between the yeast N-terminal and C-terminal domains (Fig. 2A). Fig. 2B shows the result of the complementation experiment. Both pDS1 containing the wild-type yeast RPA1 (sector b), and pDS9 containing the rice-yeast chimera (sector c) rescued the yeast mutant, whereas pJM241 containing yeast RPA2 (sector d) did not. After selection, the colonies in sectors b and c were not able to grow on synthetic complete medium without uracil, which indicates loss of the original yeast RPA1 gene on the YCp50 vector (data not shown). Also, dot blot analysis showed the presence of the yeast-rice chimera in colonies from sector c (data not shown). These results confirmed that the protein encoded by DD12 is an ortholog of RPA1. Therefore, DD12 henceforth will be called Os-RPA1.
Figure 2.
Yeast complementation. (A) Schematic representation of the chimeric protein encoded by pDS9 containing the rice SBD-A and SBD-B domains (shaded) between the yeast N-terminal and C-terminal regions. The numbers above the diagram represent the amino acid positions of Os-RPA1. (B) Transformed yeast was plated out on complete synthetic medium (minus leucine, plus 5-fluoroorotic acid). Sector a, SBY102 nontransformed; sector b, SBY102 transformed with pDS1, which contains the wild-type RPA1 of yeast; sector c, SBY102 transformed with pDS9; sector d, SBY102 transformed with pJM241, which contains the wild-type RPA2 of yeast on pRS415 (LEU2).
Tissue-Specific Expression of Os-RPA1.
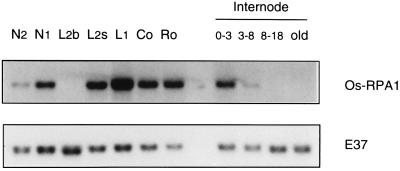
mRNA was detected in tissues that contain dividing cells, namely in the highest node, which includes the apical meristem, in the sheath of the second youngest leaf, in the youngest leaf, in root tips, and in the coleoptile (Fig. 3). Os-RPA1 transcript also was expressed in the intercalary meristem of the internode; a trace of Os-RPA1 mRNA was also evident in the internodal region just above it, which probably still contains some meristematic cells at its base but which, otherwise, consists of elongating cells. No Os-RPA1 transcript was detected in the differentiation zone and in the oldest part of the internode.
Figure 3.
Northern blot analysis of RNA from different parts of the rice plant. N2, second highest node; N1, highest node containing the apical meristem; L2b, basal 2 cm of the second leaf blade; L2s, basal 2 cm of second leaf sheath; L1, youngest leaf; Co, coleoptile, 3 days after germination; Ro, root, 3 days after germination; 0–3, internodal region 0–3 mm above N2, containing the intercalary meristem; 3–8, internodal region 3–8 mm above N2, containing mostly the elongation zone; 8–18, internodal region 8–18 mm above N2, containing the upper part of the elongation zone and the differentiation zone; old, oldest part of the internode. (Upper) The hybridization signals with Os-RPA1 as probe. (Lower) The hybridization signals with E37 used as internal loading control.
The Time Course of Os-RPA1 Expression and its Correlation with the Cell Cycle.
The level of Os-RPA1 transcript in the intercalary meristem increased after 2 to 3 hr of treatment with GA3 and reached a maximum after 8 hr (Fig. 4A, Top). The increase in transcript level of Os-RPA1 was not observed in control stem sections (data not shown). Submergence of whole plants also caused an increase in accumulation of Os-RPA1 mRNA (Fig. 4B, Top). These same blots also were hybridized to E37 as loading control (Fig. 4 A and B, Bottom).
Because RPA is involved in DNA replication, we were interested in correlating the increase in transcript level of Os-RPA1 to that of histone H3, which is a marker for the S-phase of the cell cycle (13). For this purpose, the Northern blots shown in Fig. 4 A and B were hybridized to the histone H3 probe (Fig. 4 A and B, Middle). The signals were quantified by PhosphorImager analysis and normalized for equal loading using E37 as internal standard. Taking a 3-fold higher mRNA level over the 0-hr time point as a significant increase, we found that the rise in Os-RPA1 transcript level preceded that of histone H3 by 2 hr in GA-treated internodes (Fig. 4C) and by 4 hr in internodes of submerged plants (Fig. 4D).
DISCUSSION
In our search for GA-regulated genes in deepwater rice, we identified a gene, DD12, whose transcript level increased early in response to GA treatment and submergence. Sequence analysis of DD12 indicated similarity to RPA1 genes from several organisms. Replacement of the SBD domain of yeast RPA1 with the homologous domain from DD12 yielded a construct that was used successfully to complement a yeast rpa1 mutant. Based on these results, we concluded that DD12 is an ortholog of RPA1 and called it Os-RPA1. To date, Os-RPA1 is the only identified plant RPA1 gene in the database, although apparent Arabidopsis and maize homologs exist in the EST database.
In synchronized yeast cells, increased transcript levels of RPA1 correlated with the late G1 to S-phase (31). This is also the case for the expression of several other yeast replication genes whose transcript levels increased before the accumulation of histone H2A-H2B mRNA (32). We found a similar trend in the intercalary meristem of rice internodes. Particularly during submergence (Fig. 4 B and D), the increase in Os-RPA1 mRNA levels preceded the onset of DNA replication, as indicated by the accumulation of histone H3 mRNA (13). On the basis of these results, it appears that in rice, as in yeast, expression of replication proteins is regulated differently than that of histones.
RPA1 encodes the largest subunit of the heterotrimeric complex RPA and contains three functional domains (Figs. 1A and 2A). The C-terminal region mediates the interaction with the other two subunits, RPA2 and RPA3 (33–35). The primary function of SBD-A and SBD-B is to bind single-stranded DNA (18, 34–36); however, SBD is also able to bind to damaged and double-stranded DNA (see below, and refs. 37–39). The N-terminal domain is important for protein-protein interactions, e.g., with the transcriptional activators GAL4 and VP16 (40, 41), with the tumor suppressor p53 (42), and possibly with other proteins of DNA metabolism. However, the precise role of RPA in these interactions is unclear (for a recent comprehensive review of RPA structure and function, see ref. 22).
In addition to its role in DNA replication, repair, and recombination, RPA is implicated in transcriptional regulation of several genes. RPA was identified as a protein factor that bound specifically to the upstream repression sequence of the promoter of the yeast CAR1 gene, which is involved in nitrogen metabolism (43), to a similar element in the promoter of the yeast MAG gene involved in DNA repair (44), and to a similar element in the promoter of the yeast FOX3 gene, which is required for peroxisome functioning (45). Based on sequence similarity, several cis elements, to which RPA binds, were identified in the promoters of many more genes involved in basic cell metabolism. Functional analysis of these cis elements showed that most of them act as upstream repression sequences, and some as upstream activation sequences. This indicates a role for RPA in both transcriptional repression and activation and in coordination of gene expression. The involvement of RPA in transcriptional regulation is found not only in yeast. The transcription of the human metallothionine IIA gene is repressed by RPA both in vitro and in vivo (46).
In conclusion, rice RPA probably is involved in submergence- and GA-enhanced DNA replication. In addition, it also may play a role in coordinating general transcription that accompanies accelerated growth.
Acknowledgments
We are greatly indebted to Dr. Steven J. Brill (Rutgers University, Piscataway, NJ) for providing yeast vectors, strains, and advice for our yeast complementation studies; to Dr. Marc S. Wold (University of Iowa, Iowa City, IA) for sending us a preprint of his review on RPA; and to both for critically reading our manuscript. This work was supported by Grant IBN 9407763 from the National Science Foundation and Grant DE-FG02-91ER20021 from the U.S. Department of Energy. S.J. was the recipient of a fellowship from the Institut National de la Recherche Agronomique (France).
ABBREVIATIONS
- GA
gibberellin
- GA3
gibberellin A3 (gibberellic acid)
- RPA
replication protein A
- SBD
single-stranded DNA-binding domain
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF009179).
References
- 1.Catling D. Rice in Deep Water. London: MacMillan; 1992. [Google Scholar]
- 2.Vergara B S, Jackson B, De Datta S K. Climate and Rice. Los Baños, Philippines: International Rice Research Institute; 1976. pp. 301–319. [Google Scholar]
- 3.Stünzi J T, Kende H. Plant Cell Physiol. 1989;30:49–56. [Google Scholar]
- 4.Raskin I, Kende H. Planta. 1984;160:66–72. doi: 10.1007/BF00392467. [DOI] [PubMed] [Google Scholar]
- 5.Raskin I, Kende H. Plant Physiol. 1984;76:947–950. doi: 10.1104/pp.76.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann-Benning S, Kende H. Plant Physiol. 1992;99:1156–1161. doi: 10.1104/pp.99.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauter M, Kende H. Planta. 1992;188:362–368. doi: 10.1007/BF00192803. [DOI] [PubMed] [Google Scholar]
- 8.Sauter M, Seagull R W, Kende H. Planta. 1993;190:354–362. [Google Scholar]
- 9.Bleecker A B, Schuette J L, Kende H. Planta. 1986;169:490–497. doi: 10.1007/BF00392097. [DOI] [PubMed] [Google Scholar]
- 10.Sauter M, Kende H. Plant Cell Physiol. 1992;33:1089–1097. [Google Scholar]
- 11.Sauter M, Mekhedov S L, Kende H. Plant J. 1995;7:623–632. doi: 10.1046/j.1365-313x.1995.7040623.x. [DOI] [PubMed] [Google Scholar]
- 12.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 13.van der Knaap E, Kende H. Plant Mol Biol. 1995;28:589–592. doi: 10.1007/BF00020405. [DOI] [PubMed] [Google Scholar]
- 14.Métraux J-P, Kende H. Plant Physiol. 1983;72:441–446. doi: 10.1104/pp.72.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puissant C, Houdeline L-M. BioTechniques. 1990;8:148–149. [PubMed] [Google Scholar]
- 16.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. pp. 4.9.1.–4.9.8.. [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. p. B15. [Google Scholar]
- 18.Philipova D, Mullen J R, Maniar H S, Lu J, Gu C, Brill S J. Genes Dev. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 19.Schiestl R H, Gietz R D. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 20.Sherman F. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 21.Boeke J D, Trueheart J, Natsoulis G, Fink G R. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 22.Wold M S. Annu Rev Biochem. 1997;66:61–91. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 23.Wobbe C R, Weissbach L, Borowiec J A, Dean F B, Marakami Y. Proc Natl Acad Sci USA. 1987;84:1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wold M S, Kelly T. Proc Natl Acad Sci USA. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fairman M P, Stillman B. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyer W-D, Rao M R S, Erdile L F, Kelly T J, Kolodner R D. EMBO J. 1990;9:2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore S P, Erdile L, Kelly T, Fishel R. Proc Natl Acad Sci USA. 1991;88:9067–9071. doi: 10.1073/pnas.88.20.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coverly D, Kenny M K, Munn M, Rupp W D, Lane D P, Wood R D. Nature (London) 1991;349:538–541. doi: 10.1038/349538a0. [DOI] [PubMed] [Google Scholar]
- 29.Coverly D, Kenny M K, Lane D P, Wood R D. Nucleic Acids Res. 1992;20:3873–3880. doi: 10.1093/nar/20.15.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brill S J, Stillman B. Nature (London) 1989;342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 31.Brill S J, Stillman B. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 32.Lowndes N F, Johnson A L, Johnston L H. Nature (London) 1991;350:247–250. doi: 10.1038/350247a0. [DOI] [PubMed] [Google Scholar]
- 33.Gomes X V, Wold M S. J Biol Chem. 1995;270:4534–4543. doi: 10.1074/jbc.270.9.4534. [DOI] [PubMed] [Google Scholar]
- 34.Kim D K, Stigger E, Lee S H. J Biol Chem. 1996;271:15124–15129. doi: 10.1074/jbc.271.25.15124. [DOI] [PubMed] [Google Scholar]
- 35.Lin Y L, Chen C, Keshav K F, Winchester E, Dutta A. J Biol Chem. 1996;271:17190–17198. doi: 10.1074/jbc.271.29.17190. [DOI] [PubMed] [Google Scholar]
- 36.Bochkarev A, Pfuetzner R A, Edwards A M, Frappier L. Nature (London) 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 37.Clugston C K, McLaughlin K, Kenny M K, Brown R. Cancer Res. 1992;52:6375–6379. [PubMed] [Google Scholar]
- 38.He Z, Hendricksen L A, Wold M S, Ingles C J. Nature (London) 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 39.Burns J L, Guzder S N, Sung P, Prakash S, Prakash L. J Biol Chem. 1996;271:11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- 40.He Z, Brinton B T, Greenblatt J, Hassell J A, Ingles C J. Cell. 1993;73:1223–1232. doi: 10.1016/0092-8674(93)90650-f. [DOI] [PubMed] [Google Scholar]
- 41.Li R, Botchan M R. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 42.Dutta A, Ruppert J M, Aster J C, Winchester E. Nature (London) 1993;365:79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- 43.Luche R M, Smart W C, Marion T, Tillman M, Sumrada R A, Cooper T G. Mol Cell Biol. 1993;13:5749–5761. doi: 10.1128/mcb.13.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh K K, Samson L. Proc Natl Acad Sci USA. 1995;92:4907–4911. doi: 10.1073/pnas.92.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Einerhand A W, Kos W, Smart W C, Kal A J, Tabak H F, Cooper T G. Mol Cell Biol. 1995;15:3405–3414. doi: 10.1128/mcb.15.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang C M, Tomkinson A E, Lane W S, Wold M S, Seto E. J Biol Chem. 1996;271:21637–21644. doi: 10.1074/jbc.271.35.21637. [DOI] [PubMed] [Google Scholar]